Key Points
T-cell repertoire deep sequencing clearly identifies the nucleotide and amino acid sequence of the immunodominant clone in T-LGL leukemia patients.
Deep-sequencing results suggest that CD8+ T-LGL leukemia is characterized by specific CDR3 clonotypes that are private to the disease.
Abstract
New massively parallel sequencing technology enables, through deep sequencing of rearranged T-cell receptor (TCR) Vβ complementarity-determining region 3 (CDR3) regions, a previously inaccessible level of TCR repertoire analysis. The CDR3 repertoire diversity reflects clonal composition, the potential antigenic recognition spectrum, and the quantity of available T-cell responses. In this context, T-large granular lymphocyte (T-LGL) leukemia is a chronic clonal lymphoproliferation of cytotoxic T cells often associated with autoimmune diseases and various cytopenias. Using CD8+ T-LGL leukemia as a model disease, we set out to evaluate and compare the TCR deep-sequencing spectra of both patients and healthy controls to better understand how TCR deep sequencing could be used in the diagnosis and monitoring of not only T-LGL leukemia but also reactive processes such as autoimmune disease and infection. Our data demonstrate, with high resolution, significantly decreased diversity of the T-cell repertoire in CD8+ T-LGL leukemia and suggest that many T-LGL clonotypes may be private to the disease and may not be present in the general public, even at the basal level.
Introduction
The T-cell receptor (TCR) is a heterodimer composed of α and β (or γ/δ) chains, both encoded by rearranged V(D)J segments and a constant region.1-5 Rearrangement between V, D, and J regions and the insertion/deletion of nucleotides by recombinases produces the incredibly diverse recognition spectrum of the T-cell compartment, thereby providing flexibility of immune responses to the wide variety of antigens that an individual encounters in a lifetime. The vast majority of T cells recognize targets of immunity through direct TCR interaction with antigen-derived peptide presented in the context of HLA, whereas a small subset (<1%, natural killer T cells) appears to recognize lipid moieties presented in the nonclassical major histocompatibility complex–like CD1d.6,7 Molecular analysis of the structure of the TCR has revealed that the complementarity-determining region 3 (CDR3) of the Vβ chain is directly involved in the recognition/binding of antigenic peptide-HLA complexes.8-10 Consequently, the CDR3 sequence can serve as a unique clonal marker when clonal expansion occurs following T-cell activation.
Until recently, analysis of the TCR repertoire using spectratyping or Sanger sequencing of the CDR3 was limited by technique because traditional sequencing methods are generally impractical for individual sequencing of a large number of T-cell clones. New, massively parallel sequencing technology enables, through deep sequencing of rearranged TCR Vβ chains, a previously inaccessible level of TCR repertoire analysis. Recent research has demonstrated the applicability of TCR deep sequencing in the detection of minimal residual disease in acute T lymphoblastic leukemia,11 led to advances in understanding of the composition of the healthy TCR repertoire,12,13 and addressed a number of technical concerns.14 The CDR3 repertoire diversity reflects clonal composition, the potential antigenic recognition spectrum, and quantity of available T-cell responses. In T cell–mediated autoimmune diseases, it is possible that uniquely composed Vβ CDR3 sequences identified by deep next-generation sequencing (NGS) can serve as surrogate markers for antigens recognized by autoreactive T cells, and efficacy of therapeutic strategies can be assessed by monitoring the level of immunodominant clones and evaluating expansions and contractions of the TCR repertoire over time.
In the majority of classical T cell–mediated autoimmune diseases, T-cell responses are characterized by complex polyclonal expansions with a variable number of leading immunodominant clones, whereas T-cell malignancies are identified by large-scale expansion of a single clone with a unique TCR rearrangement and CDR3 sequence. In this context, T-cell large granular lymphocyte (T-LGL) leukemia is a chronic clonal lymphoproliferation of cytotoxic T cells (CTLs) often associated with autoimmune diseases and various cytopenias and therefore may be considered a reactive process with extreme clonal skewing.15-17 In contrast, patients with CD4+ T-LGL leukemia, much less common than CD8+ T-LGL leukemia, generally do not have autoimmune disease or cytopenia.18 Recent identification of recurrent activating signal transducer and activator of transcription (STAT) 3 and STAT5b mutations at various frequencies in T-LGL19-21 suggests that in a proportion of these cases mutations play a role in the persistence of clonal expansions, whereas mutation-negative cases may represent an extreme pole of an immunologic response to chronic antigen stimulation. The mechanism behind cytopenia in T-LGL leukemia remains undetermined but has been hypothesized to involve clonal T-cell expansions directed against autoantigens, with T-cell specificity determining the clinical phenotype of the disease, be it rheumatoid arthritis, neutropenia, or red cell aplasia.15
Using CD8+ T-LGL leukemia as a model disease, we have set out to evaluate and compare the Vβ TCR deep-sequencing spectra of both patients and healthy controls to better understand how TCR deep sequencing could be used in the diagnosis and monitoring of not only T-LGL leukemia but also reactive processes such as autoimmune disease and infection.
Methods
Patients
Peripheral blood sample collection from patients was performed at clinically indicated testing following informed consent, according to the protocols approved by the Institutional Review Board of the Cleveland Clinic, and in accordance with the Declaration of Helsinki. A retrospective chart review was carried out under approval by the Institutional Review Board of the Cleveland Clinic. Based on World Health Organization guidelines, the following criteria were used to diagnose T-LGL leukemia: monoclonal TCR γ-chain rearrangement; an LGL count by peripheral blood smear of >2000/uL (not a critical criterion; patients who met all other criteria but had an LGL count <2000/uL were included); flow cytometric evidence of an abnormal CTL population characterized by expression of CD2, CD3, TCRαβ (or γδ), CD4 (in a few cases), CD5dim, CD8, CD16/56, or CD57 with negativity of CD28; and persistence of this condition for more than 6 months. Each patient must have met at least 3 of these criteria to be included in the study. In addition, TCR Vβ expansions were detected and quantitated by flow cytometry according to criteria previously described.14,22 Cytopenias were classified as neutropenia (absolute neutrophil count <1.5 × 103/μL), anemia (hemoglobin <13 g/dL), and thrombocytopenia (platelet count <150 × 103/μL). Clinical responses were determined according to the modified International Working Group criteria for myelodysplastic syndromes, as previously reported.23,24
Flow cytometry
Fresh peripheral blood was stained for Vβ flow cytometry analysis to quantitate the percentage of each Vβ family in the CD4 and CD8 lymphocyte populations, as previously described.14
Polymerase chain reaction (PCR)
RNA isolation and complementary DNA synthesis, CDR3 region amplification, CDR3 size analysis, CDR3 cloning, spectratyping, sequencing, and subsequent analysis of “clone size” were performed as previously described.25
Statistical analysis
JMP 10.0 (SAS Institute Inc., Cary, NC) was used for statistical analysis. Because distributions were not normal in the parameters analyzed, nonparametric methods (Wilcoxon) were employed to compare groups. Homology analysis was performed using RegExp in MATLAB 7.12.0; only 100% matches in both directions (A to B and B to A) were considered positive.
Diversity index
The diversity index was calculated according to the following formula, based on the Simpson index of diversity (D) where ni is the total number of amino acid sequences belonging to type i, and N is the total number of sequences in the dataset for each individual:
TCR repertoire deep sequencing
DNA was isolated from whole blood, and the diversity of the TCR repertoire was profiled using high-throughput sequencing of rearranged TCRβ loci from genomic DNA. We sequenced the CDR3 region of TCRβ genes from ∼40 000 T-cell genomes from each sample. The TCRβ CDR3 region was defined according to the International Immunogenetics Information System collaboration, beginning with the second conserved cysteine encoded by the 3′ portion of the Vβ gene segment and ending with the conserved phenylalanine encoded by the 5′ portion of the Jβ gene segment. TCRβ CDR3 regions were amplified and sequenced using protocols previously described.12 Briefly, a multiplexed PCR method was employed to amplify all possible rearranged genomic TCRβ sequences using 52 forward primers, each specific to a TCR Vβ segment, and 13 reverse primers, each specific to a TCR Jβ segment. Reads of 60-bp length were obtained using the Illumina HiSeq System. Raw HiSeq sequence data were preprocessed to remove errors in the primary sequence of each read and to compress the data. A nearest neighbor algorithm was used to collapse the data into unique sequences by merging closely related sequences, to remove both PCR and sequencing errors. Adaptive Biotechnologies (Seattle, WA) performed TCR repertoire NGS and provided data from controls. Adaptive Biotechnologies was blinded to disease status and all clinical information of patients.
STAT3 mutational analysis.
Amplicon-based sequencing.
Locus-specific primers with Illumina adapter tails were designed to cover STAT3 exon 21. Each amplicon was amplified in a multiplexed PCR reaction containing locus-specific PCR primers carrying Illumina-compatible adapter sequences, Illumina TruSeq Universal Adapter primer and Illumina TruSeq Adapter primer with a sample-specific 6-bp index sequence. The PCR reaction was cycled in the DNA Engine Tetrad 2 (Bio-Rad Laboratories) or in the G-Storm GS4 (G-Storm) thermal cycler. Following PCR amplification, samples were purified using Performa V3 96-Well Short Plate (EdgeBio) and QuickStep2 SOPE Resin (EdgeBio) and then pooled together. Sequencing of PCR amplicons was performed using the Illumina MiSeq instrument with MiSeq Control Software v.1.2 or earlier version (Illumina). Samples were sequenced as 151-bp paired-end reads and 2 8-bp index reads using amplicon workflow. Data analysis was done on Illumina MiSeq Reporter Software v1.3 or an earlier version (Illumina). The method and primer sequences have been described previously in more detail.26
Amplification refractory mutation system–PCR
The presence of D661Y and Y640F STAT3 mutations determined by NGS amplicon-based methods were confirmed using a DNA tetraprimer amplification refractory mutation system assay as previously described.17
Results
NGS TCR deep sequencing allows for global analysis of the TCR repertoire
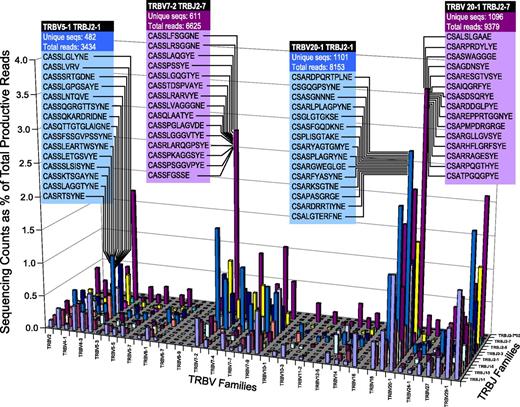
Deep sequencing of the TCR repertoire (Figure 1; representative healthy control) provides a precise estimation of the entire repertoire present in blood. Using this method, nonproductive TCR rearrangements appear out of frame and can be excluded in the analysis. By grouping Vβ and Jβ families and examining the number of unique sequences vs the number of reads within each Vβ/Jβ group, the diversity of the healthy T-cell repertoire is illuminated. Analysis in this fashion reveals a preferential usage of observed Vβ/Jβ combinations, with the exclusion of others. As such, Vβ/Jβ plots from healthy controls form a characteristic “citylike” landscape when arranged in numerical order, with appreciable distance observed between “downtown,” “midtown,” and the “suburbs.”
Diversity of the healthy T-cell repertoire. NGS of the T-cell repertoire in a healthy volunteer identifies specific CDR3 amino acid sequences within T-cell receptor beta variable (TRBV) and T-cell receptor beta joining (TRBJ) families. The 15 most frequent CDR3 sequences and number of unique sequences/total productive reads are shown for selected TRBV vs TRBJ groupings. The ratio between unique sequences and total reads illustrates the level of diversity.
Diversity of the healthy T-cell repertoire. NGS of the T-cell repertoire in a healthy volunteer identifies specific CDR3 amino acid sequences within T-cell receptor beta variable (TRBV) and T-cell receptor beta joining (TRBJ) families. The 15 most frequent CDR3 sequences and number of unique sequences/total productive reads are shown for selected TRBV vs TRBJ groupings. The ratio between unique sequences and total reads illustrates the level of diversity.
We began our investigations by analyzing 25 controls. The average depth of sequencing was 4 349 238 ± 4 675 652 (range 120 485-16 799 066) reads. Of these, the average number of productive reads (ie, the number of reads that appear in-frame during analysis) was 3 565 855 ± 3 815 784 (range 103 026-14 024 627), accounting for 82% of the total reads. The number of unique sequences relative to total productive reads provides a general assessment of diversity within a patient; in controls, this percentage was 5.1 ± 5.6% (range 0.43-19.4). Calculation of the immunodominant clone in healthy controls (ie, the DNA sequence present in highest abundance) revealed a mean of 4.2 ± 5.2%, with the relatively high standard deviation primarily driven by 1 outlier with an expansion of 24.7% (range 0.2%-24.7%). There was no statistical difference in the size of the mean immunodominant expansion between older (>65, mean 3.5%) and younger (<65, mean 6.5%) controls.
As we were interested in how much overlap was present in the TCR repertoires of various individuals, we hypothesized that the number of shared sequences would correspond with the level of shared HLAs. Analysis of a pair of identical twins, which provided a complete HLA match at all loci, revealed that the number of shared sequences was low, only 647 out of a possible 14 392 (supplemental Figure 1A; see the Blood Web site). However, the discovery of the presence of autoimmune disease in 1 of the twins prompted us to exclude this individual from the control group and perform a more in-depth examination of a possible relationship between HLA haplotype and TCR repertoire. Controls matched at 2 of 12 HLA loci shared a lower number of sequences and, in some cases, none (supplemental Figure 1B-C, respectively). Linear regression analysis of TCR repertoire overlap by both absolute number and percentage against the number of shared HLA alleles failed to reveal a statistically significant relationship (R = 0.00009 and 0.0009, respectively; supplemental Figure 1D-E).
T-LGL, a clonal model of extremely polarized CTL responses
Our initial experimental cohort consisted of 134 patients with T-LGL leukemia, identified by the 2008 World Health Organization diagnostic criteria19,27 (clinical characteristics in Table 1). As evidence of the association with autoimmune processes, 65 of 134 (49%) patients had neutropenia, 90 of 134 (67%) had reticulocytopenic anemia, and 24 of 134 (18%) had other classical autoimmune conditions. For each patient, we identified the immunodominant CTL clone by flow cytometry and determined the HLA class I background as a putative restrictive element for CTL responses. Analysis of clone size against specific HLA alleles yielded a strong correlation between the degree of clonal expansion and the presence of HLA B7 (P = .016, N = 111; supplemental Figure 2). Somatic STAT3 mutations were present in only 18% (19/105) of patients, and no STAT5b mutations were found in this cohort. Furthermore, the presence of a STAT3 mutation was associated with a more predominant clonal expansion (P = .0008, N = 102; supplemental Figure 3A). This association remained significant when absolute clone counts were considered as well (P = .014, N = 102; supplemental Figure 3B). In the absence of STAT3 mutation, extreme monoclonal skewing of the TCR repertoire (defined as a flow cytometry expansion >60% of the CD8 compartment) was found in 39% (33/83) compared with 78% (15/19) with mutated STAT3.
Clinical characteristics (N = 134)
| Parameter . | Value . |
|---|---|
| Mean age at diagnosis (y) | 62 (range 17-85) |
| Gender | 76 M:58 F |
| LGL count (103/μL) | 2324 ± 2932 |
| Splenomegaly | 29% (39/134) |
| TCR rearrangement by PCR | 97% (123/126) |
| STAT3 mutation | 18% (19/105) |
| Hematologic manifestation | 87% (116/134) |
| Anemia | 67% (90/134) |
| Neutropenia | 49% (65/134) |
| Thromobocytopenia | 31% (42/134) |
| Multilineage cytopenia | 27% (36/134) |
| Pancytopenia | 18% (24/134) |
| Lymphocytosis | 30% (40/134) |
| Lymphopenia | 16% (22/134) |
| B-cell or antibody disorder | 49% (65/134) |
| Monoclonal gammopathy of unknown significance/multiple myeloma | 16% (22/134) |
| Hypergammaglobunemia | 16% (22/134) |
| Hypogammaglobunemia | 8% (11/134) |
| Chronic lymphocytic leukemia | 10% (13/134) |
| Hairy cell leukemia | <1% (1/134) |
| Non-Hodgkin lymphoma | 1% (2/134) |
| B-cell lymphoma/lymphoproliferative disorder | 1% (2/134) |
| Autoimmune disease | 18% (24/134) |
| Rheumatoid arthritis | 14% (19/134) |
| Myasthenia gravis, Guillain-Barre | 1% (2/134) |
| Pernicious anemia, autoimmune hemolytic anemia | 1% (2/134) |
| Sjögren, Crohn, Felty | 2% (3/134) |
| Parameter . | Value . |
|---|---|
| Mean age at diagnosis (y) | 62 (range 17-85) |
| Gender | 76 M:58 F |
| LGL count (103/μL) | 2324 ± 2932 |
| Splenomegaly | 29% (39/134) |
| TCR rearrangement by PCR | 97% (123/126) |
| STAT3 mutation | 18% (19/105) |
| Hematologic manifestation | 87% (116/134) |
| Anemia | 67% (90/134) |
| Neutropenia | 49% (65/134) |
| Thromobocytopenia | 31% (42/134) |
| Multilineage cytopenia | 27% (36/134) |
| Pancytopenia | 18% (24/134) |
| Lymphocytosis | 30% (40/134) |
| Lymphopenia | 16% (22/134) |
| B-cell or antibody disorder | 49% (65/134) |
| Monoclonal gammopathy of unknown significance/multiple myeloma | 16% (22/134) |
| Hypergammaglobunemia | 16% (22/134) |
| Hypogammaglobunemia | 8% (11/134) |
| Chronic lymphocytic leukemia | 10% (13/134) |
| Hairy cell leukemia | <1% (1/134) |
| Non-Hodgkin lymphoma | 1% (2/134) |
| B-cell lymphoma/lymphoproliferative disorder | 1% (2/134) |
| Autoimmune disease | 18% (24/134) |
| Rheumatoid arthritis | 14% (19/134) |
| Myasthenia gravis, Guillain-Barre | 1% (2/134) |
| Pernicious anemia, autoimmune hemolytic anemia | 1% (2/134) |
| Sjögren, Crohn, Felty | 2% (3/134) |
Identification of clonal and oligoclonal expansions in disease: T-LGL as a malignant model of autoimmune diseases
For further analysis, we selected patients with HLA restriction elements HLA A2, HLA B7, similar presentation, and Vβ17 as the immunodominant clone by flow cytometry (N = 11; Table 2). Controls (N = 24) were matched at HLA A2 and B7 when HLA data were available.
CD8+ T-LGL leukemia patient characteristics for TCR repertoire deep sequencing (N = 11)
| Cohort ID . | STAT3 mutation . | Hematologic presentation . | AAD . | Sex . | Viral serology . | Autoimmune disease . | Therapy . | Flow VB ID . | Clone size* . | Absolute major clone size† . | HLA A . | HLA A2 . | HLA B . | HLA B2 . | HLA Cw . | HLA Cw 2 . | HLA DRB1 . | HLA DRB1 2 . | HLA DQB1 . | HLA DQB1 2 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LGL01 | Y640F | Anemia, neutropenia | 63 | F | n/a | No | CSA, MTX, ALEM | TRBV12-3, 12-4 | 84.0 | 1210 | 1 | 31 | 7 | 8 | 7 | 7 | 3 | 13 | 2 | 6 |
| LGL02 | Negative | Anemia, neutropenia | 59 | F | n/a | No | FIL, CSA, MTX | NIP | 74.7 | 463 | 2 | 24 | 7 | 15 | 3 | 7 | 1 | 3 | 2 | 5 |
| LGL03 | Negative | Anemia | 60 | M | n/a | No | CSA, MTX, DAR | TRBV19 | 85.1 | 1464 | 2 | 25 | 18 | 44 | 12 | 16 | 11 | 15 | 3 | 6 |
| LGL04 | D661Y | Anemia | 50 | M | n/a | No | CSA | TRBV20-1 | 64.3 | 1092 | 1 | 3 | 7 | 7 | 7 | 7 | 3 | 15 | 2 | 6 |
| LGL05 | D661Y | Anemia | 62 | M | n/a | No | SPL | TRBV19 | 52.7 | 1066 | 2 | 29 | 7 | 45 | 6 | 7 | 1 | 15 | 5 | 6 |
| LGL06 | Negative | Anemia | 72 | M | n/a | No | CSA, MTX, CYC, ATG | TRBV6-6 | 92.7 | 4305 | 1 | 2 | 7 | 8 | 7 | 7 | 3 | 13 | 2 | 6 |
| LGL07 | D661Y | Anemia | 54 | M | n/a | No | CYC | TRBV9 | 40.1 | 237 | 2 | 2 | 40 | 49 | 2 | 7 | 11 | 13 | 3 | 6 |
| LGL08 | Negative | Anemia | 68 | M | Parvovirus B19 | No | CSA, CYC, MTX | NIP | 65.3 | 922 | 2 | 24 | 7 | 13 | 2 | 7 | 3 | 15 | 2 | 6 |
| LGL09 | D661V | Neutropenia, thrombo-cytopenia | 81 | M | n/a | RA | MTX | TRBV9 | 71.5 | 745 | 1 | 2 | 7 | 15 | 3 | 7 | 4 | 15 | 3 | 6 |
| LGL10 | Negative | Anemia, neutropenia | 56 | F | n/a | No | CYC, MTX | TRBV12-3, 12-4 | 88.7 | 844 | 29 | 33 | 14 | 14 | 8 | 8 | 1 | 11 | 3 | 5 |
| LGL11 | Negative | Anemia | 51 | F | ANA, SSA, SSB, RNP, chromatin | RA, Sjögren | PEG, MTX | TRBV19 | 71.6 | 440 | 1 | 3 | 8 | 35 | 4 | 7 | 3 | 15 | 2 | 6 |
| Cohort ID . | STAT3 mutation . | Hematologic presentation . | AAD . | Sex . | Viral serology . | Autoimmune disease . | Therapy . | Flow VB ID . | Clone size* . | Absolute major clone size† . | HLA A . | HLA A2 . | HLA B . | HLA B2 . | HLA Cw . | HLA Cw 2 . | HLA DRB1 . | HLA DRB1 2 . | HLA DQB1 . | HLA DQB1 2 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LGL01 | Y640F | Anemia, neutropenia | 63 | F | n/a | No | CSA, MTX, ALEM | TRBV12-3, 12-4 | 84.0 | 1210 | 1 | 31 | 7 | 8 | 7 | 7 | 3 | 13 | 2 | 6 |
| LGL02 | Negative | Anemia, neutropenia | 59 | F | n/a | No | FIL, CSA, MTX | NIP | 74.7 | 463 | 2 | 24 | 7 | 15 | 3 | 7 | 1 | 3 | 2 | 5 |
| LGL03 | Negative | Anemia | 60 | M | n/a | No | CSA, MTX, DAR | TRBV19 | 85.1 | 1464 | 2 | 25 | 18 | 44 | 12 | 16 | 11 | 15 | 3 | 6 |
| LGL04 | D661Y | Anemia | 50 | M | n/a | No | CSA | TRBV20-1 | 64.3 | 1092 | 1 | 3 | 7 | 7 | 7 | 7 | 3 | 15 | 2 | 6 |
| LGL05 | D661Y | Anemia | 62 | M | n/a | No | SPL | TRBV19 | 52.7 | 1066 | 2 | 29 | 7 | 45 | 6 | 7 | 1 | 15 | 5 | 6 |
| LGL06 | Negative | Anemia | 72 | M | n/a | No | CSA, MTX, CYC, ATG | TRBV6-6 | 92.7 | 4305 | 1 | 2 | 7 | 8 | 7 | 7 | 3 | 13 | 2 | 6 |
| LGL07 | D661Y | Anemia | 54 | M | n/a | No | CYC | TRBV9 | 40.1 | 237 | 2 | 2 | 40 | 49 | 2 | 7 | 11 | 13 | 3 | 6 |
| LGL08 | Negative | Anemia | 68 | M | Parvovirus B19 | No | CSA, CYC, MTX | NIP | 65.3 | 922 | 2 | 24 | 7 | 13 | 2 | 7 | 3 | 15 | 2 | 6 |
| LGL09 | D661V | Neutropenia, thrombo-cytopenia | 81 | M | n/a | RA | MTX | TRBV9 | 71.5 | 745 | 1 | 2 | 7 | 15 | 3 | 7 | 4 | 15 | 3 | 6 |
| LGL10 | Negative | Anemia, neutropenia | 56 | F | n/a | No | CYC, MTX | TRBV12-3, 12-4 | 88.7 | 844 | 29 | 33 | 14 | 14 | 8 | 8 | 1 | 11 | 3 | 5 |
| LGL11 | Negative | Anemia | 51 | F | ANA, SSA, SSB, RNP, chromatin | RA, Sjögren | PEG, MTX | TRBV19 | 71.6 | 440 | 1 | 3 | 8 | 35 | 4 | 7 | 3 | 15 | 2 | 6 |
AAD, age at diagnosis; ALEM, alemtuzumab; ANA, anti-nuclear antibodies; ATG, antithymocyte globulin; CSA, cyclosporine; CYC, cyclophosphamide; DAR, darbepoetin; FIL, filgrastim; MTX, methotrexate; NIP, not in panel; PEG, pegfilgrastim; SPL, splenectomy; SSA, Sjögren's Syndrome A; SSB, Sjögren's Syndrome B.
Clone size: % CD8+ cells expressing a specific Vβ region by flow cytometry.
Absolute major clone size = (white blood cells) (% lymph) (% CD8) (% specific Vβ), shown in cells per microliter of blood.
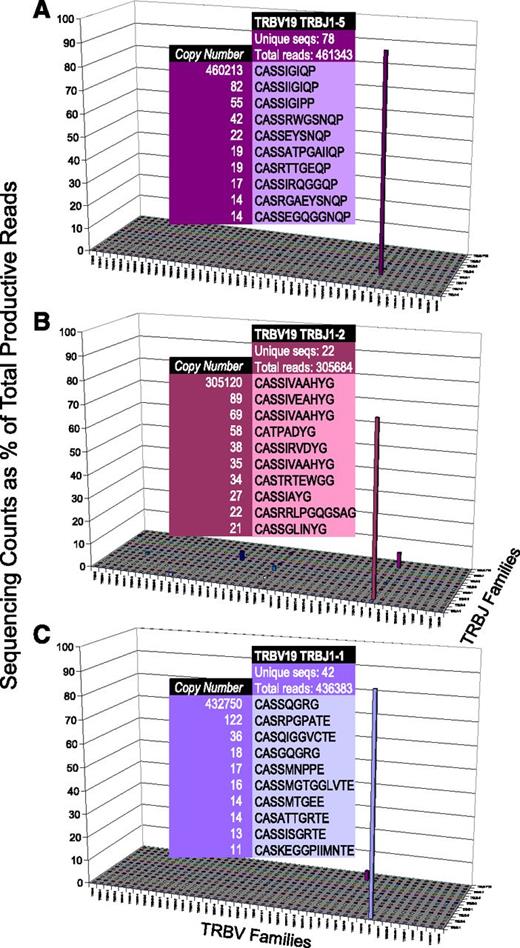
We next applied TCR repertoire deep sequencing to this subcohort of T-LGL leukemia patients, with markedly clear results (Figure 2). When expanded Vβ/Jβ counts were observed, we analyzed the sequencing counts as a percentage of overall reads; plots of this data revealed the dominance of specific CDR3 sequences, flattening the landscape of the healthy repertoire to account for the scale of the clonal expansion. Unlike the healthy repertoire, analysis of specific Vβ/Jβ pairings demonstrated the extreme expansions characteristic of T-LGL leukemia. For example, Figure 2A shows a clonal expansion of TRBV19 TRBJ1-5, which accounts for 460 213/461 343 (99.8%) sequences within this grouping. Also present, albeit at a much lower frequency, are accessory clones that differ only slightly in amino acid sequence or have identical amino acid sequences with different nucleotide sequences. All 3 representative examples shown here, and 10 of 11 patients overall, exhibited this phenomenon.
T-cell repertoire in T-LGL leukemia. In contrast to the healthy T-cell repertoire, T-LGL leukemia patients display markedly less diversity. Although the top 10 CDR3 amino acid sequences are shown for indicated TRBV vs TRJB groupings, the most abundant sequence accounts for 460 213/461 343 (A), 305 120/305 684 (B), and 432 750/436 383 (C).
T-cell repertoire in T-LGL leukemia. In contrast to the healthy T-cell repertoire, T-LGL leukemia patients display markedly less diversity. Although the top 10 CDR3 amino acid sequences are shown for indicated TRBV vs TRJB groupings, the most abundant sequence accounts for 460 213/461 343 (A), 305 120/305 684 (B), and 432 750/436 383 (C).
In 7 of 11 patients, immunodominant clonotypes accounted for >50% of the TCR repertoire and correlated well with flow cytometry results. Two of the 11 patients who appeared to have monoclonal expansions by flow cytometry demonstrated biclonal expansions by deep sequencing, illustrating the finer resolution of this more modern technology. Finally, accounting for the 2 patients whose expansions were not recognized by flow cytometry because of limitations in the antibody panel (NIP), 1 was shown to have a clear monoclonal expansion by NGS, and the other demonstrated only minor polyclonal expansions, again reinforcing the advantage of deep sequencing over flow cytometry in assessing the T-cell repertoire (Table 3).
Sequencing results of immunodominant clones
| LGL cohort ID . | Nucleotide sequence . | Amino acid sequence . | NGS copy number %* . | Vgene name . | Dgene name . | Jgene name . |
|---|---|---|---|---|---|---|
| LGL01 | TCTCAGTGACTCTGGCTTCTATCTCTGTGCCTGGCCCGACTAGCGGGAATACAATGAGCA | CAWPD*REYNE | 88.8 | TRBV30 | TRBD1-2 | TRBJ2-1 |
| LGL02 | CCAGACAGCTCTTTACTTCTGTGCCACCAGTGGGACAGGGGTCCCCCATTACAATGAGCA | CATSGTGVPHYNE | 7.1 | TRBV24-1 | TRBD1-1 | TRBJ2-1 |
| LGL03 | GGCCCAAAAGAACCCGACAGCTTTCTATCTCTGTGCCAGTAGTATAGGGATTCAGCCCCA | CASSIGIQP | 89.1 | TRBV19 | TRBD1-1 | TRBJ1-5 |
| LGL04 | GGACTCGGCCATGTATCTCTGTGCCAGCAGCTTAATAGGGGTAAGCTCCTACAATGAGCA | CASSLIGVSSYNE | 10.9 | TRVB7-9 | TRBD1-1 | TRBJ2-1 |
| LGL05 | AAAGAACCCGACAGCTTTCTATCTCTGTGCCAGTAGTATAGTAGCAGCCCACTATGGCTA | CASSIVAAHYG | 62.1 | TRBV19 | TRBD1-2 | TRBJ1-2 |
| LGL06 | TGCTCCCTCCCAGACATCTGTGTACTTCTGTGCCAGCAAATCGGGGGACCCCGGGGAGCT | CASKSGDPGE | 99.1 | TRBV6-6 | TRBD1-2 | TRBJ2-2 |
| LGL07pre | GCTGGGGGACTCAGCTTTGTATTTCTGTGCCAGCAGCGTCGGGCGGTTCCAAGAGACCCA | CASSVGRFQET | 73.9 | TRBV9 | TRBD1-1 | TRBJ2-5 |
| LGL07post | GAAGCTCCTTCTCAGTGACTCTGCTTCTATCTCTGTGCCTGGAGTCCGAACACTGAAGC | CAWSPNTE | 8.6 | TRBV30 | TRBJ1-1 | |
| LGL08 | CAACCAGACATCTATGTACCTCTGTGCCAGCAGTTTGCTAGCGGGAGGGTACAATGAGCA | CASSLLAGGYNE | 55.7 | TRBV28 | TRBD1-2 | TRBJ2-1 |
| LGL09 | GAACCCGACAGCTTTCTATCTCTGTGCCAGTAGTTCTCTCGGAGTCCCATACTACGAGCA | CASSSLGVPYYE | 34.2 | TRBV19 | TRBD1-1 | TRBJ2-7 |
| LGL09 | GGAGCTGGGGGACTCAGCTTTGTATTTCTGTGCCAGCAGCGTGGGACAGGGCTCACCCCT | CASSVGQGSP | 20.5 | TRBV9 | TRBD1-1 | TRBJ1-6 |
| LGL10 | CTCAGCTGTGTACTTCTGTGCCAGCAGTTTAGTCCCCGGGACACTCAACACCGGGGAGCT | CASSLVPGTLNTGE | 17.8 | TRBV12-4 | TRBD1-1 | TRBJ2-2 |
| LGL10 | GGACTCGGCCGTGTATCTCTGTGCCAGCAGCCGGTCCGGCTGGTCCTCGGATTCACCCCT | CASSRSGWSSDSP | 14.4 | TRVB7-2 | TRBD1-1 | TRBJ1-6 |
| LGL11 | ATCGGCCCAAAAGAACCCGACAGCTTTCTATCTCTGTGCCAGTAGTCAGGGACGGGGGGC | CASSQGRG | 88.4 | TRBV19 | TRBD1-2 | TRBJ1-1 |
| LGL cohort ID . | Nucleotide sequence . | Amino acid sequence . | NGS copy number %* . | Vgene name . | Dgene name . | Jgene name . |
|---|---|---|---|---|---|---|
| LGL01 | TCTCAGTGACTCTGGCTTCTATCTCTGTGCCTGGCCCGACTAGCGGGAATACAATGAGCA | CAWPD*REYNE | 88.8 | TRBV30 | TRBD1-2 | TRBJ2-1 |
| LGL02 | CCAGACAGCTCTTTACTTCTGTGCCACCAGTGGGACAGGGGTCCCCCATTACAATGAGCA | CATSGTGVPHYNE | 7.1 | TRBV24-1 | TRBD1-1 | TRBJ2-1 |
| LGL03 | GGCCCAAAAGAACCCGACAGCTTTCTATCTCTGTGCCAGTAGTATAGGGATTCAGCCCCA | CASSIGIQP | 89.1 | TRBV19 | TRBD1-1 | TRBJ1-5 |
| LGL04 | GGACTCGGCCATGTATCTCTGTGCCAGCAGCTTAATAGGGGTAAGCTCCTACAATGAGCA | CASSLIGVSSYNE | 10.9 | TRVB7-9 | TRBD1-1 | TRBJ2-1 |
| LGL05 | AAAGAACCCGACAGCTTTCTATCTCTGTGCCAGTAGTATAGTAGCAGCCCACTATGGCTA | CASSIVAAHYG | 62.1 | TRBV19 | TRBD1-2 | TRBJ1-2 |
| LGL06 | TGCTCCCTCCCAGACATCTGTGTACTTCTGTGCCAGCAAATCGGGGGACCCCGGGGAGCT | CASKSGDPGE | 99.1 | TRBV6-6 | TRBD1-2 | TRBJ2-2 |
| LGL07pre | GCTGGGGGACTCAGCTTTGTATTTCTGTGCCAGCAGCGTCGGGCGGTTCCAAGAGACCCA | CASSVGRFQET | 73.9 | TRBV9 | TRBD1-1 | TRBJ2-5 |
| LGL07post | GAAGCTCCTTCTCAGTGACTCTGCTTCTATCTCTGTGCCTGGAGTCCGAACACTGAAGC | CAWSPNTE | 8.6 | TRBV30 | TRBJ1-1 | |
| LGL08 | CAACCAGACATCTATGTACCTCTGTGCCAGCAGTTTGCTAGCGGGAGGGTACAATGAGCA | CASSLLAGGYNE | 55.7 | TRBV28 | TRBD1-2 | TRBJ2-1 |
| LGL09 | GAACCCGACAGCTTTCTATCTCTGTGCCAGTAGTTCTCTCGGAGTCCCATACTACGAGCA | CASSSLGVPYYE | 34.2 | TRBV19 | TRBD1-1 | TRBJ2-7 |
| LGL09 | GGAGCTGGGGGACTCAGCTTTGTATTTCTGTGCCAGCAGCGTGGGACAGGGCTCACCCCT | CASSVGQGSP | 20.5 | TRBV9 | TRBD1-1 | TRBJ1-6 |
| LGL10 | CTCAGCTGTGTACTTCTGTGCCAGCAGTTTAGTCCCCGGGACACTCAACACCGGGGAGCT | CASSLVPGTLNTGE | 17.8 | TRBV12-4 | TRBD1-1 | TRBJ2-2 |
| LGL10 | GGACTCGGCCGTGTATCTCTGTGCCAGCAGCCGGTCCGGCTGGTCCTCGGATTCACCCCT | CASSRSGWSSDSP | 14.4 | TRVB7-2 | TRBD1-1 | TRBJ1-6 |
| LGL11 | ATCGGCCCAAAAGAACCCGACAGCTTTCTATCTCTGTGCCAGTAGTCAGGGACGGGGGGC | CASSQGRG | 88.4 | TRBV19 | TRBD1-2 | TRBJ1-1 |
NGS copy number % indicates percentage of the total number of reads.
Diversity analysis of the T-cell repertoire
We next set out to statistically describe the overall diversity of each patient’s repertoire by calculating a diversity score from the data. First, analysis of the number of unique sequences relative to total productive reads differentiates T-LGL from healthy controls, 0.6 ± 0.6% vs 5.1 ± 4.7%, respectively (P < .0001; data not shown). For more in-depth analysis, we turned to the Simpson index of diversity, D,28 a mathematical formula developed for abundance assessment of ecological species that provides an objective measurement of the degree of diversity within a data set. We applied this formula to each subject’s repertoire, essentially calculating the probability of obtaining identical CDR3 sequences when selecting 2 sequences at random from the individual’s data set. When compared with controls, T-LGL leukemia patients again demonstrate significantly less diversity (P = .0004; Figure 3A), with a stratification of high, intermediate, and low diversity indices.
Index of diversity. Diversity scores were calculated for deep-sequencing TCR repertoire data from patients with T-LGL leukemia (n = 11; A, right) compared with healthy controls (n = 24; A, left) and analyzed by the Wilcoxon nonparametric method (P < .0001). Index of diversity and TCR repertoire plots for a T-LGL patient before therapy, D = 0.060 (B), and following complete remission, D = 0.966 (C). Note differences in y-axis values between panels B and C.
Index of diversity. Diversity scores were calculated for deep-sequencing TCR repertoire data from patients with T-LGL leukemia (n = 11; A, right) compared with healthy controls (n = 24; A, left) and analyzed by the Wilcoxon nonparametric method (P < .0001). Index of diversity and TCR repertoire plots for a T-LGL patient before therapy, D = 0.060 (B), and following complete remission, D = 0.966 (C). Note differences in y-axis values between panels B and C.
To illustrate the potential clinical benefit of this approach, we show here a patient who initially presented with anemia and an extreme monoclonal expansion with D = 0.06 (LGL 7; Figure 3B). After successful treatment of his disease with cyclophosphamide, which induced a complete hematologic remission, we observed restoration of T-cell diversity (D = 0.96). However, there was no statistical difference in the diversity index between patients harboring the STAT3 mutation and those without (supplemental Figure 4), yet a larger cohort with longitudinal and mutational subgroup analysis may be needed to fully answer this question.
Public vs private sequences: homology assessment
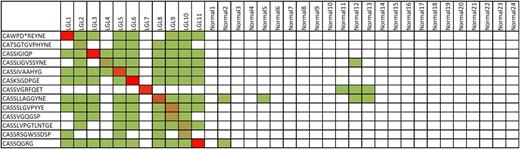
The description of public TCR CDR3 amino acid sequences in the literature29-33 and our own research22 prompted bioinformatic analysis of the immunodominant clonotypes found in our cohort of T-LGL leukemia patients. Although none of the immunodominant clones were present at high frequency in other patients, these clones were present at the basal level in patients with T-LGL leukemia in almost all cases (Figure 4). Notably, these clonotypes were largely absent from healthy controls, even those sharing HLA A2 and B7, raising the possibility that specific TCR sequences exist that are private, not at the individual level, but only to those with T-LGL leukemia. Furthermore, LGL 7 is the only patient whose immunodominant clonotype is not present in the repertoires of other patients; this patient is also the only patient in our entire cohort of 134 patients that achieved complete remission (Figure 3B-C).
Homology assessment. All immunodominant CDR3 sequences were analyzed for the presence of 100% homologous sequences in healthy controls (n = 24). Green squares indicate a sequence match at the basal level (<50 copies per million), whereas increasing intensity to red corresponds with a higher number of reads. Dominant sequences found in T-LGL patients were generally found in other T-LGL patients but were largely absent in healthy controls.
Homology assessment. All immunodominant CDR3 sequences were analyzed for the presence of 100% homologous sequences in healthy controls (n = 24). Green squares indicate a sequence match at the basal level (<50 copies per million), whereas increasing intensity to red corresponds with a higher number of reads. Dominant sequences found in T-LGL patients were generally found in other T-LGL patients but were largely absent in healthy controls.
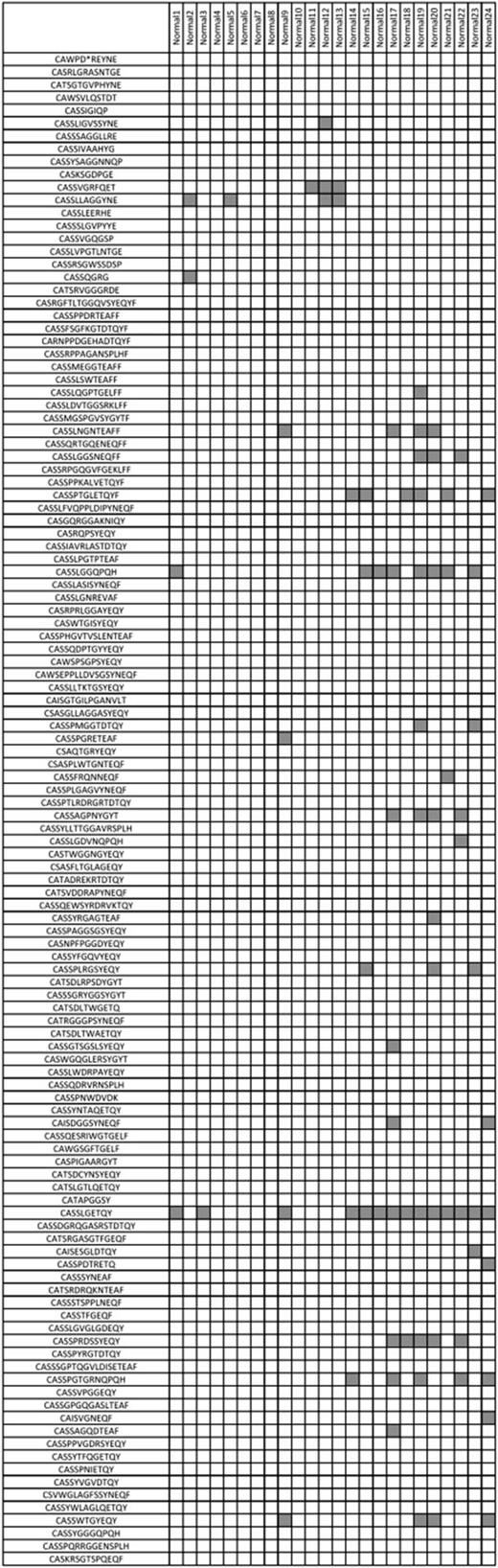
To further test the hypothesis that T-LGL leukemia clonotypes are private to the disease, we then compiled a list of all immunodominant clonotypes of T-LGL leukemia patients sequenced by traditional methods in our laboratory and queried if these were present in our healthy control group (Figure 5). Consistent with our earlier finding, these clonotypes were largely absent from the 24 healthy controls analyzed. In addition, we queried a database composed of more than 6000 immunodominant CDR3 sequences published in a variety of diseases, including various common viral infections, and did not find a match.
T-LGL leukemia immunodominant clonotypes are generally not found in healthy controls. Further analysis of dominant CDR3 clonotypes discovered by traditional sequencing methods as well as deep sequencing supports the notion of clonotypes private among patients with T-LGL leukemia because these sequences were, with a few exceptions, not found in healthy controls. Gray squares indicate a sequence match at 100% homology.
T-LGL leukemia immunodominant clonotypes are generally not found in healthy controls. Further analysis of dominant CDR3 clonotypes discovered by traditional sequencing methods as well as deep sequencing supports the notion of clonotypes private among patients with T-LGL leukemia because these sequences were, with a few exceptions, not found in healthy controls. Gray squares indicate a sequence match at 100% homology.
We were curious as to whether simply specific clonotypes or the global repertoire varied between patients and controls. Analysis of overall repertoire overlap between T-LGL patients and other T-LGL patients, between T-LGL patients and controls, and between healthy controls and other healthy controls failed to reveal distinct patterns (supplemental Figure 5), although it is apparent that LGL 7 had the lowest degree of overlap when compared with other T-LGL patients.
Discussion
In the past, molecular analysis of the T-cell repertoire using cloning and sequencing approaches allowed only a quantitatively limited insight into the clonotypic spectrum. With the advent of NGS, the clonotypic repertoire can be analyzed at an incredible depth. Here, for the first time, we report the application of deep insight into the normal and diseased clonal repertoire in patients with T-LGL leukemia and concurrent immune-mediated cytopenias.
Our initial, perhaps simplistic, hypothesis asked if T-LGL leukemia patients with similar immunogenetic and clinical characteristics would share a common CDR3 region as determined by deep sequencing. “Public” T-cell responses have been described in the literature as examples of “convergent recombination,”27 and such a finding would provide evidence for a common antigen that may drive the extreme monoclonal proliferations characteristic of T-LGL leukemia that occur even in the absence of STAT3 or STAT5b mutations. The rarity of T-LGL leukemia points toward an uncommon elusive chronic antigen, or immunologic weakness that prevents clearance of antigen. Our data suggest the presence of an undefined mechanism whereby certain clonotypes may predispose an individual toward the extreme monoclonal expansions commonly found in T-LGL leukemia because these clonotypes were found in other T-LGL leukemia patients but not in controls. Although this finding is initially striking, especially considering the overall low level of TCR repertoire overlap even in HLA matched individuals, it should be noted that we did not investigate hypothetical repertoire differences between CD4+ and CD8+ cells, nor various effector/memory subsets. It is possible that the homology, or lack thereof, found in our work reflects a biased repertoire because of either the nature of the disease or some as yet undetermined phenomenon. The considerable depth of control sequencing may compensate for possible differences between patients and controls, but we did not formally address this question. Furthermore, the diversity and complexity of infectious history and corresponding immune responses precludes simple insight into the similarity of the CDR3 spectra. Although TCR repertoire deep sequencing has tremendous promise as an investigative and diagnostic approach, new tools currently in development to comprehensively assess serology may also provide complementary information as to an individual’s history of immunity and aid in the identification of the putative antigen(s) driving such extreme T-cell responses.
As a fascinating example of the selection process that occurs upon antigen encounter, the elucidation of accessory clones by deep sequencing suggests that, even in the case of a STAT3 mutation, it is likely that T-LGL leukemia is an antigen-driven process, at least initially, and it can be hypothesized that the STAT3 mutation occurs in a subset of clonally dividing cells that obtain a growth or survival advantage over time. Quantitative sequencing of STAT3 mutations supports this hypothesis because, in some cases, the proportion of cells with the STAT3 mutation does not always correlate with the percentage of cells that have clonally expanded as determined by flow cytometry or by deep sequencing.34 In addition, the association between STAT3 mutations and larger clone sizes, both by proportion and absolute counts, provides additional evidence for the notion of STAT3 mutations as a key factor promoting extreme monoclonal expansions, at least in cases where a mutation has been discovered. The mechanism behind extreme expansions in STAT3-negative cases remains unclear, as does the relationship between clinical phenotype and the presence of a STAT3 mutation. Also, the proportion of patients harboring the STAT3 mutation in this cohort is reflective of the patient population at our single institution and is lower than more comprehensive multicenter studies, likely attributable in part to the inclusion of both monoclonal and oligoclonal T-LGL leukemia patients in this work.
Although it was beyond the scope of this study to compare all methods of assessing clonal expansions in T-cell malignancies, deep sequencing has the potential to redefine investigations into human T-cell immunology by offering significant advantages over traditional methods. First, virtually the entire repertoire can be analyzed and precisely quantitated. Second, the amino acid identity of the expanded clone can be clearly identified. Third, bioinformatic analysis of data obtained from larger cohorts has the potential to show repertoire landscape patterns that will assist in monitoring response to therapy and progression of disease as well as aid in the subclassification of many T cell–mediated diseases, not just T-LGL. Last, the diversity index provides a means to reduce a complex data set to a single number that may be of use in the clinic, although these results need to be explored in a much larger cohort to fully assess the benefit.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the National Institutes of Health, National Heart, Lung, and Blood Institute (R01 HL082983); the National Institutes of Health, National Center for Research Resources (U54 RR019391), the National Institutes of Health, National Heart, Lung, and Blood Institute (K24 077522), and the National Institutes of Health, National Cancer Institute (R01 CA113972) (J.P.M.); and by the Academy of Finland (S.M.) and the Finnish Cancer Society (S.M.).
Authorship
Contribution: M.J.C. and J.P.M. wrote the manuscript and designed the research; M.J.C., A.J., B.E.D., H.L.M.R., M.W.W., and S.M. performed research and analyzed data; B.P. performed bioinformatic analysis; M.J.C., M.G.A., and H.H. collected and analyzed clinical data; and J.P.M. was the principal investigator.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jaroslaw P. Maciejewski, Taussig Cancer Center/R40, 9500 Euclid Ave, Cleveland, OH 44195; e-mail: maciejj@ccf.org.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal