Key Points
Efficacy of transplanting allele-level HLA-matched cord blood units.
Abstract
We studied the effect of allele-level matching at human leukocyte antigen (HLA)-A, -B, -C, and -DRB1 in 1568 single umbilical cord blood (UCB) transplantations for hematologic malignancy. The primary end point was nonrelapse mortality (NRM). Only 7% of units were allele matched at HLA-A, -B, -C, and -DRB1; 15% were mismatched at 1, 26% at 2, 30% at 3, 16% at 4, and 5% at 5 alleles. In a subset, allele-level HLA match was assigned using imputation; concordance between HLA-match assignment and outcome correlation was confirmed between the actual and imputed HLA-match groups. Compared with HLA-matched units, neutrophil recovery was lower with mismatches at 3, 4, or 5, but not 1 or 2 alleles. NRM was higher with units mismatched at 1, 2, 3, 4, or 5 alleles compared with HLA-matched units. The observed effects are independent of cell dose and patient age. These data support allele-level HLA matching in the selection of single UCB units.
Introduction
The importance of high-level donor–recipient matching at various human leukocyte antigen (HLA) loci for the success of unrelated adult donor hematopoietic stem cell transplantation is well documented.1-3 The inability to identify HLA-matched volunteer unrelated adult donors has led to increasing use of unrelated umbilical cord blood (UCB) as an alternative graft because greater degrees of donor–recipient HLA mismatch are tolerated when UCB grafts are used. UCB unit cell dose is a limitation; consequently, the majority of UCB transplants are performed in children and adolescents. High nonrelapse mortality after HLA-mismatched UCB transplantation is also an obstacle.4-8 An important difference when selecting volunteer unrelated adult donors and UCB units is the criteria for HLA-matching donors to recipients. Unrelated adult donors are selected to be closely matched to recipients at HLA-A, -B, -C, and -DRB1 at the allele level,3 whereas UCB units are selected using lower resolution HLA typing (antigen-level) for HLA-A and -B and at the allele level for HLA-DRB1; HLA-C is not typically considered.9-11 We have previously identified the benefit of matching at the HLA-C locus for the outcomes of UCB transplants in a population that included children, adolescents, and adults.12 Among transplantations matched at HLA-A, -B, and -DRB1, nonrelapse mortality was higher after transplantations mismatched at HLA-C than transplantations matched at HLA-C. Similarly, among transplantations with a single mismatch at HLA-A, -B, or -DRB1, transplantations mismatched at HLA-C had higher nonrelapse mortality than transplantations matched at HLA-C. A limitation of that report was that HLA matching at HLA-A, -B, and -C was defined at the antigen level. The current analysis sought to establish the relative importance of allele-level HLA matching at HLA-A, -B, -C, and -DRB1 when selecting single UCB units for transplantation in children, adolescents and adults with acute leukemia and myelodysplastic syndrome.
Patients and methods
Patients
Data were obtained from the Center for International Blood and Marrow Transplant Research, or Eurocord, or the European Group for Blood and Marrow Transplantation. Patients received a single UCB unit after myeloablative conditioning regimens for treatment of acute leukemia or myelodysplastic syndrome using cyclosporine or tacrolimus-containing graft-versus-host disease (GVHD) prophylaxis. All transplantations were performed between 2000 and 2010. The Institutional Review Boards of the Medical College of Wisconsin, the National Marrow Donor Program, and the Eurocord-Netcord scientific committee approved this study. This study was conducted in accordance with the Declaration of Helsinki.
HLA typing
Donor and recipient HLA typing at HLA-A, -B, -C, and -DRB1 was completed using molecular techniques with a minimum of antigen split-level resolution for HLA-A, -B, and -C, and allele-level resolution at DRB1. For transplantations in the United States, recipient HLA typings were provided by the transplant center and UCB typings were from a centralized confirmatory typing laboratory or through retrospective typing of stored research samples, as previously described.13 For transplantations reported to Eurocord, donor–recipient typings were obtained from the cord blood banks or from transplant centers. A subset of the typings available included less than high-resolution typing at the HLA-A, -B, and/or -C loci (n = 784). A validated HLA high-resolution imputation algorithm, Haplogic III, developed by the National Marrow Donor Program, was used to impute allele-level match status for these donor and recipient pairs.13 Examination of the characteristics of donor–recipient pairs with actual and imputed HLA-match assignments showed the majority of these transplants occurred between the years 2000 and 2004 and that in vivo T-cell depletion was more common for this group. Of the 491 transplants between 2000 and 2004, 70% did not have allele-level HLA typing. In the later period, 40% of donor–recipient pairs did not have allele-level HLA typing. Donor–recipient HLA match assignments based on actual and imputed high-resolution typings were analyzed independently for associations with nonrelapse mortality; none were found (supplemental Table 1, available on the Blood Web site).
Outcomes
The primary outcome investigated was nonrelapse mortality defined as the time from transplantation to death not related to relapse. Other outcomes evaluated were: neutrophil recovery (inverse of primary graft failure, defined as achieving an absolute neutrophil count ≥0.5 × 109/L for 3 consecutive measurements on different days), grades 2-4 acute GVHD,14 chronic GVHD,15 relapse, and overall survival.
Statistical methods
The probabilities of neutrophil recovery, GVHD, nonrelapse mortality, and relapse were calculated using the cumulative incidence estimator.16 Death was the competing risk for neutrophil recovery and GVHD. For relapse, nonrelapse mortality was the competing risk; for nonrelapse mortality, relapse was the competing risk. The probability of overall survival was calculated using the Kaplan-Meier estimator.17 The 95% confidence intervals (CI) were calculated with log transformation.
To analyze the association between clinical outcomes and donor–recipient HLA match, a multivariate pseudo-observation logistic regression model18 was built for neutrophil recovery at day +28, and Cox regression models19 were built for acute and chronic GVHD, nonrelapse mortality, relapse, and overall mortality. The result generated from the logistic regression model is expressed as odds ratio (OR) and from Cox regression models as hazard ratio (HR).
Donor–recipient HLA match was examined for the effect of overall HLA match (matched [8/8] vs 1-allele mismatch [7/8] vs 2-allele mismatch [6/8] vs 3-allele mismatch [5/8] vs 4-allele mismatch [4/8] vs 5-allele mismatch [3/8]), and for the effect of a single mismatch at the allele level for individual HLA loci. The other variables tested are shown in Table 1. The sieve method was used to determine the optimal cutpoint for precryopreserved total nucleated cell (TNC) of 3 × 107/kg for nonrelapse mortality and overall mortality.20 Models were built with the use of a forward stepwise selection procedure and confirmed with a backward selection procedure. Proportional hazards assumption was tested for each covariate individually; all covariates met this assumption. First-order interactions between each covariate and HLA match were tested and none found. All P values are 2-sided and P values ≤ .01 were considered statistically significant. Analyses were performed using SAS 9.3 (SAS Institute, Cary, NC).
Patient, disease, and transplantation characteristics
| . | Allele-level HLA match . | ||||||
|---|---|---|---|---|---|---|---|
| . | Matched . | 1-allele mismatch . | 2-allele mismatch . | 3-allele mismatch . | 4-allele mismatch . | 5-allele mismatch . | P value . |
| Number | 117 | 230 | 413 | 466 | 256 | 86 | |
| Gender, male | 67 (57%) | 116 (50%) | 223 (54%) | 272 (58%) | 134 (52%) | 49 (57%) | .33 |
| Age at transplantation | <.001 | ||||||
| ≤16 years | 101 (86%) | 178 (77%) | 322 (78%) | 313 (67%) | 152 (59%) | 53 (62%) | |
| >16 years | 16 (14%) | 52 (23%) | 91 (22%) | 153 (33%) | 104 (41%) | 33 (38%) | |
| Recipient cytomegalovirus seropositivity | .56 | ||||||
| Positive | 61 (52%) | 123 (53%) | 196 (47%) | 248 (53%) | 125 (49%) | 47 (55%) | |
| Negative | 53 (45%) | 97 (42%) | 202 (49%) | 206 (44%) | 124 (48%) | 34 (40%) | |
| Not reported | 3 (3%) | 10 (4%) | 15 (4%) | 12 (3%) | 7 (3%) | 5 (5%) | |
| Disease | .77 | ||||||
| Acute myeloid leukemia | 47 (40%) | 86 (37%) | 141 (34%) | 172 (37%) | 113 (44%) | 32 (37%) | |
| Acute lymphoblastic leukemia | 60 (51%) | 117 (51%) | 233 (56%) | 256 (55%) | 110 (43%) | 44 (51%) | |
| Myelodysplastic syndrome | 10 (9%) | 27 (12%) | 39 (9%) | 38 (8%) | 33 (13%) | 10 (12%) | |
| Disease status | .52 | ||||||
| First complete remission | 50 (43%) | 75 (33%) | 160 (39%) | 162 (35%) | 77 (30%) | 27 (31%) | |
| Second complete remission | 47 (40%) | 95 (41%) | 158 (38%) | 217 (47%) | 97 (38%) | 36 (42%) | |
| Relapse/RAEB | 20 (17%) | 59 (26%) | 95 (23%) | 86 (18%) | 81 (32%) | 23 (27%) | |
| Not reported | __ | 1 (<1%) | __ | 1 (<1%) | 1 (<1%) | __ | |
| Conditioning regimen | .88 | ||||||
| TBI containing regimen | 60 (51%) | 122 (53%) | 226 (55%) | 260 (56%) | 132 (52%) | 49 (62%) | |
| Non-TBI containing regimen | 57 (49%) | 108 (47%) | 187 (45%) | 206 (44%) | 124 (48%) | 37 (38%) | |
| In vivo T-cell depletion | .14 | ||||||
| Yes | 87 (74%) | 165 (72% | 290 (70%) | 344 (74%) | 201 (79%) | 69 (80%) | |
| None | 28 (24%) | 52 (23%) | 106 (26%) | 111 (24%) | 47 (18%) | 14 (16%) | |
| Not reported | 2 (2%) | 13 (6%) | 17 (4%) | 11 (2%) | 8 (3%) | 3 (3%) | |
| Total nucleated cell dose, prefreeze | <.001 | ||||||
| <3 × 107/kg | 14 (12%) | 33 (14%) | 56 (14%) | 66 (14%) | 40 (16%) | 10 (12%) | |
| ≥3-5 × 107/kg | 18 (15%) | 52 (23%) | 99 (24%) | 156 (33%) | 99 (39%) | 31 (36%) | |
| >5 × 107/kg | 81 (69%) | 138 (60%) | 242 (59%) | 233 (50%) | 106 (41%) | 43 (50%) | |
| Not reported | 4 (4%) | 7 (3%) | 16 (4%) | 11 (2%) | 11 (4%) | 2 (2%) | |
| Transplant period | .04 | ||||||
| 2000-2004 | 28 (24%) | 74 (32%) | 136 (33%) | 128 (27%) | 91 (36%) | 34 (40%) | |
| 2005-2010 | 89 (76%) | 156 (68%) | 277 (67%) | 338 (73%) | 165 (64%) | 52 (60%) | |
| Follow-up, median (range), months | 37 (3-131) | 46 (5-132) | 46 (3-147) | 37 (3-127) | 45 (3-107) | 35 (3-122) | |
| . | Allele-level HLA match . | ||||||
|---|---|---|---|---|---|---|---|
| . | Matched . | 1-allele mismatch . | 2-allele mismatch . | 3-allele mismatch . | 4-allele mismatch . | 5-allele mismatch . | P value . |
| Number | 117 | 230 | 413 | 466 | 256 | 86 | |
| Gender, male | 67 (57%) | 116 (50%) | 223 (54%) | 272 (58%) | 134 (52%) | 49 (57%) | .33 |
| Age at transplantation | <.001 | ||||||
| ≤16 years | 101 (86%) | 178 (77%) | 322 (78%) | 313 (67%) | 152 (59%) | 53 (62%) | |
| >16 years | 16 (14%) | 52 (23%) | 91 (22%) | 153 (33%) | 104 (41%) | 33 (38%) | |
| Recipient cytomegalovirus seropositivity | .56 | ||||||
| Positive | 61 (52%) | 123 (53%) | 196 (47%) | 248 (53%) | 125 (49%) | 47 (55%) | |
| Negative | 53 (45%) | 97 (42%) | 202 (49%) | 206 (44%) | 124 (48%) | 34 (40%) | |
| Not reported | 3 (3%) | 10 (4%) | 15 (4%) | 12 (3%) | 7 (3%) | 5 (5%) | |
| Disease | .77 | ||||||
| Acute myeloid leukemia | 47 (40%) | 86 (37%) | 141 (34%) | 172 (37%) | 113 (44%) | 32 (37%) | |
| Acute lymphoblastic leukemia | 60 (51%) | 117 (51%) | 233 (56%) | 256 (55%) | 110 (43%) | 44 (51%) | |
| Myelodysplastic syndrome | 10 (9%) | 27 (12%) | 39 (9%) | 38 (8%) | 33 (13%) | 10 (12%) | |
| Disease status | .52 | ||||||
| First complete remission | 50 (43%) | 75 (33%) | 160 (39%) | 162 (35%) | 77 (30%) | 27 (31%) | |
| Second complete remission | 47 (40%) | 95 (41%) | 158 (38%) | 217 (47%) | 97 (38%) | 36 (42%) | |
| Relapse/RAEB | 20 (17%) | 59 (26%) | 95 (23%) | 86 (18%) | 81 (32%) | 23 (27%) | |
| Not reported | __ | 1 (<1%) | __ | 1 (<1%) | 1 (<1%) | __ | |
| Conditioning regimen | .88 | ||||||
| TBI containing regimen | 60 (51%) | 122 (53%) | 226 (55%) | 260 (56%) | 132 (52%) | 49 (62%) | |
| Non-TBI containing regimen | 57 (49%) | 108 (47%) | 187 (45%) | 206 (44%) | 124 (48%) | 37 (38%) | |
| In vivo T-cell depletion | .14 | ||||||
| Yes | 87 (74%) | 165 (72% | 290 (70%) | 344 (74%) | 201 (79%) | 69 (80%) | |
| None | 28 (24%) | 52 (23%) | 106 (26%) | 111 (24%) | 47 (18%) | 14 (16%) | |
| Not reported | 2 (2%) | 13 (6%) | 17 (4%) | 11 (2%) | 8 (3%) | 3 (3%) | |
| Total nucleated cell dose, prefreeze | <.001 | ||||||
| <3 × 107/kg | 14 (12%) | 33 (14%) | 56 (14%) | 66 (14%) | 40 (16%) | 10 (12%) | |
| ≥3-5 × 107/kg | 18 (15%) | 52 (23%) | 99 (24%) | 156 (33%) | 99 (39%) | 31 (36%) | |
| >5 × 107/kg | 81 (69%) | 138 (60%) | 242 (59%) | 233 (50%) | 106 (41%) | 43 (50%) | |
| Not reported | 4 (4%) | 7 (3%) | 16 (4%) | 11 (2%) | 11 (4%) | 2 (2%) | |
| Transplant period | .04 | ||||||
| 2000-2004 | 28 (24%) | 74 (32%) | 136 (33%) | 128 (27%) | 91 (36%) | 34 (40%) | |
| 2005-2010 | 89 (76%) | 156 (68%) | 277 (67%) | 338 (73%) | 165 (64%) | 52 (60%) | |
| Follow-up, median (range), months | 37 (3-131) | 46 (5-132) | 46 (3-147) | 37 (3-127) | 45 (3-107) | 35 (3-122) | |
TBI dose ranged from 800-1440 cGy; 75% received 1200 cGy or 1320 cGy; n = 4 received 800 cGy and n = 1 received 900 cGy. In vivo T-cell depletion: 1156 transplant regimens included in vivo T-cell depletion. Of these, 1146 received ATG (548 rabbit ATG, 284, equine ATG, and information was not available for 314); 10 patients received alemtuzumab.
ATG, anti-thymocyte globulin; RAEB, refractory anemia with excess blast; TBI, total body irradiation.
Results
Patient, disease, and transplant characteristics
Table 1 describes patients, their disease, and transplant characteristics by their HLA-match groups. The median age of the study population was 9 years and 71% of patients were 16 years or younger at transplantation. The distribution of diseases and disease status at transplantation, conditioning regimens, and GVHD prophylaxis regimens did not differ significantly among HLA-match groups. The median precryopreserved TNC dose of units was 5 × 107/kg. Overall, only 14% of transplantations were performed using units with TNC <3 × 107/kg. There were no significant differences in the use of UCB units with TNC <3 × 107/kg among HLA-match groups. However, there were differences among groups in the use of higher dose TNC units. Seventy percent of HLA-matched and 60% of 1- and 2-allele mismatched transplants used units with TNC >5 × 107/kg compared with 47% and 50% of 3- and 4-allele and 5-allele mismatched transplants, respectively. There were differences in the characteristics of patients by their age (supplemental Table 2). Older patients were more likely to have acute myeloid leukemia or myelodysplastic syndrome, receive non-TBI containing conditioning, UCB units mismatched at 2, 3, or 4 HLA loci and contain <3 × 107/kg or 3-5 × 107/kg total nucleated cells.
Only 7% of donor-recipient pairs were HLA matched at A, B, C, and DRB1 by allele-level typing. Most donor–recipient pairs were mismatched at 2 or 3 alleles, accounting for 56% of transplantations. Fifteen percent of donor–recipient pairs were mismatched at 1 allele and 16% at 4 alleles. The remaining 5% were mismatched at 5 alleles. Table 2 shows the distribution of allele-level HLA-match compared with lower resolution HLA typing, which represents the current standard for unit selection. Only 54% of transplants considered matched at HLA-A, -B, and -DRB1 using conventional standards were actually matched at the allele level at 8 loci. Similarly, only 25% of transplants mismatched at 1 HLA locus and 10% of transplants mismatched at 2 HLA loci according to conventional standards were mismatched at 1 and 2 alleles, respectively.
Allele-level HLA typing (allele-level HLA match at HLA-A, -B, -C, and -DRB1) compared with lower resolution HLA typing (antigen-level HLA match at HLA-A and -B and allele-level at -DRB1)
| Lower resolution HLA typing . | Allele-level HLA typing . | |||||
|---|---|---|---|---|---|---|
| . | 5-allele mismatch . | 4-allele mismatch . | 3-allele mismatch . | 2-allele mismatch . | 1-allele mismatch . | Matched . |
| 3-antigen mismatch (n = 26) | 16 (62%) | 8 (31%) | 2 (7%) | __ | __ | __ |
| 2-antigen mismatch (n = 605) | 65 (11%) | 187 (31%) | 294 (48%) | 59 (10%) | __ | __ |
| 1-antigen mismatch (n = 720) | 5 (1%) | 60 (8%) | 161 (22%) | 315 (44%) | 179 (25%) | __ |
| HLA-matched antigen-level (n = 217) | __ | 1 (<1%) | 9 (4%) | 39 (18%) | 51 (24%) | 117 (54%) |
| Lower resolution HLA typing . | Allele-level HLA typing . | |||||
|---|---|---|---|---|---|---|
| . | 5-allele mismatch . | 4-allele mismatch . | 3-allele mismatch . | 2-allele mismatch . | 1-allele mismatch . | Matched . |
| 3-antigen mismatch (n = 26) | 16 (62%) | 8 (31%) | 2 (7%) | __ | __ | __ |
| 2-antigen mismatch (n = 605) | 65 (11%) | 187 (31%) | 294 (48%) | 59 (10%) | __ | __ |
| 1-antigen mismatch (n = 720) | 5 (1%) | 60 (8%) | 161 (22%) | 315 (44%) | 179 (25%) | __ |
| HLA-matched antigen-level (n = 217) | __ | 1 (<1%) | 9 (4%) | 39 (18%) | 51 (24%) | 117 (54%) |
Nonrelapse mortality
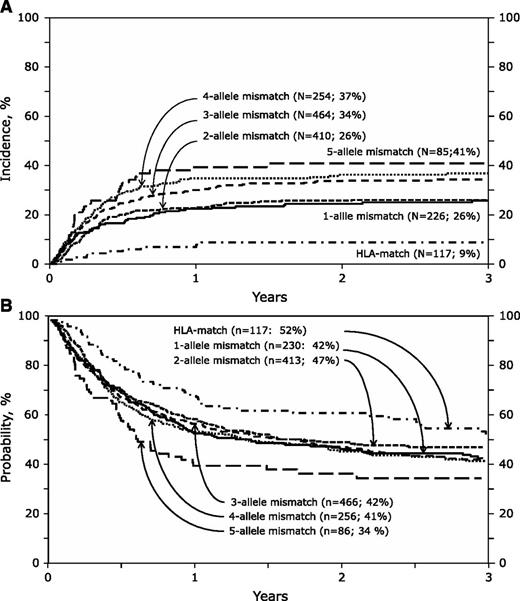
The risk of nonrelapse mortality was independently associated with the overall degree of HLA mismatch (Table 3; Figure 1A). Compared with HLA-matched transplants, nonrelapse mortality risks were not significantly different between transplantations mismatched at 1 and 2 alleles (HR 2.79 and 2.69, respectively) or between transplantations mismatched at 3 or 4 alleles (HR 3.60 and 3.48, respectively). However, compared with transplantations mismatched at 1 or 2 alleles, risks were higher after transplantations mismatched at 3 and 4 alleles (HR 1.31, 95% CI 1.07-1.60, P = .01) and at 5 alleles (HR 1.69, 95% CI 1.62-2.46, P = .006). There was no significant difference in the risk estimate of mismatching at 5 alleles compared with mismatching at 3 and 4 alleles (HR 1.29, 95% CI 0.90-1.86, P = .16). Examination of the risk estimates of mismatching at specific loci suggest isolated allele-level mismatches at HLA-A, -C, or -DRB1 but not at HLA-B are associated with higher nonrelapse mortality risks (Table 3).
Association of HLA match on nonrelapse and overall mortality
| . | . | Nonrelapse mortality* . | Overall mortality† . | ||
|---|---|---|---|---|---|
| . | Number . | HR (95% CI) . | P value . | HR (95% CI) . | P value . |
| Overall HLA match | |||||
| 1-allele mismatched vs matched | 226 vs 117 | 2.79 (1.46-5.34) | .002 | 1.31 (0.95-1.81) | .09 |
| 2-allele mismatched vs matched | 410 vs 117 | 2.69 (1.44-5.03) | .002 | 1.20 (0.89-1.63) | .23 |
| 3-allele mismatched vs matched | 464 vs 117 | 3.60 (1.94-6.69) | <.0001 | 1.36 (1.01-1.84) | .04 |
| 4-allele mismatched vs matched | 254 vs 117 | 3.48 (1.84-6.66) | .0001 | 1.15 (0.83-1.58) | .39 |
| 5-allele mismatched vs matched | 85 vs 117 | 4.61 (2.31-9.17) | <.0001 | 1.63 (1.12-2.39) | .01 |
| 3- to 4-allele vs 1- to 2-allele mismatched | 718 vs 637 | 1.31 (1.07-1.59) | .01 | 1.03 (0.89-1.19) | .69 |
| 5- vs 1- to 2-allele mismatched | 85 vs 637 | 1.69 (1.16-2.46) | .006 | 1.32 (0.99-1.76) | .06 |
| 5- vs 3- to 4-allele mismatched | 85 vs 718 | 1.29 (0.90-1.86) | .16 | 1.28 (0.96-1.70) | .09 |
| Single allele mismatch | |||||
| HLA-A mismatched vs matched | 117 vs 117 | 3.05 (1.52-6.14) | .002 | 1.27 (0.88-1.83) | .19 |
| HLA-B mismatched vs matched | 31 vs 117 | 1.26 (0.35-4.55) | .72 | 1.06 (0.75-1.97) | .77 |
| HLA-C mismatched vs matched | 40 vs 117 | 3.04 (1.28-7.20) | .01 | 1.41 (0.86-2.31) | .17 |
| HLA-DRB1 mismatched vs matched | 66 vs 117 | 2.93 (1.38-6.25) | .005 | 1.31 (0.96-1.78) | .17 |
| . | . | Nonrelapse mortality* . | Overall mortality† . | ||
|---|---|---|---|---|---|
| . | Number . | HR (95% CI) . | P value . | HR (95% CI) . | P value . |
| Overall HLA match | |||||
| 1-allele mismatched vs matched | 226 vs 117 | 2.79 (1.46-5.34) | .002 | 1.31 (0.95-1.81) | .09 |
| 2-allele mismatched vs matched | 410 vs 117 | 2.69 (1.44-5.03) | .002 | 1.20 (0.89-1.63) | .23 |
| 3-allele mismatched vs matched | 464 vs 117 | 3.60 (1.94-6.69) | <.0001 | 1.36 (1.01-1.84) | .04 |
| 4-allele mismatched vs matched | 254 vs 117 | 3.48 (1.84-6.66) | .0001 | 1.15 (0.83-1.58) | .39 |
| 5-allele mismatched vs matched | 85 vs 117 | 4.61 (2.31-9.17) | <.0001 | 1.63 (1.12-2.39) | .01 |
| 3- to 4-allele vs 1- to 2-allele mismatched | 718 vs 637 | 1.31 (1.07-1.59) | .01 | 1.03 (0.89-1.19) | .69 |
| 5- vs 1- to 2-allele mismatched | 85 vs 637 | 1.69 (1.16-2.46) | .006 | 1.32 (0.99-1.76) | .06 |
| 5- vs 3- to 4-allele mismatched | 85 vs 718 | 1.29 (0.90-1.86) | .16 | 1.28 (0.96-1.70) | .09 |
| Single allele mismatch | |||||
| HLA-A mismatched vs matched | 117 vs 117 | 3.05 (1.52-6.14) | .002 | 1.27 (0.88-1.83) | .19 |
| HLA-B mismatched vs matched | 31 vs 117 | 1.26 (0.35-4.55) | .72 | 1.06 (0.75-1.97) | .77 |
| HLA-C mismatched vs matched | 40 vs 117 | 3.04 (1.28-7.20) | .01 | 1.41 (0.86-2.31) | .17 |
| HLA-DRB1 mismatched vs matched | 66 vs 117 | 2.93 (1.38-6.25) | .005 | 1.31 (0.96-1.78) | .17 |
Adjusted for patient age, disease, disease status, precryopreserved TNC, and transplant period: nonrelapse mortality risks were higher for patients >16 years compared with younger patients (HR 1.60, 95% CI 1.26-2.03, P < .001); acute lymphoblastic leukemia (HR 1.35, 95% CI 1.09-1.69, P = .007) compared with acute myeloid leukemia; risks were not significantly different for myelodysplastic syndrome (HR 1.23, 95% CI 0.88-1.72, P = .26) compared with acute myeloid leukemia; for those transplanted in relapse/refractory anemia with excess blasts (HR 1.68, 95% CI 1.28-2.20, P < .001) but not those transplanted in second complete remission (HR 0.97, 95% CI 0.77-1.22, P = .82) compared with those in first complete remission; TNC <3 × 107/kg (HR 1.52, 95% CI 1.16-1.96, P = .011) and transplants between 2000 and 2004 (HR 1.52, 95% CI 1.23-1.85, P < .001) compared with transplants between 2005 and 2010.
Adjusted for patient age, disease, disease status, TNC, and transplant period: overall mortality risks were higher for patients >16 years compared with younger patients (HR 1.43, 95% CI 1.21-1.70, P < .001); acute lymphoblastic leukemia (HR 1.26, 95% CI 1.08-1.48, P = .004) compared with acute myeloid leukemia; mortality risks were lower for those with myelodysplastic syndrome (HR 0.65, 95% CI 0.50-0.84, P = .001) compared with acute myeloid leukemia; for those transplanted in relapse/refractory anemia with excess blasts (HR 2.98, 95% CI 2.44-3.64, P < .001) and in second complete remission (HR 1.51, 95% CI 1.21-1.88, P < .001) compared with those in first remission; TNC <3 × 107/kg (HR 1.37, 95% CI 1.14-1.67, P = .001) compared with TNC ≥3 × 107/kg and transplants between 2000 and 2004 (HR 1.35, 95% CI 1.18-1.56, P < .001) compared with transplants between 2005 and 2010.
Nonrelapse mortality and overall survival. (A) The cumulative incidence of nonrelapse mortality by HLA match: the 3-year incidence of nonrelapse mortality after HLA-matched, 1-allele mismatch, 2-allele mismatch, 3-allele mismatch, 4-allele mismatch, and 5-allele mismatch transplants were 9% (95% CI 4-14), 26% (95% CI 20-32), 26% (95% CI 22-30), 34% (95% CI 30-39), 37% (95% CI 31-43), and 41% (95% CI 30-51), respectively. (B) The probability of overall survival by HLA match: the 3-year probability of overall survival after HLA-matched, 1-allele mismatch, 2-allele mismatch, 3-allele mismatch, 4-allele mismatch, and 5-allele mismatch transplants were 52% (95% CI 42-62), 42% (95% CI 36-49), 47% (95% CI 42-52), 42% (95% CI 37-47), 41% (95% CI 35-47), and 34% (95% CI 23-45), respectively.
Nonrelapse mortality and overall survival. (A) The cumulative incidence of nonrelapse mortality by HLA match: the 3-year incidence of nonrelapse mortality after HLA-matched, 1-allele mismatch, 2-allele mismatch, 3-allele mismatch, 4-allele mismatch, and 5-allele mismatch transplants were 9% (95% CI 4-14), 26% (95% CI 20-32), 26% (95% CI 22-30), 34% (95% CI 30-39), 37% (95% CI 31-43), and 41% (95% CI 30-51), respectively. (B) The probability of overall survival by HLA match: the 3-year probability of overall survival after HLA-matched, 1-allele mismatch, 2-allele mismatch, 3-allele mismatch, 4-allele mismatch, and 5-allele mismatch transplants were 52% (95% CI 42-62), 42% (95% CI 36-49), 47% (95% CI 42-52), 42% (95% CI 37-47), 41% (95% CI 35-47), and 34% (95% CI 23-45), respectively.
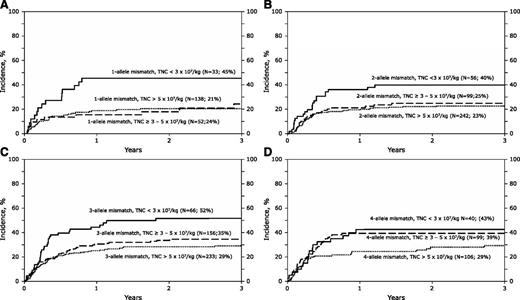
Higher nonrelapse mortality was also associated with age older than 16 years, the diagnosis of acute lymphoblastic leukemia, transplantations done in relapse, TNC <3 × 107/kg, and transplants performed before 2005. The effect of TNC (P = .78) and age (P = .97) on nonrelapse mortality was independent of HLA match. Further, the effects of HLA match were tested separately in younger and older recipients to confirm the independent effect of HLA match on nonrelapse mortality (supplemental Table 3). We also examined for an effect of TNC within HLA groups mismatched at 1, 2, 3, and 4 alleles. For transplantations with units containing TNC 3 × 107/kg or higher, nonrelapse mortality rates were not different with further increases in TNC (Figures 2A-D). Similarly, nonrelapse mortality rates were not different among recipients of HLA-matched transplants with higher TNC; the 3-year probabilities of nonrelapse mortality were 21%, 6%, and 6% after transplantation of units with TNC <3 × 107/kg, ≥3-5 × 107/kg, and >5 × 107/kg, respectively.
Nonrelapse mortality by total nucleated cell dose. (A) The cumulative incidence of nonrelapse mortality by precryopreserved TNC in recipients of 1-allele mismatched transplants: 45% (95% CI 29-62), 24% (95% 13-38), and 21% (14-28) for units with TNC <3 × 107/kg, 3-5 × 107/kg, and >5 × 107/kg, respectively. (B) The cumulative incidence of nonrelapse mortality by precryopreserved TNC in recipients of 2-allele mismatched transplants: 40% (95% CI 27-53), 25% (95% 17-34), and 23% (18-28) for units with TNC <3 × 107/kg, 3-5 × 107/kg, and >5 × 107/kg, respectively. (C) The cumulative incidence of nonrelapse mortality by precryopreserved TNC in recipients of 3-allele mismatched transplants: 52% (95% CI 39-64), 35% (95% 27-42), and 29% (23-35) for units with TNC <3 × 107/kg, 3-5 × 107/kg, and >5 × 107/kg, respectively. (D) The cumulative incidence of nonrelapse mortality by precryopreserved TNC in recipients of 4-allele mismatched transplants: 43% (95% CI 28-58), 39% (95% 30-49), and 29% (21-39) for units with TNC <3 × 107/kg, 3-5 × 107/kg and >5 × 107/kg, respectively.
Nonrelapse mortality by total nucleated cell dose. (A) The cumulative incidence of nonrelapse mortality by precryopreserved TNC in recipients of 1-allele mismatched transplants: 45% (95% CI 29-62), 24% (95% 13-38), and 21% (14-28) for units with TNC <3 × 107/kg, 3-5 × 107/kg, and >5 × 107/kg, respectively. (B) The cumulative incidence of nonrelapse mortality by precryopreserved TNC in recipients of 2-allele mismatched transplants: 40% (95% CI 27-53), 25% (95% 17-34), and 23% (18-28) for units with TNC <3 × 107/kg, 3-5 × 107/kg, and >5 × 107/kg, respectively. (C) The cumulative incidence of nonrelapse mortality by precryopreserved TNC in recipients of 3-allele mismatched transplants: 52% (95% CI 39-64), 35% (95% 27-42), and 29% (23-35) for units with TNC <3 × 107/kg, 3-5 × 107/kg, and >5 × 107/kg, respectively. (D) The cumulative incidence of nonrelapse mortality by precryopreserved TNC in recipients of 4-allele mismatched transplants: 43% (95% CI 28-58), 39% (95% 30-49), and 29% (21-39) for units with TNC <3 × 107/kg, 3-5 × 107/kg and >5 × 107/kg, respectively.
Overall mortality
Despite higher nonrelapse mortality, differences in overall mortality did not reach statistical significance except for transplants mismatched at 5 alleles compared with HLA-matched transplants (Table 3). Mortality risks were not significantly different between transplantations mismatched at 1 and 2 alleles (HR 1.31 and 1.20, respectively) or between transplantations mismatched at 3 and 4 alleles (HR 1.36 and 1.15, respectively). Other variables associated with overall mortality were age older than 16 years, cytomegalovirus seropositivity, diagnosis of acute lymphoblastic and myeloid leukemia, transplantation in relapse, TNC <3 × 107/kg, and transplantation before 2005.
Neutrophil recovery
Compared with HLA-matched transplants, the likelihood of neutrophil recovery at day +28 was lower for transplants mismatched at 3, 4, or 5 alleles (Table 4). Recovery was not significantly different for transplants mismatched at 1 and 2 alleles (OR 0.72 and 0.86, respectively) and 3 and 4 alleles (OR 0.56 and 0.55, respectively). Compared with transplants mismatched at 1 and 2 alleles, recovery was lower after transplants mismatched at 3 or 4 alleles (OR 0.69, 95% 0.55-0.86, P = .001) and 5 alleles (OR 0.56, 95% CI 0.35-0.89, P = .01). However, recovery did not differ between 3- and 4-allele and 5-allele mismatched transplants (OR 0.81, 95% CI 0.51-1.29, P = .38). We did not observe significant differences in the risk estimates considering mismatching at specific HLA loci (supplemental Table 4).
GVHD
The risks of grades 2-4 acute GVHD were higher after transplantation of mismatched UCB units compared with matched transplants but the differences in risk estimates did not reach the level of significance set for the current analyses (Table 4; P value .04-.06). There were no significant differences in the risk estimates considering mismatching at specific HLA loci (supplemental Table 4). The risks of chronic GVHD were not significantly associated with HLA matching, considered as overall degree of HLA disparity (Table 4) or when mismatching at specific loci were considered (supplemental Table 4).
Relapse
The risk of relapse after transplantation was not significantly associated with HLA matching, considered as overall degree of HLA mismatch except for transplantations mismatched at 4 alleles (Table 4) or when mismatching at specific HLA loci were considered (supplemental Table 4). However, relapse risks were higher with advanced disease (transplantation in second complete remission or relapse), diagnosis of acute myeloid leukemia, and with units containing TNC <3 × 107/kg. The observed effect of disease status and cell dose on leukemia relapse was independent of HLA match.
Availability of UCB units considering allele-level HLA match
The National Marrow Donor Program maintains a registry of 180 000 banked, unrelated UCB units. Using a population genetics model that calculated the population-specific HLA-match likelihoods of identifying an HLA-matched or a 1- or 2-allele mismatched units, we predicted the availability of such units given current inventory size.21,22 The current inventory would ensure that one-third of Caucasians will have a fully matched unit at the allele level. For the other races in the United States, the current registry would provide a much lower success rate with only 5% to 10% identifying a fully matched unit. However, many more patients needing a transplant will have a unit mismatched at 1 or 2 alleles. For Caucasians, the probability of identifying a unit mismatched at 1 or 2 alleles is 80% and 98%, respectively. Although substantially lower, 33% to 45% of US minorities will identify a unit mismatched at 1 allele and 80% to 85% at 2 alleles. With the worldwide inventory of 600 000 units, it is plausible that an even higher proportion of patients will have access to better HLA-matched units.
Discussion
We found that allele-level matching at HLA-A, -B, -C, and -DRB1 between the donor (UCB unit) and recipient is associated with the lowest nonrelapse mortality after UCB transplantation for acute leukemia and myelodysplastic syndrome. Thus reliance on HLA typings at low or intermediate resolution or the use of selection algorithms that do not consider matching at the HLA-C locus is not fully adequate for selecting optimal UCB units. When a fully allele-level HLA-matched UCB unit is not available, mismatches at 1 and 2 alleles are better tolerated than mismatches at 3, 4, and 5 alleles. Indeed, nonrelapse mortality rates are 10% to 15% lower after transplantations mismatched at 1 and 2 alleles compared with mismatches at 3 or more alleles. We did not observe significant differences in nonrelapse mortality risks between transplantations mismatched at 3, 4, and 5 alleles, suggesting the absence of an additive effect for nonrelapse mortality beyond mismatching at 2 alleles. The absence of an additive effect with mismatching in excess of 2 HLA loci is described after adult unrelated donor transplantations.3 Whereas nonrelapse mortality risks are virtually identical after 3- and 4-allele mismatched transplants, the risks are about one-third higher after 5-allele mismatched transplants compared with 3- and 4-allele mismatched transplants. It is plausible that, in a larger population, there could be significant differences in nonrelapse mortality risks between 5-allele and 3- and 4-allele mismatched transplants.
When considering locus-specific effects, a single HLA mismatch at HLA-A, -C, or -DRB1 was associated with a threefold increase in nonrelapse mortality risk. An isolated mismatch at HLA-B appears to be better tolerated, but the likelihood of identifying such a unit is relatively low because HLA-B and -C loci are in linkage disequilibrium and a mismatch at 1 locus is likely to occur with a mismatch at the other. This differential effect of HLA-B mismatching must also be viewed with caution because there were only 31 donor–recipient pairs with an isolated HLA-B mismatch in our data set.
TNC content of the unit was the only other donor characteristic associated with nonrelapse mortality, and its effect was independent of HLA match. We found that nonrelapse mortality rates are 15% to 20% higher when the UCB units contained <3 × 107 TNC/kg. Selecting units with TNC in excess of the required minimum was not associated with lower nonrelapse mortality, providing further support for the critical importance of HLA match above a minimum cell dose threshold. In the current analyses, 70% of children received units that contained TNC in excess of 5 × 107/kg and about half of adults received units with TNC ranging from 3 to 5 × 107/kg. By lowering the cell dose threshold to a minimum of 3 × 107/kg, it is plausible that better matched units may have been available for these patients. Two recent reports, one in children and the other in adults that compared the transplantation of 1 adequately dosed UCB unit (TNC >2.5 × 107/kg) to the transplantation of 2 UCB units, also failed to demonstrate differences in nonrelapse mortality risks and overall survival, lending support to our observation that TNC in excess of the minimum required does not lower mortality risks.23,24 Together these data confirm the need for a minimum TNC to ensure engraftment and thereafter prioritize UCB unit selection on HLA match considering allele-level HLA typing and incorporating matching at HLA-C locus.
Ours is a predominantly pediatric population, with 29% of recipients older than age 16 years. Older recipients were more likely to receive UCB units mismatched at 2, 3, or 4 HLA loci and with TNC <3 × 107/kg. Because both older age and units with TNC <3 × 107/kg were also associated with higher mortality, we confirmed that the observed effect of HLA match on nonrelapse mortality in older recipients was independent of age and TNC. Consequently, the importance of better HLA matching on lowering nonrelapse mortality is applicable to both children and adults. However, nonmortality risks are also mitigated by older age and lower TNC, which dampens the effect of HLA match in older recipients. Our findings differ from another recent report from Japan that concluded HLA match was not associated with nonrelapse mortality or overall survival in older patients.25 However, in that report,25 HLA-C was not considered, allele level typing was used only for DRB1, and 80% of transplants were mismatched at 2 or 3 loci at a lower resolution HLA match. It is likely that the small (n = 76) control group of “matched” transplantations in that report included a high proportion of allelic disparities and/or disparities at HLA-C.
The observed higher nonrelapse mortality risks in our study did not translate into lower overall survival except for transplantations mismatched at 5 alleles. The absence of an adverse effect on overall survival after mismatched UCB transplantation, except for transplants with TNC dose <2.5 × 107/kg, has been recorded by others.26 The absence of a survival advantage in the current analyses may be attributed to the high competing risk of death from leukemia, obscuring the ability of a decrease in nonrelapse mortality to lead to an improvement in overall survival in unselected populations, as in the current analyses. We did not find significant differences in relapse risk after HLA-matched and HLA-mismatched transplantations except after transplantation mismatched at 4 alleles, which is likely a chance finding. The absence of lower relapse risks after HLA-mismatched transplants is consistent with a previous report24 and challenges the common practice of tolerating mismatches in the belief that higher mortality is offset by lower leukemia relapse.
Association of HLA-match on neutrophil recovery, acute and chronic GVHD and leukemia recurrence
| Overall HLA-match . | Number . | OR/ HR (95% CI) . | P value . |
|---|---|---|---|
| Neutrophil recovery* | |||
| 1-allele mismatched vs matched | 227 vs 116 | 0.72 (0.44-1.70) | .18 |
| 2-allele mismatched vs matched | 410 vs 116 | 0.86 (0.54-1.36) | .52 |
| 3-allele mismatched vs matched | 464 vs 116 | 0.56 (0.36-0.88) | .01 |
| 4-allele mismatched vs matched | 254 vs 116 | 0.55 (0.34-0.88) | .01 |
| 5-allele mismatched vs matched | 84 vs 116 | 0.45 (0.25-0.82) | .009 |
| 3-4 allele vs 1-2 allele mismatched | 718 vs 637 | 0.69 (0.55-0.86) | .001 |
| 5-allele vs 1-2 allele mismatched | 85 vs 637 | 0.56 (0.35-0.89) | .01 |
| 5-allele vs 3-4 allele mismatched | 85 vs 718 | 0.81 (0.51-1.29) | .38 |
| Grade 2-4 acute GVHD† | |||
| 1-allele mismatched vs matched | 226 vs 117 | 1.27 (0.83-1.93) | .26 |
| 2-allele mismatched vs matched | 410 vs 117 | 1.45 (0.99-2.12) | .06 |
| 3-allele mismatched vs matched | 464 vs 117 | 1.53 (1.05-2.23) | .03 |
| 4-allele mismatched vs matched | 254 vs 117 | 1.51 (1.00-2.27) | .05 |
| 5-allele mismatched vs matched | 85 vs 117 | 1.49 (0.89-2.49) | .13 |
| 3-4 allele vs 1-2 allele mismatched | 718 vs 637 | 1.09 (0.92-1.32) | .31 |
| 5-allele vs 1-2 allele mismatched | 85 vs 637 | 1.08 (0.72-1.61) | .72 |
| 5-allele vs 3-4 allele mismatched | 85 vs 718 | 0.98 (0.66-1.46) | .92 |
| Chronic GVHD‡ | |||
| 1-allele mismatched vs matched | 226 vs 117 | 0.76 (0.47-1.23) | .27 |
| 2-allele mismatched vs matched | 410 vs 117 | 1.16 (0.77-1.75) | .48 |
| 3-allele mismatched vs matched | 464 vs 117 | 1.02 (0.67-1.54) | .93 |
| 4-allele mismatched vs matched | 254 vs 117 | 1.20 (0.80-1.87) | .48 |
| 5-allele mismatched vs matched | 85 vs 117 | 1.27 (0.70-2.31) | .27 |
| 3-4 allele vs 1-2 allele mismatched | 718 vs 637 | 1.07 (0.85-1.34) | .59 |
| 5 allele vs 1-2 allele mismatched | 85 vs 637 | 1.25 (0.75-2.07) | .39 |
| 5 allele vs 3-4 allele mismatched | 85 vs 718 | 1.17 (0.71-1.94) | .53 |
| Relapse§ | |||
| 1-allele mismatched vs matched | 226 vs 117 | 0.93 (0.65-1.33) | .69 |
| 2-allele mismatched vs matched | 410 vs 117 | 0.77 (0.55-1.07) | .12 |
| 3-allele mismatched vs matched | 464 vs 117 | 0.75 (0.53-1.05) | .09 |
| 4-allele mismatched vs matched | 254 vs 117 | 0.50 (0.33-0.73) | .001 |
| 5-allele mismatched vs matched | 84 vs 117 | 0.83 (0.50-1.36) | .45 |
| 3-4 allele vs 1-2 allele mismatched | 718 vs 637 | 0.78 (0.64-0.96) | .02 |
| 5 allele vs 1-2 allele mismatched | 85 vs 637 | 1.01 (0.66-1.54) | .98 |
| 5 allele vs 3-4 allele mismatched | 85 vs 718 | 1.28 (0.84-1.97) | .25 |
| Overall HLA-match . | Number . | OR/ HR (95% CI) . | P value . |
|---|---|---|---|
| Neutrophil recovery* | |||
| 1-allele mismatched vs matched | 227 vs 116 | 0.72 (0.44-1.70) | .18 |
| 2-allele mismatched vs matched | 410 vs 116 | 0.86 (0.54-1.36) | .52 |
| 3-allele mismatched vs matched | 464 vs 116 | 0.56 (0.36-0.88) | .01 |
| 4-allele mismatched vs matched | 254 vs 116 | 0.55 (0.34-0.88) | .01 |
| 5-allele mismatched vs matched | 84 vs 116 | 0.45 (0.25-0.82) | .009 |
| 3-4 allele vs 1-2 allele mismatched | 718 vs 637 | 0.69 (0.55-0.86) | .001 |
| 5-allele vs 1-2 allele mismatched | 85 vs 637 | 0.56 (0.35-0.89) | .01 |
| 5-allele vs 3-4 allele mismatched | 85 vs 718 | 0.81 (0.51-1.29) | .38 |
| Grade 2-4 acute GVHD† | |||
| 1-allele mismatched vs matched | 226 vs 117 | 1.27 (0.83-1.93) | .26 |
| 2-allele mismatched vs matched | 410 vs 117 | 1.45 (0.99-2.12) | .06 |
| 3-allele mismatched vs matched | 464 vs 117 | 1.53 (1.05-2.23) | .03 |
| 4-allele mismatched vs matched | 254 vs 117 | 1.51 (1.00-2.27) | .05 |
| 5-allele mismatched vs matched | 85 vs 117 | 1.49 (0.89-2.49) | .13 |
| 3-4 allele vs 1-2 allele mismatched | 718 vs 637 | 1.09 (0.92-1.32) | .31 |
| 5-allele vs 1-2 allele mismatched | 85 vs 637 | 1.08 (0.72-1.61) | .72 |
| 5-allele vs 3-4 allele mismatched | 85 vs 718 | 0.98 (0.66-1.46) | .92 |
| Chronic GVHD‡ | |||
| 1-allele mismatched vs matched | 226 vs 117 | 0.76 (0.47-1.23) | .27 |
| 2-allele mismatched vs matched | 410 vs 117 | 1.16 (0.77-1.75) | .48 |
| 3-allele mismatched vs matched | 464 vs 117 | 1.02 (0.67-1.54) | .93 |
| 4-allele mismatched vs matched | 254 vs 117 | 1.20 (0.80-1.87) | .48 |
| 5-allele mismatched vs matched | 85 vs 117 | 1.27 (0.70-2.31) | .27 |
| 3-4 allele vs 1-2 allele mismatched | 718 vs 637 | 1.07 (0.85-1.34) | .59 |
| 5 allele vs 1-2 allele mismatched | 85 vs 637 | 1.25 (0.75-2.07) | .39 |
| 5 allele vs 3-4 allele mismatched | 85 vs 718 | 1.17 (0.71-1.94) | .53 |
| Relapse§ | |||
| 1-allele mismatched vs matched | 226 vs 117 | 0.93 (0.65-1.33) | .69 |
| 2-allele mismatched vs matched | 410 vs 117 | 0.77 (0.55-1.07) | .12 |
| 3-allele mismatched vs matched | 464 vs 117 | 0.75 (0.53-1.05) | .09 |
| 4-allele mismatched vs matched | 254 vs 117 | 0.50 (0.33-0.73) | .001 |
| 5-allele mismatched vs matched | 84 vs 117 | 0.83 (0.50-1.36) | .45 |
| 3-4 allele vs 1-2 allele mismatched | 718 vs 637 | 0.78 (0.64-0.96) | .02 |
| 5 allele vs 1-2 allele mismatched | 85 vs 637 | 1.01 (0.66-1.54) | .98 |
| 5 allele vs 3-4 allele mismatched | 85 vs 718 | 1.28 (0.84-1.97) | .25 |
Adjusted for transplant period: higher likelihood of recovery for transplants in 2005-2010 compared to 2000-2004 (OR 1.41, 95% CI 1.12-1.76, P = .003).
Adjusted for patient age and in vivo T-cell depletion: acute GVHD risks were lower for patients >16 years compared to those ≤16 years (HR 0.62, 95% CI 0.50-0.77, P < .0001) and with in vivo T-cell depletion (HR 0.82, 95% CI 0.51-0.74, P < .0001).
Adjusted for TNC and in vivo T-cell depletion: chronic GVHD risks were lower with units containing TNC 3 x 107/kg or higher compared to units containing TNC <3 x 107/kg (HR 0.59, 95% CI 0.45-0.79, P < .001) and with in vivo T-cell depletion (HR 0.65, 95% CI 0.52-0.82, P < .001).
Adjusted for disease, disease status, interval from diagnosis to transplant, and TNC: relapse risks were lower for myelodysplastic syndrome (HR 0.32, 95% CI 0.22-0.47, P < .0001) compared to acute myeloid leukemia, and with units containing TNC 3 x 107/kg or higher compared to units containing TNC <3 x 107/kg (HR 0.71, 95% CI 0.54-0.92, P = .009) and when the interval between diagnosis and transplant was >24 months compared to <12 months (HR 0.51, 95% CI 0.37-0.70, P < .0001). Relapse risks were higher for transplants in relapse (HR 5.57, 95% CI 4.25-7.30, P < .0001) and in second remission (HR 2.28, 95% CI 1.69-3.07, P < .0001) compared to transplants in first remission.
Mismatching at 3 or more alleles was associated with higher risks of primary graft failure. Prolonged neutropenia leads to opportunistic infections, which adds to the burden of morbidity and mortality. This may in part explain the recorded differences in nonrelapse mortality between transplantations mismatched at 1 and 2 alleles and those mismatched at 3 or more. Others have found an association between TNC and neutrophil recovery.26-28 We did not observe such an effect, but the median TNC delivered was 5 × 107/kg and more than 85% of transplantations used units with TNC in excess of 3 × 107/kg. Further, the current analyses considered allele-level HLA matching and matching at the HLA-C locus, whereas all previous reports considered lower resolution HLA matching and did not consider matching at HLA-C. We were unable to test for donor-specific anti-HLA antibodies known to be associated with graft failure29-31 or viability of cells after the unit was thawed. Similar to that recorded for adult donor transplantations, acute GVHD risks were higher after HLA-mismatched UCB transplants even though the risk estimates did not reach the level of significance set for the current analyses.1,3 Acute GVHD rates are low after UCB transplantations, and to detect significant differences attributable to mismatching, many more donor-recipient pairs mismatched at 1, 2, 3, or 4 HLA loci would be required than the number of pairs included in the current analysis. We did not observe differences in chronic GVHD rates after mismatched UCB transplantations and consistent with that reported after adult donor transplantation.1,3
Our ability to study the effect of mismatching at specific HLA loci is limited by the relatively low patient numbers and the limited number of events in some subgroups. Another limitation was the use of the validated imputation algorithm, Haplogic III, to assign allele-level match status for donor–recipient pairs with lower resolution HLA typing. The imputation process may be more biased toward common HLA alleles for assignments because it is driven by population frequencies. This could lead to a potential underestimation of the HLA disparity in the imputed group. However, the results between the imputed and the actual are not significantly different, suggesting that any underestimation of HLA disparity had a minimal effect on the results. Of note, the magnitude of the relative hazards for nonrelapse mortality for the imputed HLA-mismatched transplantations is lower compared with that observed for the group for which allele-level HLA typing was available. About 70% of transplantations with imputed HLA match occurred before 2005, a period during which mortality risks were high compared with the later period. We hypothesize that nonrelapse mortality risks in the earlier period may be attributed to several other factors and the effect of HLA mismatch becomes more apparent after 2005 because several of the other known risk factors have been optimized.
Despite the limitations, ours is the first report that identifies higher risks of primary graft failure and nonrelapse mortality after allele-level HLA-mismatched UCB transplants. The results of our analyses suggest current practice for UCB selection should be revised; single-unit UCB transplants must have a minimum precryopreserved TNC of 3 × 107/kg and thereafter the best allele-level HLA-matched unit should be selected. In the absence of a fully matched UCB unit, units mismatched at 1 or 2 alleles are acceptable. We acknowledge that only a small proportion of patients who may benefit from transplantation will have a fully matched UCB unit. With similar nonrelapse mortality risks after 1- and 2-allele mismatched transplants (HR 2.7-2.8), selecting a unit mismatched at 2 alleles will extend access to 80% to 85% of minorities and almost all Caucasians without an added risk to nonrelapse mortality compared with transplantations mismatched at 1 allele. Higher graft failure and nonrelapse mortality associated with mismatches at 3 or more alleles imply such transplants should be recommended with caution. The recent successes of haploidentical transplantation warrant the relative merits of transplantation with a haploidentical donor weighed against the relative merits of 3 or more allele-mismatched single UCB transplantation.32 To ensure wider access to all ethnic groups to UCB units with optimal HLA matching (ie, no more than 2 alleles disparate), additional investments are needed to expand the worldwide inventory.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
The authors acknowledge Professor Zhang who stepped in and assisted with the revisions for this article.
This work was supported by grants from the National Cancer Institute (Public Health Service grant U24-CA76518), the National Heart Lung and Blood Institute, and the National Institute of Allergy and Infectious Diseases; a Scholar in Clinical Research Award from the Leukemia and Lymphoma Society (M.E.); the Office of Naval Research, Department of Navy to the National Marrow Donor Program (N00014-11-01-0339); and the National Institute for Health Research, Oxford Biomedical Center, Oxford University Hospitals National Health Service Trust and University of Oxford (V.R.). Opinions, findings, and conclusions or recommendations expressed herein are those of the authors and do not reflect the views of the Office of Naval Research or the National Marrow Donor Program.
Authorship
Contribution: M.E., J.P.K., S.S., E.G., and V.R. designed the study; S.S., M.M., and M.B. reviewed and assigned donor-recipient HLA match; W.H. and M.E. prepared the study file; J.P.K. analyzed and interpreted data; J.F. and M.M. predicted donor availability based on the National Marrow Donor Program’s umbilical cord blood inventory; M.E. drafted the manuscript; A.R., S.J.L., C.A., W.A., J.N.B., L.A.B.-L., M.A.F.-V., A.P.I., M.M.H., F.L., S.M., G.M., G.F.S., E.G., and V.R. critically reviewed the manuscript; and all authors approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
John P. Klein died on July 20, 2013.
Correspondence: Mary Eapen, Center for International Blood and Marrow Transplant Research, Department of Medicine, Medical College of Wisconsin, 8701 Watertown Plank Rd, Milwaukee, WI 53226; e-mail: meapen@mcw.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal