Key Points
Runx proteins and Cbfβ are required for the development of dendritic cells, which can be rescued by Irf8.
Pan-hematopoietic Cbfb-deficient mice develop a myeloproliferative disease and severe anemia and die at 3 to 4 months of age.
Abstract
Runx1 and Cbfβ are critical for the establishment of definitive hematopoiesis and are implicated in leukemic transformation. Despite the absolute requirements for these factors in the development of hematopoietic stem cells and lymphocytes, their roles in the development of bone marrow progenitor subsets have not been defined. Here, we demonstrate that Cbfβ is essential for the development of Flt3+ macrophage-dendritic cell (DC) progenitors in the bone marrow and all DC subsets in the periphery. Besides the loss of DC progenitors, pan-hematopoietic Cbfb-deficient mice also lack CD105+ erythroid progenitors, leading to severe anemia at 3 to 4 months of age. Instead, Cbfb deficiency results in aberrant progenitor differentiation toward granulocyte-macrophage progenitors (GMPs), resulting in a myeloproliferative phenotype with accumulation of GMPs in the periphery and cellular infiltration of the liver. Expression of the transcription factor Irf8 is severely reduced in Cbfb-deficient progenitors, and overexpression of Irf8 restors DC differentiation. These results demonstrate that Runx proteins and Cbfβ restrict granulocyte lineage commitment to facilitate multilineage hematopoietic differentiation and thus identify their novel tumor suppressor function in myeloid leukemia.
Introduction
Diversified subsets of mature blood cells develop from hematopoietic stem cells (HSCs) via intermediate progenitor populations with limited self-renewal and lineage potential. HSCs are enriched in the Lin–Sca1+c-kit+ (LSK) fraction of bone marrow (BM) cells and differentiate into Flt3+CD115+ (Macrophage-colony-stimulating factor receptor [MCSF-R]) macrophage/dendritic cell (DC) progenitors (MDPs), Flt3+IL7R+ common lymphoid progenitors (CLPs), and CD16/32+ granulocyte/macrophage progenitors (GMPs) via an intermediate Flt3+ LSK population, called lymphoid-primed multipotent progenitors (LMPPs).1-3 Balanced multilineage differentiation from HSCs to lineage-restricted progenitors requires gene expression programs regulated by transcription factors, such as Sfpi1 (PU.1), Ikzf1 (Ikaros), Gfi1, and Cebpa.4,5
The Runx family of transcription factors plays key roles in the development of HSCs, lymphocytes, and megakaryocytes.6-9 The Runx family consists of 3 α subunits, Runx1, Runx2, and Runx3, and 1 β subunit, Cbfβ, which functions as the obligatory dimerization partner for each α subunit to form DNA-binding complexes. Alteration of Runx1 or Cbfβ function caused by chromosomal translocations is frequently found in human acute myelogenous leukemia (AML),10 whereas Runx3 deficiency has been implicated in myeloproliferative diseases (MPDs) in aged mice.11 Although Runx1 is essential for HSC formation from hemogenic endothelial cells during embryogenesis, conditional deletion of Runx1 in HSCs neither influences the survival of mice nor the maintenance of long-term HSCs.12,13 By contrast, Runx1 is required for the development of B and T lymphocytes and megakaryocytes,14-17 suggesting that it functions in cell fate decisions during BM progenitor differentiation. Although Runx1 is essential for the development of pro-B cells and double-negative thymocytes,14,16,17 its requirement in early BM progenitors has not been defined. Furthermore, compound disruption of multiple Runx genes results in stronger phenotypes than single Runx gene inactivation, suggesting that the 3 Runx proteins have partially redundant functions.14,18,19 This redundancy may underestimate the importance of Runx proteins in HSCs or early hematopoiesis, and therefore it remains unclear whether Runx proteins are important for HSC differentiation and prevention of MPD or leukemia.
In this study, we demonstrate that Runx1 and Cbfb are absolutely required for the development of Flt3+ DC progenitors and all mature DC lineages. Pan-hematopoietic Cbfb-deficient mice show aberrant differentiation of HSCs to GMPs, leading to MPD and severe anemia. The expression of Irf8 is downregulated in Cbfb-deficient progenitors, and forced expression of Irf8, which is known to inhibit the development of granulocytes from HSCs,20 rescues defective DC differentiation. These findings suggest that the Runx-Irf8 axis is critical for DC lineage differentiation and the prevention of MPD by restricting the granulocyte lineage.
Methods
Mice
C57BL/6 and B6-CD45.1 (B6-Ly5.2) mice were purchased from the National Cancer Institute. Cbfb-flox (Cbfbtm1Itan), Runx1-flox (Runx1tm3Spe), Runx2-KO (Runx2tm1Mjo), and Runx3-flox (Runx3tm1Itan) alleles were described previously.12,21,22 Vav1-iCre transgenic mice23 were purchased from The Jackson Laboratory. All mice except Runx1-flox mice were maintained under the specific pathogen-free conditions at Washington University. Analysis of Cbfb-CKO mice used Vav1-iCre CbfbF/+ heterozygous mice as controls, unless otherwise specified. We did not observe abnormal hematopoiesis in Vav1-iCre CbfbF/+ mice compared with age-matched Cre(–) CbfbF/F mice except for a small increase in the GMP frequency in BM (supplemental Table 1, see supplemental Data available at the Blood Web site). For analysis of samples from Runx1-CKO mice, tissues were collected at the University of Pennsylvania and shipped overnight in complete media 1 day before the analysis at Washington University. Comparative tissues from Cbfb-CKO mice were prepared 1 day before analysis and kept in complete media at 4°C overnight at Washington University. All mice were maintained on a C57BL/6 background. All experiments were performed according to protocols approved by Washington University’s Animal Studies Committee and the University of Pennsylvania’s Animal Resources Center.
Generation of mixed BM chimeras
BM cells from wild type (WT) (CD45.1/2 or CD45.1) and those from either Vav1-iCre+CbfbF/+ or Vav1-iCre+CbfbF/F (CD45.2) were mixed at a 1:4 ratio and a total of 1 × 107 mixed BM cells were IV transferred to lethally irradiated (10 Gy) B6-CD45.1 or CD45.1/2 recipients. The recipients were fed with sulfamethoxazole-trimethoprim containing drinking water for 4 weeks. Hematopoietic reconstitution by donor cells was examined 5 to 6 weeks after transplantation.
Flow cytometry
Fluorescently labeled monoclonal antibodies were purchased from eBioscience or Biolegend. Samples were analyzed with FACSCantoII, LSRII, or LSR Fortessa cytometers (BD Biosciences), and data were analyzed using Flowjo (Tree Star). Cell sorting was performed with FACSAria II (BD Biosciences).
Cell culture
Total BM cells were cultured in Iscove Modified Dulbecco Medium/10% fetal bovine serum supplemented with granulocyte macrophage–colony-stimulating factor (GM-CSF) (Peprotec) or Flt3L as previously described.24 DC differentiation was assessed on day 7 or 8, respectively. For retroviral transduction, BM cells were cultured in the presence of Flt3L and stem cell factor (10 ng/mL; Peprotec) for 3 days, then cultured with Flt3L for 7 days. Retroviral supernatant was prepared by transfecting PlatE packaging cells25 using TransIT (Mirus Bio) and concentrated by centrifugation at 21 000g for 1 hour at 4°C. Concentrated retroviral supernatant was added on day 1 of culture and kept for 2 days in the presence of 2 μg/mL polybrene (Sigma-Aldrich). Clonal assays were performed as previously described.26
Gene expression analysis
B220–CD11b–major histocompatibility class II (MHC-II)–c-kit+Sca1+ cells and B220–CD11b–MHC-II–c-kit+Sca1–CD16/32+ cells were sorted to >98% purity from 2 Cbfb-CKO and 2 control mice. RNA was purified from 2 × 105 cells using a NucleoSpin RNA XS kit (Macherey-Nagel), amplified using a PicoSL RNA amplification kit (Nugen) and biotinylated with Encore biotin module (Nugen) in a single experiment to minimize technical variation. Labeled RNA was hybridized to Mouse Gene 1.0ST microarrays (Affymetrix) according to the manufacturer’s instruction. Data were analyzed using Arraystar (DNA Star) with robust multiarray average normalization.27 CEL files for GMP (GSM791119, GSM791120, GSM791121) and MDP (GSM791105, GSM791106, GSM791107) subsets generated by the Immunological Genome Project28 were used to make “GMP-specific” and “MDP-specific” gene lists. For quantitative reverse transcription–polymerase chain reaction (qRT-PCR) analysis, RNA was purified from separately sorted GMPs using TRIzol (Life Technologies), and complementary DNA (cDNA) was prepared using qScript cDNA Supermix (QuantaBio). Quantitative real-time PCR was performed with the DyNAmo SYBR Green qPCR kit (Thermo Scientific) and LightCycler 480 (Roche). The following primers were used:
Hprt1: (forward [F]) AGGTTGCAAGCTTGCTGGT, (reverse [R]) TGAAGTACTCATTATAGTCAAGGGCA; Cbfb: (F) TCTCCACAGATTGGATGGTATG, (R) GTTCTTCTTCGAGCCTCTTCAA; Irf8: (F) GACCATGTTCCGTATCCCCTGGAAG, (R) GGGACCGGTCAGTCACTTCTTCA; Sfpi1: (F) TGACTACTACTCCTTCGTGGGC, (R) TCTCAAACTCGTTGTTGTGGAC; Cebpa: (F) TGGATAAGAACAGCAACGAG, (R) TCACTGGTCAACTCCAGCAC; Csf1r: (F) GAGGGAGACTCCAGCTACAAGA, (R) TGTCTACACCCTGACTGGAGAA.
Statistical analysis
Data from multiple independent samples were analyzed with Prism 6 (GraphPad) and significance of statistical difference was tested using the unpaired 2-tailed Student t test, unless otherwise specified.
Results
Cbfb is essential for DC development
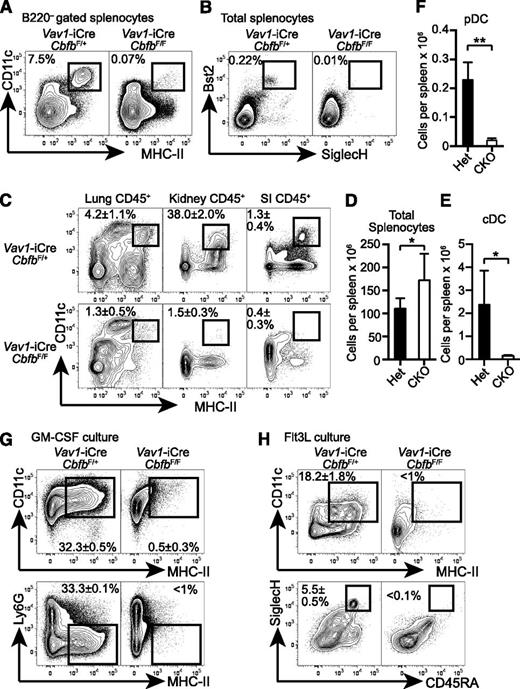
To determine the requirement for Cbfb in hematopoietic progenitor differentiation, we conditionally deleted Cbfb in HSCs using a Vav1-iCre deleter (Cbfb-CKO). Although germline deletion of Cbfb or Runx1 results in embryonic lethality and a complete lack of definitive hematopoiesis,6-9 Cbfb-CKO mice were viable and showed no gross phenotypes until 6 weeks after birth. In addition to the previously reported defects in B- and T-cell differentiation,17,21 Cbfb-CKO mice had severely reduced DCs, including CD11c+MHC-II+B220– classical DCs (cDCs), as well as Bst2+SiglecH+ plasmacytoid DCs (pDCs) (Figure 1A-B). The absolute numbers of cDCs and pDCs in the spleen were reduced by ∼25-fold and 15-fold, respectively, in Cbfb-CKO mice as compared to control mice (Figure 1E-F). cDCs were also severely reduced in peripheral tissues such as the lung, small intestine lamina propria, and kidney in Cbfb-CKO mice (Figure 1C). Because DCs can be generated from BM cells supplemented with GM-CSF or Flt3L in vitro,29 we next determined whether Cbfb is required for DC differentiation in vitro. CD11c+ MHC-II+ DCs and CD45RA+ SiglecH+ pDC were generated from control BM in the presence of either GM-CSF or Flt3L (Figure 1G-H). In contrast, we observed drastically reduced cDC and pDC differentiation from Cbfb-CKO BM cells (Figure 1G-H). These results indicate that Cbfb is required for the differentiation of DCs both in vivo and in vitro.
Cbfb is required for the development of DCs. (A) Splenocytes from 6- to 8-week-old Vav1-iCre+CbfbF/+ (Het) and Vav1-iCre+CbfbF/F (CKO) mice were analyzed for CD11c, MHC-II, and B220 expression. CD11c+MHC-II+ cDCs in B220– gated splenocytes are shown with rectangle gates and percentages. Statistical analysis is shown in panel E as mean and standard deviation (n = 8). (B) Bst2 and SiglecH expression in total splenocytes from Vav1-iCre+CbfbF/+ and Vav1-iCre+CbfbF/F mice. Bst2+SiglecH+ pDCs are shown with rectangle gates and percentages, and statistical analysis is shown as mean and standard deviation in panel F (n = 4). (C) Mononuclear cells from lung, kidney, and SI in 6- to 8-week-old Vav1-iCre+CbfbF/+ and Vav1-iCre+CbfbF/F mice were analyzed for expression of CD45, CD11c, and MHC-II. CD11c+MHC-II+ cDCs in CD45+ gated cells are shown with rectangle gates. Mean percentages and standard deviation of total CD45+ cells from 4 mice are shown (n = 4). (D) Statistical analysis of total splenocyte numbers from Vav1-iCre+CbfbF/+ and Vav1-iCre+CbfbF/F mice shown as mean and standard deviation (n = 8). (G-H) Total BM cells were cultured in the presence of GM-CSF (G, n = 2) or Flt3L (H, n = 4) in vitro, and DC differentiation was examined by expression of CD11c, MHC-II, Ly6G, CD45RA, and SiglecH using flow cytometry. Statistical difference was assessed with the unpaired 2-tailed Student t test. *P < .05; **P < .01. SI, small intestine.
Cbfb is required for the development of DCs. (A) Splenocytes from 6- to 8-week-old Vav1-iCre+CbfbF/+ (Het) and Vav1-iCre+CbfbF/F (CKO) mice were analyzed for CD11c, MHC-II, and B220 expression. CD11c+MHC-II+ cDCs in B220– gated splenocytes are shown with rectangle gates and percentages. Statistical analysis is shown in panel E as mean and standard deviation (n = 8). (B) Bst2 and SiglecH expression in total splenocytes from Vav1-iCre+CbfbF/+ and Vav1-iCre+CbfbF/F mice. Bst2+SiglecH+ pDCs are shown with rectangle gates and percentages, and statistical analysis is shown as mean and standard deviation in panel F (n = 4). (C) Mononuclear cells from lung, kidney, and SI in 6- to 8-week-old Vav1-iCre+CbfbF/+ and Vav1-iCre+CbfbF/F mice were analyzed for expression of CD45, CD11c, and MHC-II. CD11c+MHC-II+ cDCs in CD45+ gated cells are shown with rectangle gates. Mean percentages and standard deviation of total CD45+ cells from 4 mice are shown (n = 4). (D) Statistical analysis of total splenocyte numbers from Vav1-iCre+CbfbF/+ and Vav1-iCre+CbfbF/F mice shown as mean and standard deviation (n = 8). (G-H) Total BM cells were cultured in the presence of GM-CSF (G, n = 2) or Flt3L (H, n = 4) in vitro, and DC differentiation was examined by expression of CD11c, MHC-II, Ly6G, CD45RA, and SiglecH using flow cytometry. Statistical difference was assessed with the unpaired 2-tailed Student t test. *P < .05; **P < .01. SI, small intestine.
Cbfb is essential for the development of Flt3+ lymphoid and DC progenitors and CD105+ erythroid progenitors
To determine the stage at which the development of DCs and lymphocytes is arrested, we examined progenitor populations in the BM. Frequencies of Lin (B220, CD11b, MHC-II,)-negative CD16/32–c-kit+Sca1+ progenitors (CD16/32– LSK) were comparable between Cbfb-CKO and control mice at 6 to 8 weeks and 12 to 16 weeks after birth (Figure 2A,E). However, the majority of LSK cells in Cbfb-CKO mice expressed CD150, a marker for long-term HSCs30 (Figure 2B). Similarly, whereas GMPs in normal mice barely express CD150,31 a large proportion of GMPs and Lin–c-kit+Sca1– progenitors in Cbfb-CKO mice also expressed CD150 (Figure 2C), suggesting aberrant CD150 expression in the absence of Cbfb. Progenitors for DCs and lymphocytes are enriched in the CD135/Flt3+ fractions of LSK cells (LMPPs) and Lin–Sca1–c-kit+ cells.1-3,32 Compared with control mice, the frequencies of Flt3+ progenitor subsets were severely reduced in Cbfb-CKO mice (Figure 2B,D-E). We also observed a substantial reduction in the frequency of Lin–Sca1–c-kit+ cells expressing CD115, an additional marker for DC lineage progenitors1,3 (Figure 2D). Furthermore, CD105+ erythroid progenitors31,33,34 were severely reduced in Cbfb-CKO mice (Figure 2F). These results indicate that Cbfb is required for the development of Flt3+ DC progenitor populations as well as for erythroid progenitors in the BM.
Cbfb is required for the development of Flt3+ progenitor cells in BM. (A-B) BM cells from 6- to 8-week-old Vav1-iCre+CbfbF/+ (Het) and Vav1-iCre+CbfbF/F (CKO) mice were analyzed for expression of lineage markers (B220, CD11b, MHC-II, CD16/32), c-kit, Sca1, CD150, CD105, CD115, and CD135 (Flt3). Frequencies of LSK HSCs/MPPs (A), and Flt3+ LSK LMPPs (B) are shown with rectangular gates. (C) CD150 and CD16/32 expression in Lin–c-kit+Sca1– progenitors. CD16/32+CD150+ and CD16/32–CD150+ populations are shown with rectangular gates with percentages. (D) c-kit and CD135 or CD115 expression in Lin–Sca1– BM progenitors. Flt3+c-kit+ (MDP/CLP), Flt3+Lin–Sca1–c-kitlo/– (CDP), and c-kit+CD115+ populations are shown with rectangular gates with percentages. (E) Statistical analysis of CD16/32– LSK, LMPP, and MDP cells at 6 to 8 (shown as 7 weeks) and 12 to 16 weeks (14 weeks) of age. (F) Expression of CD105 and CD16/32 in B220–CD11b–Sca1– BM cells in Vav1-iCre+CbfbF/+ (het) and Vav1-iCre+CbfbF/F (CKO) mice. CD105+ erythroid progenitor cells are shown with rectangular gates and frequencies (mean and standard deviation). *P < .05; ***P < .005. n.s., not significant. Flow cytometry data represent analysis of 3 to 6 mice.
Cbfb is required for the development of Flt3+ progenitor cells in BM. (A-B) BM cells from 6- to 8-week-old Vav1-iCre+CbfbF/+ (Het) and Vav1-iCre+CbfbF/F (CKO) mice were analyzed for expression of lineage markers (B220, CD11b, MHC-II, CD16/32), c-kit, Sca1, CD150, CD105, CD115, and CD135 (Flt3). Frequencies of LSK HSCs/MPPs (A), and Flt3+ LSK LMPPs (B) are shown with rectangular gates. (C) CD150 and CD16/32 expression in Lin–c-kit+Sca1– progenitors. CD16/32+CD150+ and CD16/32–CD150+ populations are shown with rectangular gates with percentages. (D) c-kit and CD135 or CD115 expression in Lin–Sca1– BM progenitors. Flt3+c-kit+ (MDP/CLP), Flt3+Lin–Sca1–c-kitlo/– (CDP), and c-kit+CD115+ populations are shown with rectangular gates with percentages. (E) Statistical analysis of CD16/32– LSK, LMPP, and MDP cells at 6 to 8 (shown as 7 weeks) and 12 to 16 weeks (14 weeks) of age. (F) Expression of CD105 and CD16/32 in B220–CD11b–Sca1– BM cells in Vav1-iCre+CbfbF/+ (het) and Vav1-iCre+CbfbF/F (CKO) mice. CD105+ erythroid progenitor cells are shown with rectangular gates and frequencies (mean and standard deviation). *P < .05; ***P < .005. n.s., not significant. Flow cytometry data represent analysis of 3 to 6 mice.
Cell-autonomous requirement for Cbfb in the development of DCs and DC progenitors
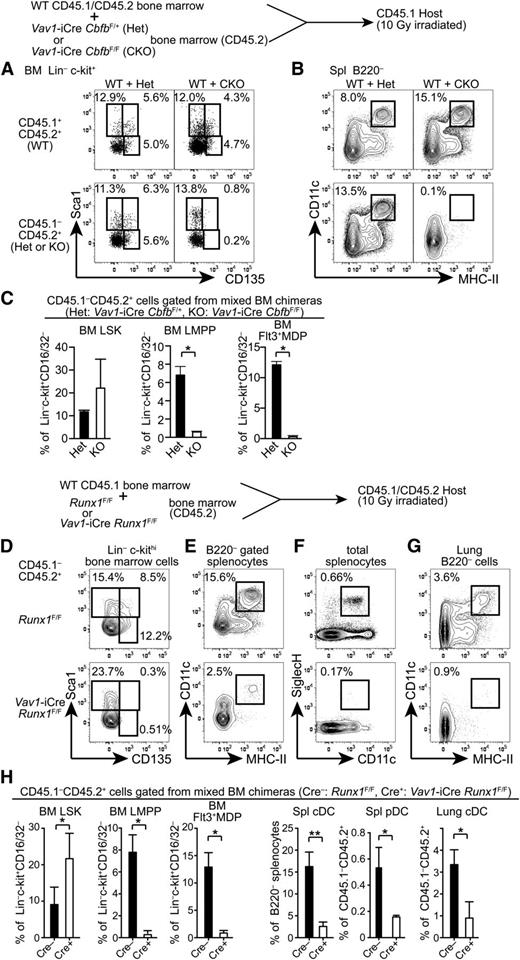
To determine whether Cbfb is cell-autonomously required for the development of Flt3+ progenitors and mature DCs, we generated mixed BM chimeras. CD45.2 Cbfb-CKO (CKO) or control Vav1-iCre+CbfbF/+ (Het) BM cells were mixed with CD45.1/2 WT BM cells and transferred into lethally irradiated CD45.1 congenic recipient mice, and hematopoietic reconstitution of donor origin was analyzed 5 to 6 weeks posttransplantation. In control chimeras reconstituted with WT+Het BM cells, Flt3+Sca1+ LMPPs and Flt3+Sca1– MDP/CLPs in the BM and cDCs in the spleen were reconstituted from both CD45.1/2 and CD45.2 progenitor cells (Figure 3A-C). In chimeras transferred with WT+CKO BM cells, we observed similar frequencies of LSK cells from both CD45.1/2 WT and CD45.2 CKO cells. However, LMPPs and Flt3+ MDP/CLP populations from CD45.2 CKO BM cells were completely absent (Figure 3A,C). Accordingly, cDC differentiation from CKO progenitors was not detected in the recipient mice (Figure 3B). These results indicate that Cbfb is cell-autonomously required for the differentiation of Flt3+ BM progenitors and mature cDCs in vivo.
Cell-autonomous requirements for Runx1 and Cbfb in the development of DCs. (A-B) A mixture of CD45.1/2 WT and either Cbfb-CKO or control Het BM cells (CD45.2) were transferred into lethally irradiated CD45.1 recipients, and reconstitution of BM progenitor populations (A) and spleen cDCs (B) was analyzed by flow cytometry. Percentages of HSCs (Sca1+CD135–), LMPPs (Sca1+CD135+), and Flt3+ MDP/CLPs (Sca1lo/–CD135+) in Lin–c-kit+ BM cells, and CD11c+MHC-II+ cDCs in the spleen are shown. Data represent 4 mice with mean and standard deviation shown in panel C. (D-H) BM cells, splenocytes, and lung mononuclear cells from BM chimeras reconstituted with a mixture of CD45.1 WT and either Runx1-CKO or control Cre(–) Runx1F/F BM cells (CD45.2) were analyzed for the presence of DC progenitors and mature DCs. Runx1-CKO or control BM-derived (CD45.1–CD45.2+) HSCs (Sca1+CD135–), LMPPs (Sca1+CD135+), and Flt3+MDP/CLPs (Sca1lo/–CD135+) in Lin–c-kit+ BM cells, CD11c+ MHC-II+ cDCs in B220– cells in the spleen and lung, and CD11c+SiglecH+ pDCs in the spleen are shown. Data shown represent 5 mice and statistical analysis is shown in panel H. *P < .05; **P < .01.
Cell-autonomous requirements for Runx1 and Cbfb in the development of DCs. (A-B) A mixture of CD45.1/2 WT and either Cbfb-CKO or control Het BM cells (CD45.2) were transferred into lethally irradiated CD45.1 recipients, and reconstitution of BM progenitor populations (A) and spleen cDCs (B) was analyzed by flow cytometry. Percentages of HSCs (Sca1+CD135–), LMPPs (Sca1+CD135+), and Flt3+ MDP/CLPs (Sca1lo/–CD135+) in Lin–c-kit+ BM cells, and CD11c+MHC-II+ cDCs in the spleen are shown. Data represent 4 mice with mean and standard deviation shown in panel C. (D-H) BM cells, splenocytes, and lung mononuclear cells from BM chimeras reconstituted with a mixture of CD45.1 WT and either Runx1-CKO or control Cre(–) Runx1F/F BM cells (CD45.2) were analyzed for the presence of DC progenitors and mature DCs. Runx1-CKO or control BM-derived (CD45.1–CD45.2+) HSCs (Sca1+CD135–), LMPPs (Sca1+CD135+), and Flt3+MDP/CLPs (Sca1lo/–CD135+) in Lin–c-kit+ BM cells, CD11c+ MHC-II+ cDCs in B220– cells in the spleen and lung, and CD11c+SiglecH+ pDCs in the spleen are shown. Data shown represent 5 mice and statistical analysis is shown in panel H. *P < .05; **P < .01.
Roles of Runx1, Runx2, and Runx3 in DC development
Cbfβ protein does not directly bind to DNA and is recruited to its target loci through association with 1 of the Runx proteins. To determine which Runx protein is required for the development of Flt3+ progenitors and DCs, we examined DC progenitors and mature DCs in Vav1-iCre Runx1F/F mice (Runx1-CKO). As reported previously,12 the LMPP frequency in the LSK fraction was reduced by threefold to fourfold in Runx1-CKO mice compared to control Runx1F/F mice, although the reduction was milder than that in Cbfb-CKO mice (supplemental Figure 1A,C). A similar reduction of Flt3+ MDP/CLPs was also observed in Runx1-CKO mice (supplemental Figure 1B-C). However, the frequency of Flt3+c-kitlo/– common DC progenitors (CDPs) was affected less as compared to LMPPs or MDP/CLPs (supplemental Figure 1B-C). In the spleen, cDC differentiation was unaffected in Runx1-CKO mice with normal proportions of CD24+ (CD8α+) and CD11b+ (CD8α–) cDCs (supplemental Figure 1D). Because Runx1 inactivation resulted in a modest, but significant, reduction in early stage DC progenitors, we more rigorously examined the requirement for Runx1 in the development of Flt3+ progenitors. We generated BM chimeras reconstituted with Runx1-CKO or control Cre(–) Runx1F/F BM cells mixed with WT BM cells and analyzed reconstitution of DC progenitors in the BM and DCs in the spleen and the lung (Figure 3D-H). Although Runx1-CKO progenitors could reconstitute the CD135– LSK fraction enriched for HSCs, they hardly contributed to CD135+ LMPPs and Flt3+ MDP/CLPs in the recipient BM under competitive conditions (Figure 3D,H). Accordingly, DCs in the spleen and the lung as well as pDCs in the spleen were severely reduced (Figure 3E-H). These results demonstrate a cell-autonomous requirement for Runx1 in the differentiation of Flt3+ BM progenitors and DCs.
Next, to determine whether Runx2 is required for DC development, we generated chimeras reconstituted with E14.5 Runx2−/− or control Runx2+/+ fetal liver cells. In the BM of the chimeric mice, Runx2−/− progenitors reconstituted the LSK fraction with a similar frequency of Flt3+ LMPPs compared to control progenitors (supplemental Figure 2A). Flt3+ MDP/CDPs in the BM and CD11chi DCs in the spleen, including both CD8α+ and CD11b+ (CD8α–) cDC subsets, were also reconstituted from Runx2−/− fetal liver cells (supplemental Figure 2A-B). Although we observed comparable frequencies of Bst2+B220+ pDCs derived from Runx2−/− and control fetal liver cells in the BM of chimeras, CD11c expression was substantially reduced in Runx2−/− pDCs (supplemental Figure 2C). These results support a role for Runx2 in the terminal differentiation of pDCs, as recently reported,35 but not in the development of DC progenitors or cDCs. Finally, we analyzed Vav1-iCre Runx3F/F mice and did not observe altered BM progenitor frequencies or splenic DC numbers (supplemental Figure 2D-E). These results indicate that Runx1 is the primary Runx protein that cooperates with Cbfβ to regulate the development of DCs and their progenitors.
Development of severe anemia in Cbfb-CKO mice
Despite a severe reduction of CD105+ erythroid progenitors, Cbfb-CKO mice did not show gross defects at birth. At 6 to 8 weeks of age, they had only slightly reduced hemoglobin levels and red blood cell (RBC) and white blood cell (WBC) counts in the peripheral blood (Figure 4A). However, at 12 to 16 weeks of age, Cbfb-CKO mice developed severe anemia and died. These mice likely succumbed to progressive anemia but also possibly to infection or hemorrhage due to pancytopenia (Figure 4A-B). Platelet counts were reduced by twofold to threefold in Cbfb-CKO mice, although the magnitude of thrombocytopenia varied between mice (Figure 4A). At 6 to 8 weeks of age, Ter119+ cells in the BM were reduced by twofold to threefold in Cbfb-CKO mice with a compensatory increase of Ter119+ cells in the spleen, suggesting defective BM erythropoiesis (Figure 4C). Strikingly, at 12 to 16 weeks of age, Ter119+ cells were almost absent in both the BM and spleen in Cbfb-CKO mice, supporting the hypothesis that they succumbed to severe anemia.
Development of severe anemia in Cbfb-CKO mice. (A) WBC and RBC counts, HGB levels, and platelet counts in the peripheral blood from Vav1-iCre+CbfbF/+ (Het) and Vav1-iCre+CbfbF/F (CKO) mice at 6 to 8 weeks (7w) and 12 to 16 weeks (14w) of age. (B) Kaplan-Meier plot showing survival of Vav1-iCre+CbfbF/+ and Vav1-iCre+CbfbF/F mice. (C) Ter119 and CD11b expression in total BM cells and splenocytes from Vav1-iCre+CbfbF/+ and Vav1-iCre+CbfbF/F mice at 6 to 8 and 12 to 16 weeks of age. Percentages of Ter119+ and CD11b+ cells are shown. Data are representative of 3 or more biological replicates. Statistical differences were assessed by unpaired Student t test (A) and the log-rank test (B). *P < .05; **P < .01. HGB, hemoglobin.
Development of severe anemia in Cbfb-CKO mice. (A) WBC and RBC counts, HGB levels, and platelet counts in the peripheral blood from Vav1-iCre+CbfbF/+ (Het) and Vav1-iCre+CbfbF/F (CKO) mice at 6 to 8 weeks (7w) and 12 to 16 weeks (14w) of age. (B) Kaplan-Meier plot showing survival of Vav1-iCre+CbfbF/+ and Vav1-iCre+CbfbF/F mice. (C) Ter119 and CD11b expression in total BM cells and splenocytes from Vav1-iCre+CbfbF/+ and Vav1-iCre+CbfbF/F mice at 6 to 8 and 12 to 16 weeks of age. Percentages of Ter119+ and CD11b+ cells are shown. Data are representative of 3 or more biological replicates. Statistical differences were assessed by unpaired Student t test (A) and the log-rank test (B). *P < .05; **P < .01. HGB, hemoglobin.
Aberrant granulopoiesis in Cbfb-CKO mice
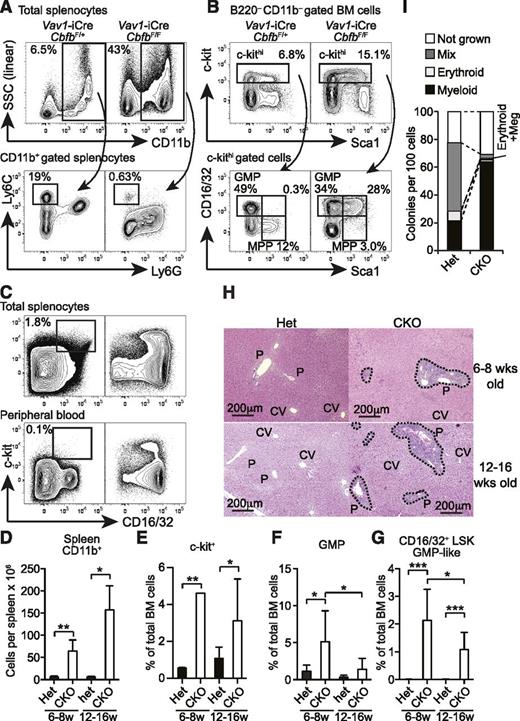
Although Cbfb-CKO mice also lacked the majority of B and T cells as previously observed in Runx1-conditional KO mice,15,17,22 spleens from Cbfb-CKO mice were significantly larger than those from their heterozygous littermate controls (Figure 1D). The elevated spleen cellularity in Cbfb-CKO mice was due to markedly increased CD11b+ cells (Figure 5A,D). The absolute number of CD11b+ cells in the spleen of Cbfb-CKO mice was increased by more than sixfold compared to control mice, and these CD11b+ cells consisted of predominantly Ly6Gint Ly6Clo/– granulocytes (Figure 5A). Despite the increased number, the majority of Cbfb-CKO CD11b+ cells did not fully upregulate Ly6G and were side scatter intermediate (Figure 5A), suggesting defective maturation after they acquired CD11b+ expression. In contrast, CD11b+Ly6Chi monocytes were severely reduced in Cbfb-CKO mice (Figure 5A). Consistent with the increased myeloid cells in the spleen, we observed an increased frequency of CD16/32+ GMPs in the c-kit+ fraction of BM cells in Cbfb-CKO mice compared to control mice (Figure 5B,E-F). In Cbfb-CKO mice, the frequency of c-kit+ cells in total BM cells was increased by twofold to fourfold (Figure 5B,E), as reported in Runx1-CKO mice.12 Notably, a large proportion of the c-kit+ cells in Cbfb-CKO mice showed the CD16/32+ GMP phenotype, although the GMP-like LSK cells were slightly reduced in 12- to 16-week-old Cbfb-CKO mice (Figures 5B,F). Approximately one-quarter of c-kit+ cells coexpressed Sca1 and CD16/32, while few cells coexpressed the 2 markers in control mice (Figures 5B,G). Furthermore, substantial numbers of cells with the GMP phenotype also accumulated in the spleen and peripheral blood in Cbfb-CKO mice (Figure 5C). We also observed marked cellular infiltration around the portal veins in the liver in both 6- to 8-week-old and 12- to 16-week-old Cbfb-CKO mice (Figure 5H). In addition to aberrant surface marker expression on progenitors, Cbfb-CKO HSCs also showed skewing toward myeloid differentiation at a clonal level (Figure 5I). These results suggest that a loss of Cbfb causes premature myeloid differentiation at an early stage between HSCs or multipotent progenitors (MPPs) and GMPs, resulting in a preleukemic state.
Development of MPD in Cbfb-CKO mice. (A) Splenocytes from 6- to 8-week-old Vav1-iCre+CbfbF/+ (Het) and Vav1-iCre+CbfbF/F (CKO) mice were analyzed for expression of CD11b, Ly6C, and Ly6G. Percentages of CD11b+ cells in total splenocytes and Ly6ChiLy6G– monocytes in CD11b+ splenocytes are shown. (B) B220–CD11b– BM cells from 6- to 8-week-old Vav1-iCre+CbfbF/+ and Vav1-iCre+CbfbF/F mice were analyzed for c-kit, Sca1, and CD16/32 expression. GMP, MPP, and CD16/32+ MPP (bottom panels) percentages in c-kithi cells (top panels) are shown. Statistical analysis from 6 to 9 mice in 6- to 8-week-old and 12- to 16-week-old groups is shown in panels D to G as mean and standard deviation. (C) c-kit and CD16/32 expression in total splenocytes (top) and peripheral blood WBCs (bottom) in 12- to 16-week-old Vav1-iCre+CbfbF/+ (Het, left) and Vav1-iCre+CbfbF/F (CKO, right) mice. Percentages of c-kit+CD16/32+ GMPs are shown with rectangular gates (n = 3, peripheral blood; and n = 6, splenocytes). (H) Hematoxylin and eosin staining of formalin-fixed liver sections from 6- to 8- and 12- to 16-week-old Vav1-iCre+CbfbF/+ (Het) and Vav1-iCre+CbfbF/F (CKO) mice. Clusters of infiltrating cells are surrounded by dotted lines. P, portal triad; CV, central vein. Images are representative of 3 mice for each genotype and age. (I) Clonal lineage potential of BM Lin–c-kit+Sca1+CD135–CD150+ cells from Cbfb-CKO (CKO) and control mice (Het) determined by a single cell culture (n = 2). SSC, side scatter. *P < .05; **P < .01; ***P < .001.
Development of MPD in Cbfb-CKO mice. (A) Splenocytes from 6- to 8-week-old Vav1-iCre+CbfbF/+ (Het) and Vav1-iCre+CbfbF/F (CKO) mice were analyzed for expression of CD11b, Ly6C, and Ly6G. Percentages of CD11b+ cells in total splenocytes and Ly6ChiLy6G– monocytes in CD11b+ splenocytes are shown. (B) B220–CD11b– BM cells from 6- to 8-week-old Vav1-iCre+CbfbF/+ and Vav1-iCre+CbfbF/F mice were analyzed for c-kit, Sca1, and CD16/32 expression. GMP, MPP, and CD16/32+ MPP (bottom panels) percentages in c-kithi cells (top panels) are shown. Statistical analysis from 6 to 9 mice in 6- to 8-week-old and 12- to 16-week-old groups is shown in panels D to G as mean and standard deviation. (C) c-kit and CD16/32 expression in total splenocytes (top) and peripheral blood WBCs (bottom) in 12- to 16-week-old Vav1-iCre+CbfbF/+ (Het, left) and Vav1-iCre+CbfbF/F (CKO, right) mice. Percentages of c-kit+CD16/32+ GMPs are shown with rectangular gates (n = 3, peripheral blood; and n = 6, splenocytes). (H) Hematoxylin and eosin staining of formalin-fixed liver sections from 6- to 8- and 12- to 16-week-old Vav1-iCre+CbfbF/+ (Het) and Vav1-iCre+CbfbF/F (CKO) mice. Clusters of infiltrating cells are surrounded by dotted lines. P, portal triad; CV, central vein. Images are representative of 3 mice for each genotype and age. (I) Clonal lineage potential of BM Lin–c-kit+Sca1+CD135–CD150+ cells from Cbfb-CKO (CKO) and control mice (Het) determined by a single cell culture (n = 2). SSC, side scatter. *P < .05; **P < .01; ***P < .001.
Irf8 restores DC differentiation from Cbfb-CKO BM progenitors
To understand the molecular mechanism by which Runx proteins and Cbfβ regulate cell fate decisions in BM progenitors, we purified LSK cells and GMPs from Cbfb-CKO and control mice and performed gene expression analysis. We identified 366 and 300 probe sets showing more than threefold differences in LSK cells or GMPs, respectively (Figure 6A, supplemental Tables 2-3). Consistent with the observed GMP-skewed differentiation, the gene expression profile of Cbfb-CKO LSK cells was also skewed toward that of GMPs rather than MDPs. We identified GMP- and MDP-specific genes by comparing Immgen datasets28 and compared the expression of these genes between Cbfb-CKO and control LSK cells. The majority of “GMP-specific” genes, including Cpa3, Hdc, Itga2, and Slc7a8, were upregulated, whereas a large number of “MDP-specific” genes, such as Flt3, Dntt, Dpp4, and Gria3, were downregulated in Cbfb-CKO LSK cells compared to control cells (Figure 6B). Among transcription factors that were differentially expressed between Cbfb-CKO and control LSKs or GMPs, Irf8 plays a key regulatory role in DC vs granulocyte differentiation.20 Irf8 expression determined by qRT-PCR was reduced by sevenfold in Cbfb-CKO GMPs compared to control cells (Figure 6C). The increase in GMP differentiation and reduction in Irf8 expression suggested that Cbfb is required to prevent premature differentiation of MPPs toward GMPs at least in part through Irf8-dependent mechanisms, which preserve the ability of MPPs to give rise to DCs. To determine whether inhibition of premature myeloid differentiation by Irf8 is required for DC differentiation, we retrovirally overexpressed Irf8 in Cbfb-CKO BM cells and cultured transduced cells in the presence of Flt3L. Forced expression of Irf8 was sufficient to restore differentiation of MHC-II+ cells to a similar extent as retroviral expression of 2 major isoforms of Cbfb (182 amino acids: NP_001154930, and 187 amino acids: NP_071704.3), although Irf8 was insufficient to restore CD11c expression in those MHC-II+ cells (Figure 6D). Furthermore, Flt3-expressing retrovirus did not rescue DC differentiation from Cbfb-CKO BM cells (Figure 6D). These results suggest that Irf8 functions downstream of Cbfβ to restrict myeloid differentiation and facilitate DC lineage specification.
Runx proteins and Cbfβ restrict GMP-skewed differentiation of MPPs. (A) Microarray analysis of gene expression in LSK cells and GMPs from Vav1-iCre+CbfbF/+ (Het) and Vav1-iCre+CbfbF/F (CKO) mice presented as scattered plots. Signals with higher than threefold differences between Het and CKO samples are shown. (B) Expression of representative “GMP-specific” and “MDP-specific” genes in LSK cells is shown as a heat map. Expression of each gene is shown relative to control (Het) LSK cells. (C) qRT-PCR analysis of Cbfb, Irf8, Sfpi1 (PU.1), Cebpa (C/EBPα), and Csf1r (MCSF-R) in GMPs. Expression levels were normalized against the levels of 18S ribosomal RNA and shown as relative to Het control. Values show mean and standard deviation (n = 4); *P < .05. (D) In vitro DC differentiation from Cbfb-CKO BM cells with retroviral overexpression of Cbfβ (2 isoforms, n = 3), Irf8 (n = 3), and Flt3 (n = 2). CD11c+MHC-II+ and CD11c–MHC-II+ cells are gated with rectangles, and percentages from a representative experiment are shown.
Runx proteins and Cbfβ restrict GMP-skewed differentiation of MPPs. (A) Microarray analysis of gene expression in LSK cells and GMPs from Vav1-iCre+CbfbF/+ (Het) and Vav1-iCre+CbfbF/F (CKO) mice presented as scattered plots. Signals with higher than threefold differences between Het and CKO samples are shown. (B) Expression of representative “GMP-specific” and “MDP-specific” genes in LSK cells is shown as a heat map. Expression of each gene is shown relative to control (Het) LSK cells. (C) qRT-PCR analysis of Cbfb, Irf8, Sfpi1 (PU.1), Cebpa (C/EBPα), and Csf1r (MCSF-R) in GMPs. Expression levels were normalized against the levels of 18S ribosomal RNA and shown as relative to Het control. Values show mean and standard deviation (n = 4); *P < .05. (D) In vitro DC differentiation from Cbfb-CKO BM cells with retroviral overexpression of Cbfβ (2 isoforms, n = 3), Irf8 (n = 3), and Flt3 (n = 2). CD11c+MHC-II+ and CD11c–MHC-II+ cells are gated with rectangles, and percentages from a representative experiment are shown.
Discussion
In this study, we examined the cell-intrinsic requirement for Runx proteins and Cbfβ in the development of DCs and Flt3+ BM progenitors. Our results demonstrate that Cbfβ is essential for the differentiation of both cDC and pDC lineages. The absence of cDCs and pDCs in Cbfb-CKO mice was due to the absence of Flt3+ MDPs and CDPs, which are required and sufficient to give rise to DCs.3,36 Among the 3 Runx proteins, Runx1 was the primary binding partner for Cbfβ in DC lineage specification since we observed a severe reduction of LMPPs, MDP/CLPs, and CDPs derived from Runx1-CKO HSCs in mixed BM chimeras. A loss of Flt3+ progenitors in Cbfb-CKO mice could result from defective Flt3 expression rather than the actual absence of Flt3+ progenitors. However, although Runx1-CKO mice develop reduced numbers of LMPPs, the Flt3 expression level in remaining LMPPs is comparable to that of WT LMPPs12 (supplemental Figure 1E). Furthermore, because Runx1 was necessary for the development of LMPP and MDP populations in the BM and for mature DCs in the spleen under competitive conditions, it is unlikely that Runx1 or Cbfβ is required only for Flt3 expression but dispensable for the development of DC progenitors. Accordingly, we also observed reduced numbers of Lin–c-kit+ cells expressing CD115, which is another marker for MDPs, and reduced expression of the MDP-specific signature in LSK cells. These results collectively suggest that the development of DC progenitors that normally express Flt3 requires Runx proteins and Cbfβ.
The more stringent requirement for Cbfb compared with Runx1 could result from compensation by Runx2, Runx3, or both. In T-cell development, Runx1 and Runx3 have partially redundant functions.14,18,19 Similarly, all 3 Runx genes are expressed in CLPs and MDPs.37 Although Runx2 and Runx3 were individually dispensable for cDC differentiation, it is likely that these genes contributed to the differentiation of LMPPs and DC lineage progenitors in the absence of Runx1 under noncompetitive conditions. Although defective DC differentiation in Cbfb-CKO and Runx1-CKO was attributed to the lack of Flt3+ BM progenitors, including MDPs, it is not yet clear whether Runx proteins and Cbfβ are also required during later stages of DC differentiation, or DC functions in immune responses. In Runx1-CKO mice, we observed reduced numbers of LMPPs and MDPs. However, mature DCs were normally formed in the absence of Runx1 under noncompetitive conditions. These results suggest that additional Runx proteins may fully compensate for Runx1 deficiency in CDPs and developing DCs. Alternatively, terminal DC differentiation may no longer require Runx protein and Cbfβ functions. The generation of novel Cre drivers that delete genes at the MDP or CDP stage would help clarify these questions.
In Cbfb-CKO mice, differentiation toward GMPs in BM progenitors was markedly increased, whereas the development of lymphoid cells, including T, B, and NK cells, was severely compromised. The defective lymphoid differentiation was due to the impaired development of CLPs from MPPs or LMPPs. The skewed differentiation toward myeloid cells was already apparent in the LSK fraction of BM cells because the majority of Cbfb-CKO cells expressed CD16/32 (FcγR) and other genes which are normally more highly expressed in GMPs than MDPs. Although Runx1-CKO mice still developed LMPPs, FcγR transcripts were upregulated in the total LSK fraction or LMPPs in Runx1-CKO mice.12 These findings suggest that aberrant myeloid-skewed differentiation is initiated in MPPs or even in HSCs in Runx1-CKO and Cbfb-CKO mice. This possibility is also consistent with severely compromised erythroid differentiation, which resulted in severe anemia in Cbfb-CKO mice. Therefore, Runx proteins and Cbfβ are required for the suppression of premature myeloid differentiation. Accordingly, a higher frequency of myeloid leukemia is observed compared to lymphoid leukemia with RUNX1-RUNX1T1 (MTG8/ETO) or CBFB-MYH11 (SMMHC) translocations in humans.10
It is not clear how Runx proteins and Cbfβ regulate the development of MPPs to DC or lymphoid progenitors. However, our data suggest that Runx-mediated Irf8 induction may play an important role. During normal differentiation, Irf8 is only weakly expressed in HSCs and LMPPs but upregulated during transition to the DC lineage, whereas GMPs and CLPs express intermediate levels of Irf8.20,28 In Cbfb-CKO mice, Irf8 expression was reduced slightly in LSK cells and markedly in GMPs. Deregulation of IRF8 has also been reported in AML patient samples with RUNX1 mutations.38 Functionally, a previous study showed that Irf8 promotes MPP differentiation toward CDPs over GMPs.20 Lower basal expression of Irf8 in HSCs or MPPs, or failure to upregulate Irf8 in Cbfb-CKO progenitors, may skew MPP differentiation toward GMPs at the expense of DC progenitor differentiation. Accordingly, forced expression of Irf8 facilitated DC differentiation from Cbfb-CKO progenitors in our experiments, although CD11c expression still required Cbfβ as we observed in Runx2-deficient pDCs. Because Flt3 overexpression did not restore DC differentiation in the absence of Cbfb, the lack of Flt3 expression alone does not account for the defective DC development in Cbfb-CKO mice. Neither Irf8 nor Flt3 is likely to be a direct target of Runx/Cbfβ complexes because there are no consensus Runx-binding sequences in conserved noncoding regions in the Irf8 and Flt3 loci. This is also supported by the absence of Runx1-binding peaks in these loci in a ChIPseq study using a hematopoietic progenitor cell line39 (supplemental Figure 3). Therefore, these data suggest that Runx proteins may act through intermediate signals to upregulate Irf8 and facilitate Flt3+ BM progenitor differentiation.
In addition to the substantially altered expression of Irf8, our gene expression profiling of LSK cells and GMPs lacking Cbfb suggests additional regulatory roles of Runx proteins and Cbfβ in hematopoietic differentiation. In the myeloid lineage, despite increased differentiation toward GMPs from HSCs, Cebpa was downregulated. Although Cebpa is dispensable for maturation of granulocytes after the GMP stage,5 reduced Cebpa in combination with reduced Csf1r and a loss of Cbfβ functions may compromise the differentiation program of CD11b+ cells or Ly6C+ monocytes in Cbfb-CKO mice. Consistent with defective lymphocyte development, basal expression of transcription factors that are important for B- and T-cell development, including Ebf1, Notch1, Pax5, and Tcf7, was reduced in Cbfb-CKO LSK cells. Intriguingly, several genes differentially expressed in Cbfb-CKO LSK cells or GMPs compared with control are also dysregulated in AML patients with RUNX1 mutations,40,41 such as CEBPA, P2RY14, IRF8, BLNK, FOXO1, RBPMS, DNTT, and GIMAP7. These results indicate that Runx1-CKO and Cbfb-CKO mice may serve as models to study the pathogenesis of leukemias with RUNX1 mutations.
In summary, our current study provides genetic evidence that Runx proteins and Cbfβ are required for maintenance of the undifferentiated state of HSCs and for the differentiation of Flt3+ DC progenitors. Our data suggest that restriction of aberrant myeloid lineage differentiation mediated by Runx proteins and Cbfβ preserves the potential of MPPs to give rise to diverse lineages of blood cells, in part through Irf8 induction. These results provide insights into the mechanisms underlying hematopoietic lineage commitment and Runx-mediated myeloid leukemic transformation.
The raw data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE55227).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ichiro Taniuchi (RIKEN) and Dan R. Littman (New York University) for Cbfb and Runx3 conditional KO mice, Gerard Karsenty (Columbia University) for Runx2 KO mice, Toshio Kitamura (University of Tokyo) for PlatE cells, and Kenneth M. Murphy (Washington University) for discussion.
This work was supported by grants from the National Institutes of Health (National Institute of Allergy and Infectious Diseases, R01AI097244 [T.E.] National Institute of Diabetes and Digestive and Kidney Disease, K01DK078318 [D.B.], and National Cancer Institute, R01CA149976 [N.A.S.]) and the Edward Mallinckrodt Foundation (T.E.). D.B. is a New York Stem Cell Foundation Robertson Investigator.
Authorship
Contribution: A.T.S. and T.E. designed experiments; A.T.S., C.G.B., C.C., S.H., D.G.M., D.B., and T.E. performed experiments; X.C. and N.A.S. provided experimental material; A.T.S. and T.E. analyzed data; and T.E. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Takeshi Egawa, Department of Pathology and Immunology, Washington University School of Medicine, 660 South Euclid Ave, Campus Box 8118, St. Louis, MO 63110; e-mail: tegawa@wustl.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal