Key Points
c-Myc is required for leukemia-initiating cell maintenance in murine models of T-ALL.
c-Myc inhibition prevents the growth of treatment-resistant primary T-ALL patient samples in vitro.
Although prognosis has improved for children with T-cell acute lymphoblastic leukemia (T-ALL), 20% to 30% of patients undergo induction failure (IF) or relapse. Leukemia-initiating cells (LICs) are hypothesized to be resistant to chemotherapy and to mediate relapse. We and others have shown that Notch1 directly regulates c-Myc, a known regulator of quiescence in stem and progenitor populations, leading us to examine whether c-Myc inhibition results in efficient targeting of T-ALL–initiating cells. We demonstrate that c-Myc suppression by small hairpin RNA or pharmacologic approaches prevents leukemia initiation in mice by eliminating LIC activity. Consistent with its anti-LIC activity in mice, treatment with the BET bromodomain BRD4 inhibitor JQ1 reduces C-MYC expression and inhibits the growth of relapsed and IF pediatric T-ALL samples in vitro. These findings demonstrate a critical role for c-Myc in LIC maintenance and provide evidence that MYC inhibition may be an effective therapy for relapsed/IF T-ALL patients.
Introduction
NOTCH1 mutations are prevalent in patients with T-cell acute lymphoblastic leukemia (T-ALL), with 55% of patients harboring mutations in the heterodimerization (HD) and/or PEST regulatory regions.1 These mutations are thought to result in ligand-independent, γ-secretase–dependent cleavage and increased stability of intracellular NOTCH1. An additional 10% to 20% of T-ALL patients contain mutations in FBW7,2 the E3 ubiquitin ligase responsible for the degradation of NOTCH1, C-MYC, Cyclin E, and C-FOS.3,,-6 NOTCH1 activation may contribute to the development of other hematopoietic malignancies, including chronic lymphocytic leukemia7,-9 and mantle cell lymphoma.10,11 These findings support the continued development of NOTCH inhibitors as cancer therapeutics.
Notch1 mutations develop spontaneously in our Tal1 and Tal1/Lmo2 mouse T-ALL models12 and treatment with γ-secretase inhibitors (GSI) prevents Notch1 activation and extends the survival of leukemic mice, demonstrating that GSIs have antileukemia activity in vivo.12,-14 Leukemia-initiating cells (LICs) contribute to T-ALL pathogenesis,13,15,,-18 and we and others have shown that a committed thymic progenitor population is enriched in the ability to initiate disease in syngeneic recipients.13,16 We then provided evidence that Notch1 inhibition can eliminate the LIC population and prevent disease initiation.13 Consistent with our studies in mice, Armstrong et al provide evidence that when primary human T-ALL cells are treated with GSI in vitro, this interferes with the ability of the leukemic cells to initiate disease in immunodeficient mice.19 Collectively, these studies suggest that the LIC population in T-ALL depends on sustained NOTCH1 activity.
Treatment of human T-ALL cell lines with a GSI primarily results in cell-cycle arrest.2,20,21 Notch1 regulates leukemic proliferation by directly stimulating c-Myc and cyclin D3 expression.20,,-23 Retroviral c-Myc expression has been shown to rescue mouse and human T-ALL cells from the effects of NOTCH1 inhibition, suggesting that MYC is essential for NOTCH1-mediated leukemogenesis.20,22 The Notch1 pathway regulates mouse thymocyte survival and metabolism,24,,,-28 and c-Myc is required for DN3 and DN4 thymic progenitor expansion.29 These findings led us to hypothesize that c-Myc drives mouse LIC expansion in vivo and that c-Myc inhibition may interfere with multiple biological processes associated with LIC activity, including extensive proliferation, survival, and self-renewal as well as metabolic and/or epigenetic changes that may be associated with persistence and drug resistance.
Materials and methods
Mice
Tal1/Lmo2 transgenic mice were maintained and monitored daily for development of leukemia as previously described.30,31 We obtained NOD.Cg-Prkdcscidll2tm1Wjl/SzJ (NSG) mice from the colonies maintained by Dr Shultz at The Jackson Laboratory. All animal procedures used in this study were approved by the University of Massachusetts Medical School Institutional Animal Care and Use Committee.
Primary mouse and patient T-ALL cells and cell lines
Primary mouse Tal1/Lmo2 T-ALL cells were plated in RPMI with 20% fetal bovine serum (FBS), 1% penicillin/streptomycin, and 1% l-glutamine (Gibco). Interleukin-7 (2 ng/mL), Flt3L (5 ng/mL), and stem cell factor (10 ng/mL) (R&D Systems) were added to the culture media every 2 to 3 days until the leukemic cells adapted to in vitro culture (approximately 2 weeks). Cells were infected with retroviruses32 encoding small hairpin RNAs (shRNAs) to c-Myc (shMyc) or Renilla luciferase (shRen), with green fluorescent protein (GFP) expression driven by a separate promoter. Human T-ALL cell lines were cultured in RPMI supplemented in 10% FBS, 1% l-glutamine, and 1% penicillin/streptomycin at 37°C under 5% CO2.
Primary human T-ALL samples were obtained from children with T-ALL enrolled in clinical trials of the Dana-Farber Cancer Institute or University of Massachusetts Memorial Hospital. Samples were collected with informed consent and with approval of the institutional review board. This study was conducted in accordance with the Declaration of Helsinki. Leukemic blasts were isolated from peripheral blood or bone marrow by Ficoll-Hypaque centrifugation and cryopreserved in FBS containing 10% dimethylsulfoxide (DMSO) and stored in liquid nitrogen. Fresh or frozen leukemic blasts were expanded in NSG mice by transplanting 0.5 to 5 × 106 viable leukemic cells via intravenous injection. Primary human T-ALL samples were isolated from the spleen and bone marrow of NSG mice and were cultured at 37°C under 5% CO2 in WIT-L media without MS5 feeder layer as described previously.33
In vivo studies
To determine the effect of Myc silencing on LIC frequency and survival, mouse Tal1/Lmo2 T-ALL cells were infected with retroviruses expressing Renilla luciferase or c-Myc–specific shRNAs32 and sorted for expression of GFP. GFP-positive mouse leukemic cells were then serially diluted and transplanted into the syngeneic recipient mice as described previously.30 Mice were monitored for disease development and LIC frequency estimated using Poisson distribution statistics with ELDA software.34 To determine the effect of c-Myc inhibition on LIC frequency, Tal1/Lmo2 mouse T-ALLs were serially diluted and transplanted into syngeneic recipient mice. Mice were then randomized and injected intraperitoneally with vehicle or 50 mg/kg JQ1 daily for 3 weeks, beginning 1 hour after transplantation. All mice were monitored for any signs of drug toxicity and euthanized when disease was evident.
Fluorescence-activated cell sorter analysis
Single-cell suspensions of Tal1/Lmo2 mouse T-ALL cells or patient leukemic cells were stained with cell surface antibodies and analyzed as described previously.30 For in vivo shRNA studies, retrovirally transduced leukemic cells were sorted for GFP expression on the FACSAria (BD Biosciences). For cell-cycle analysis, leukemic cells were fixed in 70% ethanol overnight and then stained with propidium iodide for DNA quantification. For Annexin V staining, Tal1/Lmo2 mouse T-ALL cells, human T-ALL cell lines, or patient leukemic cells were treated with DMSO, GSI (Compound E or DBZ), or JQ1 for 96 hours and stained with Annexin V and 7AAD following the manufacturer’s protocol (BD Bioscience).
Immunoblotting
To monitor c-Myc expression levels, mouse leukemic cells were lysed in radioimmunoprecipitation assay buffer and total protein run on a 4% to 12% Bis-Tris gel (Invitrogen). Protein was transfered to a nitrocellulose membrane and probed with antibodies to c-Myc (clone N262; Santa Cruz Biotechnology), Notch1 Val1744, or Erk1/2 (Cell Signaling). Blots were imaged and quantified using ImageLab Software (Bio-Rad).
DNA and RNA analyses
RNA was extracted and complementary DNA synthesized as previously described; complementary DNA was quantified using specific c-Myc primers.30 Additional primer sets were also used against the following c-Myc target genes: Apex1 F: 5′-ACGGGGAAGAACCCAAGTC-3′; R: 5′- GGTGAGGTTTTCTGATCTGGAG-3′, Cdk4 F: 5′- ATGGCTGCCACTCGATATGAA-3′; R: 5′- TCCTCCATTAGGAACTCTCACAC-3′, Fbl F: CAAAATTGAGTACAGAGCCTGGA-3′; R: 5′- CGGGCCGACAATATCAGAGA-3′, Tk1 F: 5′-AAGTGCCTGGTCATCAAGTATG-3′; R: 5′- GCTGCCACAATTACTGTCTTGC-3′. Human T-ALL cell lines or primary T-ALL samples were cultured in the presence of DMSO, 1 μM GSI (DBZ), or 1 μM JQ1 for 24 hours and quantitative polymerase chain reaction (PCR) performed using the following primer sets: cMYC F: 5′-GCAGCTGCTTAGACGCTGGATTTT-3′; R: 5′-GCAGCAGCTCGAATTTCTTCGAGA-3′ and β-ACTIN F: 5′- CGCGAGAAGATGACCCAGAT- 3′; R: 5′-GATAGCACAGCCTGGATAGCAAC- 3′. Gene expression was determined using the ΔΔCT method normalized to β-ACTIN, and the DMSO control was set to 1.
To determine if mouse leukemic cells retained the retrovirus encoding the Myc shRNAs, DNA was isolated from leukemic cells using TRIzol (Invitrogen). Primers were designed against GFP (F: 5′-TATATCATGGCCGACAAGCA-3′; R: 5′-CCTACAGGTGGGGTCTTTCA-3′) and standard qualitative PCR was performed with Taq polymerase.
Cell viability and death assays
Tal1/Lmo2 mouse T-ALL cell lines were treated with 1 μM Compound E (Alexis) or with 250 nM JQ1 (Bradner Laboratory, Dana-Farber Cancer Institute). The number of viable leukemic cells was calculated after 48 and 96 hours using a trypan blue exclusion assay. To determine whether exogenous Myc expression rescues leukemic cells from the effects of JQ1, mouse T-ALL cells were infected with the MSCV2.2-IRES-GFP retrovirus only or with one expressing mouse c-Myc. Cells were then cultured in the absence or presence of GSI or JQ1 and the effects on proliferation and survival determined by cell-cycle analysis and trypan blue staining. Human T-ALL cell lines or primary relapsed or induction failure (IF) T-ALL samples were cultured for 5 days in the presence of DMSO, GSI (DBZ; Bradner Laboratory, Dana-Farber Cancer Institute), or increasing concentrations of JQ1. Metabolic activity was assayed after 5 days by the addition of CellTiter-Glo chemiluminescence reagent (Promega) and measured using a Beckman Coulter DTX 880 plate reader. Absorbance values were normalized to DMSO control for each patient sample and cell line. Nonlinear dose-response curves were fitted, from which the GI50 (the concentration of JQ1 at which 50% of the cells are affected) and the Emax (the percentage of cells affected at the highest concentration of JQ1) were calculated using GraphPad Prism 5 software (GraphPad).
Statistical measures
Kaplan-Meier survival curves and statistical analyses were performed using GraphPad Prism software (version 5.0). The hazard ratio and its 95% confidence interval (CI) were also measured, comparing the c-Myc and Renilla shRNA groups or vehicle- and JQ1-treated groups and adjusting for the dilutions of leukemic cells using the Cox proportional hazards model analysis. A 2-sided P < .05 was considered statistically significant. Student t tests were performed on proliferation and Annexin V data using GraphPad Prism software (version 5.0). LIC frequency was calculated using Poisson distribution analysis and ELDA software.34 Correlation was determined using a nonparametric Spearman correlation, and a 2-sided 95% CI was used to determine significance.
Results
c-Myc silencing prolongs survival and reduces the frequency of LICs
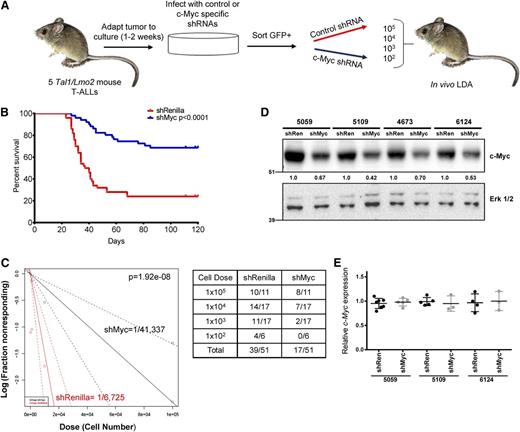
To test a requirement for c-Myc in LIC expansion and self-renewal in vivo, we infected primary mouse T-ALLs with GFP-expressing retroviruses encoding shRNAs to c-Myc or Renilla luciferase. GFP-positive leukemic cells (supplemental Figure 1, available on the Blood Web site) were transplanted into syngeneic recipients and an in vivo limiting dilution assay performed to estimate LIC frequency (Figure 1A). Five independent mutant Notch1, Tal1/Lmo2 mouse T-ALLs were examined (4673, 6124, 5059, 5109, and 9421). Overall, silencing of c-Myc resulted in statistically significant increases in overall survival (Figure 1B; P < .0001) and an average 6.14-fold decrease in mouse LIC frequency (Figure 1C; P = 1.92 × 10−8; LIC frequency shRen = 0.0149% [CI: 0.0254%-0.0087%] vs shMyc = 0.0024% [CI: 0.0044-0.0013]). At the time of transplant, all 5 mouse T-ALLs examined showed a similar reduction in c-Myc protein levels (Figure 1D; average 42%, range 30%-58%) and sort purity was between 90% and 100% (supplemental Figure 1 and data not shown). Myc silencing also interfered with mouse T-ALL growth in vitro, resulting in the depletion of viable leukemic cells (supplemental Figure 2). Thus, the mouse T-ALLs examined appeared similarly dependent on Myc for their growth in vitro and in vivo.
Silencing of c-Myc prolongs survival and reduces LIC frequency in mice transplanted with murine T-ALL cells. (A) Experimental design. Five independent murine Tal1/Lmo2 T-ALL cells were infected with retroviruses encoding an shRNA to c-Myc or Renilla luciferase. Cells were sorted for GFP expression, serially diluted, and transplanted into syngeneic recipients via intraperitoneal injection. Transplanted mice were monitored for evidence of disease. (B) The survival curve for each group of mice was estimated using the Kaplan-Meier method and the difference in overall survival between the 2 groups assessed by the log-rank test (P < .0001). (C) c-Myc silencing reduces LIC frequency in mice transplanted with murine T-ALL cells. A log-log plot and LIC frequency was calculated using ELDA software for ShRenilla (red, 1/7308) and shMyc (black, 1/41 337). A portion of secondary recipients develop leukemia when transplanted with limiting dilutions of shRenilla- and shMyc-infected leukemic cells. (D) Reduced c-Myc protein levels in shMyc-transduced leukemic cells at time of transplant. Protein was isolated from GFP-positive leukemic cells prior to transplant and lysates probed with a c-Myc and Erk1/2 antibodies. (E) Transplanted mice that develop disease do not exhibit reduced Myc expression. RNA was isolated from leukemic cells transduced with the shRen or shMyc retroviruses and c-Myc mRNA levels determined by quantitative real-time PCR. Copy number was normalized to β-actin using the ΔΔCT method. Each point represents c-Myc mRNA levels of leukemic cells isolated from a single mouse.
Silencing of c-Myc prolongs survival and reduces LIC frequency in mice transplanted with murine T-ALL cells. (A) Experimental design. Five independent murine Tal1/Lmo2 T-ALL cells were infected with retroviruses encoding an shRNA to c-Myc or Renilla luciferase. Cells were sorted for GFP expression, serially diluted, and transplanted into syngeneic recipients via intraperitoneal injection. Transplanted mice were monitored for evidence of disease. (B) The survival curve for each group of mice was estimated using the Kaplan-Meier method and the difference in overall survival between the 2 groups assessed by the log-rank test (P < .0001). (C) c-Myc silencing reduces LIC frequency in mice transplanted with murine T-ALL cells. A log-log plot and LIC frequency was calculated using ELDA software for ShRenilla (red, 1/7308) and shMyc (black, 1/41 337). A portion of secondary recipients develop leukemia when transplanted with limiting dilutions of shRenilla- and shMyc-infected leukemic cells. (D) Reduced c-Myc protein levels in shMyc-transduced leukemic cells at time of transplant. Protein was isolated from GFP-positive leukemic cells prior to transplant and lysates probed with a c-Myc and Erk1/2 antibodies. (E) Transplanted mice that develop disease do not exhibit reduced Myc expression. RNA was isolated from leukemic cells transduced with the shRen or shMyc retroviruses and c-Myc mRNA levels determined by quantitative real-time PCR. Copy number was normalized to β-actin using the ΔΔCT method. Each point represents c-Myc mRNA levels of leukemic cells isolated from a single mouse.
When transplanted animals appeared moribund, GFP and c-Myc expression levels were examined. In most transplanted mice, the leukemic cells remained GFP positive and harbored the retroviral provirus. A total of 17 of 51 mice transplanted with shMyc-transduced cells developed leukemia but appeared to express c-Myc at levels similar to that observed in the control shRNA-transduced leukemic cells (Figure 1E and data not shown), suggesting that c-Myc suppression was not maintained in these leukemic mice, even though the leukemic cells retained GFP expression (data not shown). The long terminal repeat driving the expression of the c-Myc shRNA may have been silenced, leaving the phosphoglycerate kinase promoter driving the expression of GFP intact. Thus, those mice transplanted with shMyc-transduced cells that developed leukemia escaped from c-Myc silencing, supporting our data that c-Myc expression is required for mouse LIC activity.
In order to determine whether Myc suppression eradicates the LIC, we euthanized the primary transplants that failed to develop disease after 120 days. GFP-positive leukemic cells in the bone marrow of these mice were low to undetectable (supplemental Figure 3). To further demonstrate that c-Myc silencing depletes the LIC population and does not induce cell-cycle arrest or quiescence, 5 × 106 pooled bone marrow cells were transplanted into secondary recipients, which were then monitored for evidence of disease for an additional period of 120 days. None of the 17 secondary recipients developed leukemia (data not shown), indicating that Myc suppression eliminates the LICs.
BET bromodomain inhibition induces apoptosis of murine T-ALL cell lines in vitro
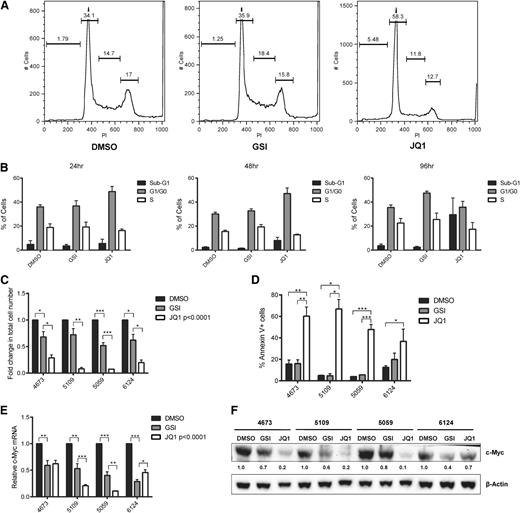
Although these c-Myc shRNAs have been reported to reduce c-Myc expression in vivo,35 shRNAs can have off-target effects,36,-38 which could alter LIC activity. Therefore, we also undertook a pharmacologic approach to inhibit c-Myc using the BET bromodomain 4 (Brd4) inhibitor JQ1 developed by our coauthors.39 We compared the effects of JQ1 to GSI treatment on mouse T-ALL proliferation and survival in vitro. We treated mouse Tal1/Lmo2 T-ALL cell lines with the GSI (Compound E) or JQ1 and monitored cell growth and viability and performed cell-cycle and apoptosis analyses. JQ1 treatment of 24 or 48 hours induced G0/G1 cell-cycle arrest (Figure 2A-B) followed by an increase in sub-G1 cells evident at 96 hours (Figure 2B). In contrast and as expected, GSI-induced cell-cycle arrest was detected at 96 hours (Figure 2B, third panel; P < .0098) with no significant increases in sub-G1 or Annexin V–positive leukemic cells (Figure 2B,D). Overall, JQ1 treatment reduced the proliferation of the murine T-ALL cell lines examined (Figure 2C) and induced apoptosis in over half the leukemic cells by 96 hours (Figure 2D). Although GSI treatment arrested Tal1/Lmo2 leukemic cell line growth, it did not induce apoptosis (Figure 2C-D).
JQ1 treatment of murine T-ALL cells results in cell-cycle arrest followed by apoptosis. Murine T-ALL cell lines were treated with JQ1 (250 nM) or with the GSI (Compound E, 1 μM), unless otherwise noted. (A) JQ1-induced cell-cycle arrest is evident at 24 hours. Murine T-ALL cell lines were left untreated or treated with JQ1 or Compound E for 24 hours and then stained with propidium iodide followed by flow cytometry. Four mouse T-ALL cell lines were analyzed; 1 representative plot from mouse T-ALL 5109 is shown. (B) JQ1 treatment induces cell-cycle arrest followed by apoptosis. Murine T-ALL cell line 5109 was treated with JQ1 or Compound E for 24, 48, and 96 hours and the percentage of cells in the subG1, G0/G1, and S phases determined. (C) Mouse T-ALL cell lines are more sensitive to treatment with JQ1 than GSI. Four mouse T-ALL cell lines (4673, 5109, 5059, and 6124) were treated with DMSO, JQ1, or the GSI (Compound E) for 96 hours and the total number of viable cells was calculated via trypan blue exclusion assay. The results are averages of 3 independent experiments and error bars represent standard error of the mean (SEM). P < .0001 for all JQ1-treated cell lines, *P < .05, **P < .01, ***P < .001. (D) JQ1 induces apoptosis of mouse T-ALL cells. Four mouse T-ALL cell lines were treated with vehicle, JQ1, or Compound E and the percentage of apoptotic cells determined by Annexin V and 7AAD staining followed by flow cytometry. The results are averages of 3 independent experiments and error bars represent SEM. *P < .05, **P < .01, ***P < .001. (E) Reduced c-Myc mRNA levels in JQ1- and GSI-treated mouse T-ALL cells. RNA was isolated from leukemic cultures treated with vehicle, JQ1, or Compound E for 48 hours and c-Myc expression was analyzed by quantitative real-time PCR. Copy number was normalized to β-actin using the ΔΔCT method. The results are averages of 3 independent experiments and error bars represent SEM. P < .0001 for all JQ1-treated cell lines *P < .05, **P < .01, ***P < .001. (F) c-Myc protein levels were reduced upon treatment with JQ1. Protein was isolated from mouse T-ALL cells treated with DMSO, JQ1, or GSI (Compound E) for 48 hours and c-Myc and β-actin protein levels determined by immunoblotting.
JQ1 treatment of murine T-ALL cells results in cell-cycle arrest followed by apoptosis. Murine T-ALL cell lines were treated with JQ1 (250 nM) or with the GSI (Compound E, 1 μM), unless otherwise noted. (A) JQ1-induced cell-cycle arrest is evident at 24 hours. Murine T-ALL cell lines were left untreated or treated with JQ1 or Compound E for 24 hours and then stained with propidium iodide followed by flow cytometry. Four mouse T-ALL cell lines were analyzed; 1 representative plot from mouse T-ALL 5109 is shown. (B) JQ1 treatment induces cell-cycle arrest followed by apoptosis. Murine T-ALL cell line 5109 was treated with JQ1 or Compound E for 24, 48, and 96 hours and the percentage of cells in the subG1, G0/G1, and S phases determined. (C) Mouse T-ALL cell lines are more sensitive to treatment with JQ1 than GSI. Four mouse T-ALL cell lines (4673, 5109, 5059, and 6124) were treated with DMSO, JQ1, or the GSI (Compound E) for 96 hours and the total number of viable cells was calculated via trypan blue exclusion assay. The results are averages of 3 independent experiments and error bars represent standard error of the mean (SEM). P < .0001 for all JQ1-treated cell lines, *P < .05, **P < .01, ***P < .001. (D) JQ1 induces apoptosis of mouse T-ALL cells. Four mouse T-ALL cell lines were treated with vehicle, JQ1, or Compound E and the percentage of apoptotic cells determined by Annexin V and 7AAD staining followed by flow cytometry. The results are averages of 3 independent experiments and error bars represent SEM. *P < .05, **P < .01, ***P < .001. (E) Reduced c-Myc mRNA levels in JQ1- and GSI-treated mouse T-ALL cells. RNA was isolated from leukemic cultures treated with vehicle, JQ1, or Compound E for 48 hours and c-Myc expression was analyzed by quantitative real-time PCR. Copy number was normalized to β-actin using the ΔΔCT method. The results are averages of 3 independent experiments and error bars represent SEM. P < .0001 for all JQ1-treated cell lines *P < .05, **P < .01, ***P < .001. (F) c-Myc protein levels were reduced upon treatment with JQ1. Protein was isolated from mouse T-ALL cells treated with DMSO, JQ1, or GSI (Compound E) for 48 hours and c-Myc and β-actin protein levels determined by immunoblotting.
To compare the effects of GSI or JQ1 treatment on c-Myc expression levels, mouse T-ALL cell lines were treated with DMSO, GSI, or increasing concentrations of JQ1 for 48 hours and c-Myc messenger RNA (mRNA) levels determined by real-time quantitative PCR. Dose-dependent reductions in c-Myc mRNA levels were observed in the JQ1-treated leukemic cells (not shown) with optimal reductions observed when cultures were treated with 250 nM JQ1 (Figure 2E). With the exception of mouse T-ALL 6124, JQ1 treatment was also more effective than GSI at suppressing c-Myc protein levels (Figure 2F).
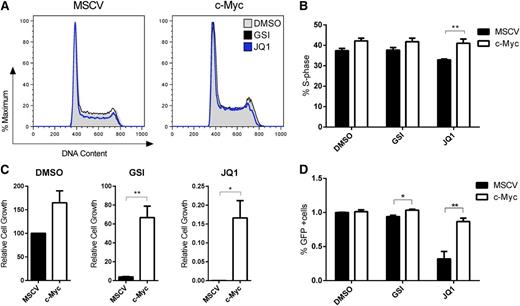
These data argue that Myc is a critical Brd4 target in mouse T-ALL. To test this directly, we transduced mouse T-ALL cells with a MSCV2.2 IRES-GFP retrovirus or with one expressing mouse c-Myc and then cultured the cells in the absence or presence of GSI or JQ1 and examined the effects on the cell cycle, leukemic growth, and survival. Cell-cycle analysis revealed a significant increase in S-phase cells in the JQ1-treated Myc-transduced cells, resulting in significant increases in leukemic cell growth and the percentage of GFP-positive cells (Figure 3A-D). Consistent with our previous work22 (and Figure 2), the Myc-transduced cells were also more resistant to growth arrest induced by GSI treatment (Figure 3C), although maximal effects on leukemic growth are observed at later time points (Figure 2C, 96 hours). JQ1 treatment resulted in reduced expression of several c-Myc–regulated genes including Cdk4, Apex1, Fbl, and Tk1, indicating that JQ1 reduces the transcriptional activity of c-Myc (supplemental Figure 4). Collectively, these data raise the possibility that JQ1 might be more effective than GSI treatment because JQ1 suppresses c-Myc and potentially other targets and more effectively interferes with the Myc transcriptional program.
Exogenous c-Myc expression partially rescues cell-cycle arrest and apoptosis of murine T-ALL cells treated with JQ1. Murine T-ALL cell lines were transduced with a MSCV2.2 IRES-GFP retrovirus or with one expressing mouse c-Myc. Murine stem cell virus– or c-Myc–expressing cells were treated with JQ1 (250 nM) or with the GSI (Compound E, 1 μM), unless otherwise noted. (A-B) c-Myc expression rescues JQ1 effects on cell-cycle progression at 24 hours. Murine T-ALL cell lines were left untreated or treated with JQ1 or Compound E for 24 hours and then stained with propidium iodide followed by flow cytometry. Mouse T-ALL cell line 5059 was analyzed; shown are 1 representative histogram overlay of DNA content (A) and collated data for the percentage of cells in S phase (B). The results are averages of 3 independent experiments and error bars represent SEM. *P < .05, **P < .01. (C) Mouse T-ALL cell line 5059 was treated with DMSO, JQ1 or GSI for 10 days and the total number of viable cells was calculated via trypan blue exclusion assay. The results are averages of 3 independent experiments and error bars represent SEM. **P < .01. (D) JQ1 induces apoptosis of mouse T-ALL cells. T-ALL cell lines were treated with vehicle, JQ1, or GSI for 10 days and the percentage of GFP-positive cells were determined by flow cytometry. The results are averages of 3 independent experiments and error bars represent SEM. *P < .05, **P < .001.
Exogenous c-Myc expression partially rescues cell-cycle arrest and apoptosis of murine T-ALL cells treated with JQ1. Murine T-ALL cell lines were transduced with a MSCV2.2 IRES-GFP retrovirus or with one expressing mouse c-Myc. Murine stem cell virus– or c-Myc–expressing cells were treated with JQ1 (250 nM) or with the GSI (Compound E, 1 μM), unless otherwise noted. (A-B) c-Myc expression rescues JQ1 effects on cell-cycle progression at 24 hours. Murine T-ALL cell lines were left untreated or treated with JQ1 or Compound E for 24 hours and then stained with propidium iodide followed by flow cytometry. Mouse T-ALL cell line 5059 was analyzed; shown are 1 representative histogram overlay of DNA content (A) and collated data for the percentage of cells in S phase (B). The results are averages of 3 independent experiments and error bars represent SEM. *P < .05, **P < .01. (C) Mouse T-ALL cell line 5059 was treated with DMSO, JQ1 or GSI for 10 days and the total number of viable cells was calculated via trypan blue exclusion assay. The results are averages of 3 independent experiments and error bars represent SEM. **P < .01. (D) JQ1 induces apoptosis of mouse T-ALL cells. T-ALL cell lines were treated with vehicle, JQ1, or GSI for 10 days and the percentage of GFP-positive cells were determined by flow cytometry. The results are averages of 3 independent experiments and error bars represent SEM. *P < .05, **P < .001.
JQ1 treatment interferes with leukemia initiation and reduces LICs
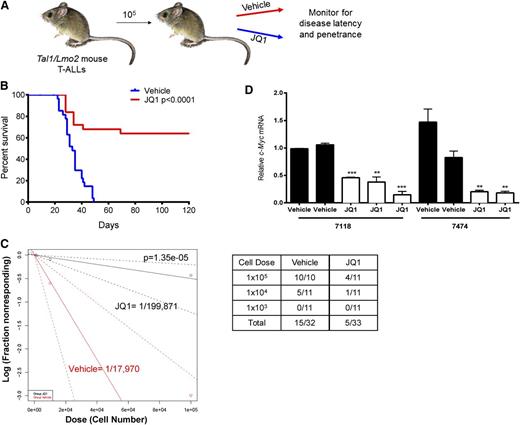
Treatment of mouse Tal1/Lmo2 T-ALL cells with JQ1 in vitro induces apoptosis (Figure 2D), suggesting that JQ1 treatment may extend survival in this mouse T-ALL model. To examine the effects of JQ1 treatment on mouse survival, mice were each transplanted with leukemic cells from 3 independent mouse T-ALLs (8236, 7624, 7118) and vehicle or JQ1 administered for 3 weeks (Figure 4A). JQ1 treatment resulted in significant increases in overall survival (Figure 4B; P < .0001). In the mouse T-ALLs examined, JQ1 treatment significantly prolonged survival and in some mice prevented disease initiation, leading us to hypothesize that JQ1 treatment may interfere with the LIC population.
JQ1 treatment in vivo reduces c-Myc expression and significantly prolongs survival. (A) Experimental design. Tal1/Lmo2 mouse T-ALLs were transplanted (cell dose 105) into syngeneic recipients and vehicle or JQ1 was administered at 50 mg/kg daily starting at the time of transplant and continuing for 3 consecutive weeks. Mice were monitored for disease and euthanized when they became moribund. (B) Survival was estimated using the Kaplan-Meier method and the difference in overall survival between the 2 groups assessed by the log-rank test. (C) c-Myc inhibition targets the LIC in Tal1/Lmo2 T-ALLs. Leukemic cells were diluted and transplanted into syngeneic mice. JQ1 was administered at 50 mg/kg daily starting at the time of transplant and continuing for 3 consecutive weeks. A log-log plot and LIC frequency was calculated using ELDA software for vehicle, (red, 1/17 970) and JQ1 (black, 1/199 871). A fraction of secondary recipients develop leukemia when transplanted with limiting dilutions of leukemic cells and treated with either vehicle or JQ1. (D) Leukemic mice remain responsive to JQ1 treatment. When disease was evident, a single dose of JQ1 was readministered and RNA isolated from mouse leukemic tissues. c-Myc expression was analyzed by quantitative real-time PCR. Copy number was normalized to β-actin using the ΔΔCT method. Each bar represents a single mouse and error bars represent SEM. **P < .01, ***P < .001, ****P < .0001.
JQ1 treatment in vivo reduces c-Myc expression and significantly prolongs survival. (A) Experimental design. Tal1/Lmo2 mouse T-ALLs were transplanted (cell dose 105) into syngeneic recipients and vehicle or JQ1 was administered at 50 mg/kg daily starting at the time of transplant and continuing for 3 consecutive weeks. Mice were monitored for disease and euthanized when they became moribund. (B) Survival was estimated using the Kaplan-Meier method and the difference in overall survival between the 2 groups assessed by the log-rank test. (C) c-Myc inhibition targets the LIC in Tal1/Lmo2 T-ALLs. Leukemic cells were diluted and transplanted into syngeneic mice. JQ1 was administered at 50 mg/kg daily starting at the time of transplant and continuing for 3 consecutive weeks. A log-log plot and LIC frequency was calculated using ELDA software for vehicle, (red, 1/17 970) and JQ1 (black, 1/199 871). A fraction of secondary recipients develop leukemia when transplanted with limiting dilutions of leukemic cells and treated with either vehicle or JQ1. (D) Leukemic mice remain responsive to JQ1 treatment. When disease was evident, a single dose of JQ1 was readministered and RNA isolated from mouse leukemic tissues. c-Myc expression was analyzed by quantitative real-time PCR. Copy number was normalized to β-actin using the ΔΔCT method. Each bar represents a single mouse and error bars represent SEM. **P < .01, ***P < .001, ****P < .0001.
Although c-Myc inhibition has been shown to inhibit bulk leukemic growth in certain hematologic malignancies,35,40,41 the effect(s) of Myc inhibition on leukemia stem or LICs has not been addressed. To quantify JQ1 effects on the mouse LIC population, we transplanted 3 additional murine T-ALLs under in vivo limiting-dilution conditions and treated recipient mice with vehicle or JQ1 for 3 weeks. A 3-week course of JQ1 treatment reduced the estimated LIC frequency by 11.2-fold (Figure 4C; P = 1.35 × 10−5; LIC frequency for vehicle 0.0056% [CI: 0.0121%-0.0026%] vs JQ1 0.0005% [CI:0.0012%-0.0002%]), revealing a central role for c-Myc in mouse LIC maintenance. JQ1-treated mice that developed leukemia remained responsive to the inhibitor, as we found c-Myc mRNA levels reduced in leukemic cells upon JQ1 readministration (Figure 4D). These findings led us to conclude that disease recurrence was not due to the development of resistance to JQ1 but rather may reflect the transient and partial c-Myc inhibition achieved in vivo.
We found that 15 of 32 (47%) vehicle-treated mice developed disease whereas 5 of 333 (15%) mice treated with JQ1 succumbed to T-ALL (Figure 4C). Secondary transplants were performed using pooled bone marrow cells from the 28 surviving mice treated with JQ1, and these mice were observed for an additional 120 days. None of the secondary transplants developed disease (data not shown), indicating that LIC frequency was reduced below the detection limit of 1 in 5 × 106 leukemic cells.
JQ1 inhibits the growth of human T-ALL cell lines and relapsed or IF pediatric T-ALL cells in vitro
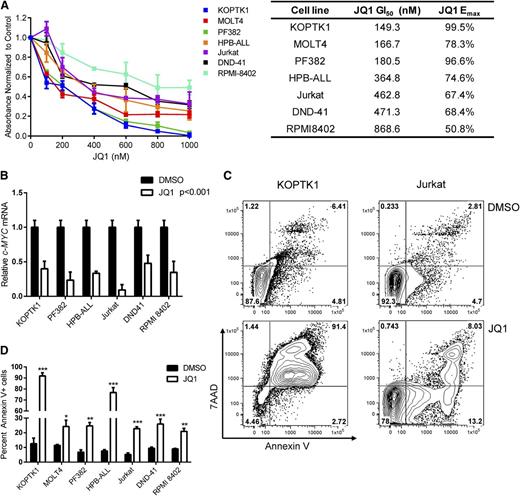
Our findings in the mouse model prompted us to test whether JQ1 has similar effects on human T-ALL growth. Treatment with JQ1 inhibited the growth of all 7 human T-ALL cell lines examined, resulting in an average GI50 of 380 nM (Figure 5A; range 149-868 nM). JQ1 treatment also significantly reduced c-MYC mRNA levels in 6 human T-ALL cell lines examined (Figure 5B; P < .001; average sixfold). Unlike GSIs, which primarily induce cell-cycle arrest,2,20 JQ1 treatment significantly increased the percentage of apoptotic cells when the human T-ALL cell lines were treated with JQ1 for 96 hours (Figure 5C-D; P < .05 and data not shown). These in vitro data suggest that C-MYC inhibitors may have therapeutic potential for treatment-resistant pediatric T-ALL patients.
JQ1 treatment impairs growth and induces apoptosis of human T-ALL cell lines. (A) JQ1 treatment inhibits human T-ALL cell line growth. Human T-ALL cell lines were cultured with vehicle or increasing concentrations of JQ1 (100-1000 nM) for 5 days and growth and metabolism assayed by MTS. The absorbance levels were normalized to the vehicle control and the GI50 of each cell line was calculated using Graph Pad Prism 5 software. The results are averages of 3 to 5 independent experiments and error bars represent SEM. (B) JQ1 treatment reduces C-MYC expression in human T-ALL lines. Human T-ALL lines were cultured with DMSO or JQ1 (1 μM) for 24 hours and C-MYC expression levels quantified by quantitative PCR. C-MYC expression was normalized to β-ACTIN and calculated using the ΔΔCT method. The results are averages of 3 independent experiments and error bars represent SEM. ***P < .001 for all JQ1-treated cell lines. (C-D) JQ1 induces apoptosis of human T-ALL lines. (C) KOPTK1 and Jurkat cell lines were treated with DMSO or JQ1 (1 μM) for 96 hours and the apoptotic cells detected by Annexin V and 7AAD staining followed by flow cytometry. A representation fluorescence-activated cell sorter plot is shown. (D) Collated data of Annexin V–positive cells from 7 human T-ALL cell lines treated with vehicle or JQ1 (1 μM) for 96 hours. The results are averages of 3 to 5 independent experiments and error bars represent SEM. *P < .05, **P < .01, ***P < .001.
JQ1 treatment impairs growth and induces apoptosis of human T-ALL cell lines. (A) JQ1 treatment inhibits human T-ALL cell line growth. Human T-ALL cell lines were cultured with vehicle or increasing concentrations of JQ1 (100-1000 nM) for 5 days and growth and metabolism assayed by MTS. The absorbance levels were normalized to the vehicle control and the GI50 of each cell line was calculated using Graph Pad Prism 5 software. The results are averages of 3 to 5 independent experiments and error bars represent SEM. (B) JQ1 treatment reduces C-MYC expression in human T-ALL lines. Human T-ALL lines were cultured with DMSO or JQ1 (1 μM) for 24 hours and C-MYC expression levels quantified by quantitative PCR. C-MYC expression was normalized to β-ACTIN and calculated using the ΔΔCT method. The results are averages of 3 independent experiments and error bars represent SEM. ***P < .001 for all JQ1-treated cell lines. (C-D) JQ1 induces apoptosis of human T-ALL lines. (C) KOPTK1 and Jurkat cell lines were treated with DMSO or JQ1 (1 μM) for 96 hours and the apoptotic cells detected by Annexin V and 7AAD staining followed by flow cytometry. A representation fluorescence-activated cell sorter plot is shown. (D) Collated data of Annexin V–positive cells from 7 human T-ALL cell lines treated with vehicle or JQ1 (1 μM) for 96 hours. The results are averages of 3 to 5 independent experiments and error bars represent SEM. *P < .05, **P < .01, ***P < .001.
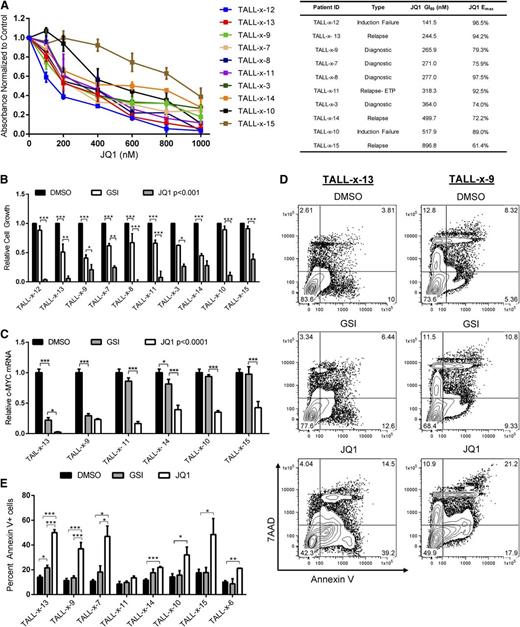
To determine whether primary pediatric T-ALL cells were sensitive to JQ1 treatment, we expanded 4 diagnostic, 4 relapsed, and 2 IF pediatric T-ALL samples in NOD-scid IL2rγ−/− (NSG) mice and adapted the cells to short-term culture. We then treated the T-ALL cells with DMSO or increasing doses of JQ1 and measured growth and metabolism using an MTS assay. JQ1 treatment inhibited the growth of the 10 pediatric T-ALL samples examined, including 2 IF samples, 4 relapsed pediatric T-ALL samples, and 1 relapsed early T-cell progenitor T-ALL sample, a particularly aggressive T-ALL subtype (Figure 6A; average GI50 of 380 nM, range 141-896 nM). All 10 patients expressed high levels of intracellular NOTCH1 and patients TALL-x-9 and TALL-x-13 harbored heterozygous deletions in the PTEN locus (supplemental Table 1), a genetic alteration associated with relapse.42 We then compared the effect of JQ1 or GSI treatment on the growth and survival of these pediatric T-ALL samples. GSI treatment arrested growth in 7 of the 10 samples examined, including the 2 patient samples with PTEN mutations (Figure 6B). However, treatment with JQ1 suppressed leukemic growth in all 10 samples examined, albeit to varying degrees (Figure 6A-B). A total of 8 of the 10 samples examined exhibited greater growth suppression with JQ1 than with GSI (Figure 6B). C-MYC mRNA levels were significantly reduced in all the JQ1-treated and in some of the GSI-treated cultures tested (Figure 6C). Consistent with the idea that JQ1 inhibits C-MYC, patient-sample sensitivity to JQ1 correlated with the fold reduction in C-MYC mRNA levels (Figure 6A,C; Spearman correlation 0.94; P = .0167). We also observed an increase in apoptotic cells when some of the treatment-resistant samples were treated in vitro for 96 hours with JQ1 (Figure 6D-E). Interestingly, 2 IF samples and 1 relapsed patient sample (TALL-x-12, TALL-x-10, and TALL-x-15) appeared resistant to GSI treatment yet remained sensitive to JQ1 (Figure 6B). Consistent with the effects on growth, C-MYC mRNA levels in the 2 GSI-resistant patient samples examined were significantly reduced with JQ1, but not GSI, treatment (Figure 6C; P < .0001). Collectively, these in vitro studies reveal the therapeutic potential of JQ1 for these refractory pediatric leukemia patients and in particular for relapsed/IF patients who fail to respond to GSIs or potentially other NOTCH inhibitors.
JQ1 treatment inhibits the growth of pediatric T-ALL cells in vitro. (A-B) Treatment of pediatric T-ALL cells are sensitive to GSI or JQ1 treatment. (A) Diagnostic, relapsed, or IF pediatric T-ALL cells were cultured with vehicle, GSI (1 μM), or increasing concentrations of JQ1 (100-1000 nM) for 5 days and growth and metabolism assayed by MTS. The absorbance levels were normalized to the vehicle control and the GI50 of each cell line was calculated using Graph Pad Prism 5 software. The results are averages of 3 to 5 independent experiments and error bars represent SEM. (B) Absorbance values determined by MTS assay are plotted for each patient sample treated with DMSO, GSI (DBZ, 1 μM), or JQ1 (1 μM). The results are averages of 3 to 5 independent experiments and error bars represent SEM. P < .0001 for all patient samples treated with JQ1; *P < .05, **P < .01, ***P < .001. (C) JQ1 treatment reduces C-MYC expression in diagnostic, relapsed, or IF patient T-ALL samples. Primary leukemic cells from patients were cultured with DMSO, GSI (DBZ, 1 μM), or JQ1 (1 μM) for 24 hours and C-MYC expression levels were measured using quantitative real-time PCR. C-MYC expression was normalized to β-ACTIN and calculated using the ΔΔCT method. The results are averages of 3 independent experiments and error bars represent SEM. P < .001 for all samples treated with JQ1; *P < .05, ***P < .001. (D-E) JQ1 treatment of some diagnostic and relapsed/IF T-ALL samples results in apoptosis. (D) Patient samples TALL-x-13 (relapse) and TALL-x-9 (diagnostic) were treated with DMSO, GSI (DBZ, 1 μM), or JQ1 (1 μM) for 96 hours and apoptotic cells detected by staining with Annexin V and 7AAD followed by flow cytometry. A representative fluorescence-activated cell sorter plot is shown. (E) Collated data of Annexin V–positive cells from relapsed/IF patients treated with DMSO, GSI (DBZ, 1 μM), or JQ1 (1 μM) for 96 hours. The results are averages of 3 independent experiments and error bars represent SEM. *P < .05, ***P < .001.
JQ1 treatment inhibits the growth of pediatric T-ALL cells in vitro. (A-B) Treatment of pediatric T-ALL cells are sensitive to GSI or JQ1 treatment. (A) Diagnostic, relapsed, or IF pediatric T-ALL cells were cultured with vehicle, GSI (1 μM), or increasing concentrations of JQ1 (100-1000 nM) for 5 days and growth and metabolism assayed by MTS. The absorbance levels were normalized to the vehicle control and the GI50 of each cell line was calculated using Graph Pad Prism 5 software. The results are averages of 3 to 5 independent experiments and error bars represent SEM. (B) Absorbance values determined by MTS assay are plotted for each patient sample treated with DMSO, GSI (DBZ, 1 μM), or JQ1 (1 μM). The results are averages of 3 to 5 independent experiments and error bars represent SEM. P < .0001 for all patient samples treated with JQ1; *P < .05, **P < .01, ***P < .001. (C) JQ1 treatment reduces C-MYC expression in diagnostic, relapsed, or IF patient T-ALL samples. Primary leukemic cells from patients were cultured with DMSO, GSI (DBZ, 1 μM), or JQ1 (1 μM) for 24 hours and C-MYC expression levels were measured using quantitative real-time PCR. C-MYC expression was normalized to β-ACTIN and calculated using the ΔΔCT method. The results are averages of 3 independent experiments and error bars represent SEM. P < .001 for all samples treated with JQ1; *P < .05, ***P < .001. (D-E) JQ1 treatment of some diagnostic and relapsed/IF T-ALL samples results in apoptosis. (D) Patient samples TALL-x-13 (relapse) and TALL-x-9 (diagnostic) were treated with DMSO, GSI (DBZ, 1 μM), or JQ1 (1 μM) for 96 hours and apoptotic cells detected by staining with Annexin V and 7AAD followed by flow cytometry. A representative fluorescence-activated cell sorter plot is shown. (E) Collated data of Annexin V–positive cells from relapsed/IF patients treated with DMSO, GSI (DBZ, 1 μM), or JQ1 (1 μM) for 96 hours. The results are averages of 3 independent experiments and error bars represent SEM. *P < .05, ***P < .001.
Discussion
Using a mouse T-ALL model that recapitulates several features of the cortical human T-ALL subtype, we found that c-Myc has an indispensable role in LIC maintenance and self-renewal. We demonstrate that partial and transient c-Myc suppression using shRNAs or the Brd4 inhibitor JQ1 interferes with leukemia initiation by targeting the LIC population. Mice transplanted with c-Myc shRNA-transduced leukemic cells that went on to develop disease no longer exhibited evidence of c-Myc knockdown, thereby indirectly supporting the importance of c-Myc in disease initiation. Collectively, these results reveal a crucial role for c-Myc in mouse LIC maintenance.
c-Myc is required for the emergence of the hematopoietic stem cell from a state of quiescence and has been shown to mediate the expansion of the DN3 progenitor in the thymus.29,43 Recent studies have revealed that under conditions of blocked thymic entry, normal thymic progenitors exhibit self-renewal capabilities.44,45 We find the leukemic DN3 thymic progenitors enriched in disease potential13 and hypothesize that sustained activation of the Notch1-c-Myc pathway in this progenitor cell type results in extensive proliferative capacity while retaining the ability to self-renew and differentiate into double-positive leukemic blasts.
Exogenous c-Myc expression has been shown to rescue human and mouse T-ALL cell lines from the effects of NOTCH1 inhibition in vitro.20,22 However, in vivo studies suggest a prominent role for Notch1 over c-Myc in leukemia maintenance in vivo.46 Consistent with this report, we have demonstrated that Notch1 inhibition prolongs the survival of leukemic mice and in some cases eliminates the LIC population.13,14 GSI and JQ1 treatment resulted in similar overall reductions in mouse LIC frequency30 (Figure 4C), leading us to speculate that the Notch1 effects on LIC activity may be mediated by c-Myc. Yet, Notch-independent mechanisms of c-Myc activation and stabilization clearly contribute to T-ALL pathogenesis. In addition to C-MYC amplification, mutations in the F-box protein FBW7 occur in 20% of T-ALL patients.2,4,6 These heterozygous mutations cluster in the WD40 substrate recognition domain and in mouse models appear to affect the stability of Notch1 and c-Myc.2,4,6,47 Using a mouse model of the Fbw7R468C substitution common in human T-ALL, a similar dependence on c-Myc was observed.47 Collectively, these data as well as reported effects of microRNA deregulation on C-MYC stabilization in T-ALL48,49 argue for direct targeting of C-MYC in this disease.
Although we demonstrate that JQ1 treatment reduces expression, it does not abrogate c-Myc expression in mouse T-ALL cells in vivo (Figure 4D), indicating that more potent JQ1 derivatives or other c-MYC inhibitors may be required to further suppress c-Myc levels and achieve complete remissions. Long-term c-Myc inhibition in vivo may result in on-target toxicities, perhaps due to effects on hematopoietic stem cell emergence from the niche,43 but because transient c-Myc inhibition was sufficient in some cases to cure leukemia, such effects may prove manageable.
An additional crucial finding illustrated here is that JQ1 inhibited the growth of treatment-resistant pediatric T-ALL cells, suggesting that JQ1 or other C-MYC inhibitors may have therapeutic benefit for these patients. In some cases, apoptotic cells were detected in the JQ1-treated leukemic cultures, raising the possibility that long-term C-MYC inhibition may result in apoptosis in vivo. Some (3 out of 10) of the relapsed/IF samples examined in this study appear resistant to GSI treatment in vitro, though these patients’ leukemic cells remained sensitive to JQ1 treatment (Figure 6B). Although the mechanism is unclear, these data suggest that the GSI-resistant relapsed samples become less dependent on NOTCH1 and more dependent on c-MYC for their growth and survival.
In the relapsed/IF patient samples examined, JQ1 sensitivity correlates with reductions in C-MYC mRNA levels, supporting the idea that JQ1 interferes with leukemic growth via effects on c-MYC. These data might reflect the fact that JQ1 interferes with C-MYC levels and activity better than treatment with GSIs. Alternatively, JQ1 treatment may result in apoptosis of the relapsed/IF T-ALL cells by inhibiting the expression of additional BRD4- or BRD2-dependent genes. A third possibility, and one consistent with C-MYC as an amplifier of gene expression,50,51 postulates that C-MYC inhibition also dampens the NOTCH1 signature and/or other T-cell survival programs, resulting in apoptosis of the human T-ALL cells.
Because LICS are thought to mediate relapse and the relapsed/IF pediatric T-ALL samples examined appear sensitive to JQ1 treatment in vitro, these findings suggest that c-MYC may also be required for the maintenance of the human LIC. Based on the widespread deregulation of c-MYC in human malignancy, these findings raise the possibility that a requirement for MYC may not be limited to T-ALL–initiating cells but may extend to other tumor-initiating populations.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health (National Cancer Institute) grants CA096899 (M.A.K.), CA034196 (L.D.S.), and (National Institute of Allergy and Infectious Diseases) grant AI04669 (D.L.G., L.D.S., and M.A.B.). J.R. was supported by a National Institutes of Health (National Cancer Institute) T32 training grant CA130807 to the Department of Cancer Biology at the University of Massachusetts Medical School and the American Cancer Society Postdoctoral Fellowship 125087-PF-13-247-01-LIB.
Authorship
Contribution: J.T. and J.E.R. performed the experiments and made the figures; A.G. and A.T.L. provided samples and institutional review board consent for patient data in Figure 6; J.Q. and J.E.B. provided JQ1 and DBZ used in Figures 2-6 and supplemental Figure 4; N.S., S.S., and M.H. provided pediatric T-ALL samples; J.E.R., L.D.S., M.A.B., and D.L.G. established the primary T-ALL engraftments in NSG mice; and M.A.K. designed the research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: D.L.G. and M.A.B. are consultants for The Jackson Laboratory. Drug-like derivatives of JQ1 have been licensed by Dana-Farber Cancer Institute to Tensha Therapeutics for clinical development; J.E.B. and Dana-Farber Cancer Institute have received equity in Tensha associated with this license. The remaining authors declare no competing financial interests.
Correspondence: Michelle A. Kelliher, Department of Cancer Biology, University of Massachusetts Medical School, Worcester, MA 01605; e-mail: Michelle.Kelliher@umassmed.edu.
References
Author notes
J.E.R. and T.T. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal