Key Points
AITL is characterized by high frequencies of overlapping mutations in epigenetic modifiers, including TET2, IDH2, and DNMT3A.
Targetable mutations are present in a subset of cases.
Abstract
The genetics of angioimmunoblastic T-cell lymphoma (AITL) are very poorly understood. We defined the mutational landscape of AITL across 219 genes in 85 cases from the United States and Europe. We identified ≥2 mutations in 34 genes, nearly all of which were not previously implicated in AITL. These included loss-of-function mutations in TP53 (n = 4), ETV6 (n = 3), CCND3 (n = 2), and EP300 (n = 5), as well as gain-of-function mutations in JAK2 (n = 2) and STAT3 (n = 4). TET2 was mutated in 65 (76%) AITLs, including 43 that harbored 2 or 3 TET2 mutations. DNMT3A mutations occurred in 28 (33%) AITLs; 100% of these also harbored TET2 mutations (P < .0001). Seventeen AITLs harbored IDH2 R172 substitutions, including 15 with TET2 mutations. In summary, AITL is characterized by high frequencies of overlapping mutations in epigenetic modifiers and targetable mutations in a subset of cases.
Introduction
Angioimmunoblastic T-cell lymphoma (AITL) accounts for 15% to 20% of peripheral T-cell lymphomas and 1% to 2% of all non-Hodgkin lymphomas. Most patients with AITL are diagnosed with advanced stage disease and present with a wide range of clinical manifestations, including generalized lymphadenopathy, hepatosplenomegaly, B-symptoms, and autoimmune phenomena.1,2 Histologic examination of AITL characteristically demonstrates a polymorphic infiltrate of malignant cells with an abundance of nonmalignant bystander cells and marked proliferation of high endothelial venules. AITL has a dismal prognosis with a median survival <3 years.2-4 Thus, there is an urgent need to define new therapeutic targets in this disease.
AITLs can be distinguished from other T-cell lymphomas based on gene expression pattern, suggesting that AITLs have a unique biology.5 Prior studies to identify recurrent mutations in AITL have focused on 1 or 2 genes,6-8 at least in part because of the low tumor cell fraction characteristic of AITL, the relative rarity of this disease, and the difficulties in analyzing formalin-fixed paraffin-embedded tissue. To overcome these issues, we performed targeted next-generation sequencing of 85 AITLs for mutations in the coding regions of 219 genes (supplemental Table 1, available on the Blood Web site) known to be recurrently altered in hematologic malignancies.
Study design
Patient enrollment
Patients with AITL and lymphoma tissue available at the time of diagnosis were identified by searching databases of stored formalin-fixed paraffin-embedded specimens at the Dana-Farber/Brigham and Women’s Hospital, Massachusetts General Hospital, Memorial Sloan-Kettering Cancer Center, and Università di Torino. The specimens were collected between 1997 and 2012, with 87% between 2003 and 2012. Medical records were reviewed for demographic and clinical data. All studies were approved by the Dana-Farber/Harvard Cancer Center Institutional Review Board. Informed consent was obtained in accordance with the Declaration of Helsinki. The diagnosis was confirmed by histology and immunophenotyping by hematopathologists (S.J.R., J.T.-F., A.L., and G.I.) at the participating centers, with some cases harboring low frequencies of Epstein-Barr virus–infected cells (supplemental Figure 1; supplemental Table 2). A single additional case with histology that overlapped AITL and peripheral T-cell lymphoma was also sequenced but not included in the AITL cohort.
Next-generation sequencing and mutant allele validation
Targeted exon capture and next-generation sequencing of all coding exons of 219 genes (supplemental Table 1)9-17 were performed as previously described.18,19 Mutations identified as germ-line variants within dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) were removed, and reads from non-dbSNP variants were manually reviewed using the IGV browser. To validate the next-generation sequencing calls, a subset of mutations were confirmed by either Sanger sequencing (if allele fraction ≥20%, n = 69) or by mass spectrometry–based genotyping (if allele fraction <20%, n = 29) and compared with paired germ-line material, when available.
Statistical analysis
Associations between categorical variables were assessed by Fisher’s exact test, and the Wilcoxon rank-sum test was performed to test for differences between continuous variables for 2-group comparisons. Overall survival (OS) was calculated from the date of diagnosis to the date of death, censored at the time last known alive using the Kaplan-Meier method, and compared with the log-rank test. P values are 2-sided and considered significant at the .05 level.
Results and discussion
A total of 85 AITLs underwent targeted sequencing. Median age was 69 years (range, 30-89 years), and 51 of 52 cases with evaluable staging information had advanced disease. Median OS was 18 months (95% confidence interval, 12-31); patients ≥70 years had significantly worse OS (median, 12 vs 59 months; P < .0001) and those with elevated lactate dehydrogenase (LDH) had a trend to a shorter OS (median, 15 vs 27 months; P = .050; supplemental Figure 2). Of cases with available material for review, 94.0% had histology consistent with either pattern II or pattern III, 100% expressed CXCL13 and PD-1, and 98.1% expressed CD4, consistent with a follicular helper T-cell phenotype (supplemental Table 2). Of 72 cases with available Epstein-Barr virus-encoded small RNAs (EBER) staining, 48 had detectable EBER-positive cells. The median number of mutations detected was identical in cases with EBER-positive cells and those that lacked EBER-positive cells (3 vs 3; P = .69 by 2-sided Student t-test).
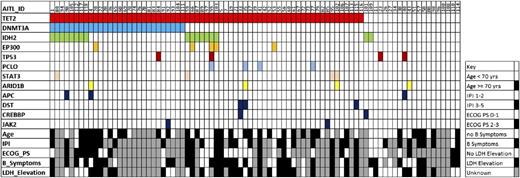
We identified 286 non-dbSNP variants (199 single nucleotide variants and 87 insertion/deletions [InDels]) within coding regions in 80 genes, including 34 genes mutated in ≥2 cases (Figure 1; supplemental Tables 3 and 4). Genes previously not known to be altered in AITL with ≥2 loss-of-function mutations (eg, premature stop, frameshift) included ARID1A, CCND3, EP300, FAS, CD58, PRDM1, TNFSF9, and ETV6. Known gain-of-function mutations in JAK2 (V617F and G571S)20 were identified in 2 cases. Although we cannot rule out the possibility that these mutations were present in cells other than the AITL tumor cells, neither case had histologic or laboratory evidence of a concurrent hematologic disorder. Three of 4 mutations in STAT3 occurred at codons 614 and 661, which are known to confer gain-of-function in chronic lymphoproliferative disorders of natural killer cells and T-cell large granular lymphocyte leukemia.21,22 TP53 mutations were present in only 4 cases (supplemental Table 3), raising the possibility that inhibitors of MDM2 could be broadly active in AITL.23
Distribution of mutations in 85 cases of AITL. Common mutations and demographic and clinical factors across the cohort are shown. Each column represents a single patient. IPI, International Prognostic Index; ECOG PS, European Cooperative Oncology Group Performance Status.
Distribution of mutations in 85 cases of AITL. Common mutations and demographic and clinical factors across the cohort are shown. Each column represents a single patient. IPI, International Prognostic Index; ECOG PS, European Cooperative Oncology Group Performance Status.
TET2 mutations were present in 65 (76%) AITLs, a higher frequency than previously reported.7,8 Of note, 43 of the 65 harbored either 2 or 3 TET2 mutations, suggesting a strong selective pressure for loss of TET2 in this disease. Of the 115 total TET2 mutations, 95 were either frameshift, splice site, or premature stop codon mutations (supplemental Tables 3 and 4). The presence of the TET2 mutation was associated with elevated LDH at diagnosis (P = .038) and a trend toward older median age at diagnosis (72 vs 63 years, P = .060; Table 1).
Patient characteristics based on TET2 and DNM3TA mutational status
| Variable . | Overall N (%) . | TET2 . | DNMT3A . | ||||
|---|---|---|---|---|---|---|---|
| Unmutated N (%) . | Mutated N (%) . | P value* . | Unmutated N (%) . | Mutated N (%) . | P value* . | ||
| N | 85 | 20 | 65 | 57 | 28 | ||
| Gender | |||||||
| Female | 41 (48) | 9 (45) | 32 (49) | .80 | 31 (54) | 10 (36) | .11 |
| Male | 43 (51) | 11 (55) | 32 (49) | 25 (44) | 18 (64) | ||
| Unknown | 1 (1) | 0 (0) | 1 (1) | 1 (2) | 0 (0) | ||
| Median age at Dx (range) | 69 (30, 89) | 63 (30, 85) | 72 (48, 89) | .060 | 67 (30, 88) | 73 (58, 89) | .037 |
| Stage | |||||||
| II | 1 (1) | 0 (0) | 1 (1) | >.99 | 1 (2) | 0 (0) | .66 |
| III-IV | 28 (33) | 8 (40) | 20 (31) | 19 (33) | 9 (32) | ||
| IV | 23 (27) | 7 (35) | 16 (25) | 18 (32) | 5 (18) | ||
| Unknown | 33 (39) | 5 (25) | 28 (43) | 19 (33) | 14 (50) | ||
| B-Symptoms | |||||||
| Yes | 33 (39) | 7 (35) | 26 (40) | .20 | 23 (40) | 10 (36) | .74 |
| No | 18 (21) | 7 (35) | 11 (17) | 14 (25) | 4 (14) | ||
| Unknown | 34 (40) | 6 (30) | 28 (43) | 20 (35) | 14 (50) | ||
| Elevated LDH | |||||||
| Yes | 31 (36) | 6 (30) | 25 (38) | .038 | 23 (40) | 8 (29) | >.99 |
| No | 21 (25) | 10 (50) | 11 (17) | 15 (26) | 6 (21) | ||
| Unknown | 33 (39) | 4 (20) | 29 (45) | 19 (33) | 14 (50) | ||
| BM involvement | |||||||
| Yes | 10 (12) | 3 (15) | 7 (11) | .18 | 8 (14) | 2 (7) | .45 |
| No | 26 (31) | 9 (45) | 17 (26) | 19 (33) | 7 (25) | ||
| Unknown | 49 (58) | 8 (40) | 41 (63) | 30 (53) | 19 (68) | ||
| ECOG PS | |||||||
| 0-1 | 22 (26) | 6 (30) | 16 (25) | .47 | 17 (30) | 5 (18) | .49 |
| 2-3 | 19 (22) | 3 (15) | 16 (25) | 12 (21) | 7 (25) | ||
| Unknown | 44 (52) | 11 (55) | 33 (51) | 28 (49) | 16 (57) | ||
| Extranodal disease | |||||||
| Yes | 19 (22) | 5 (25) | 14 (22) | .75 | 16 (28) | 3 (11) | .33 |
| No | 30 (35) | 10 (50) | 20 (31) | 21 (37) | 9 (32) | ||
| Unknown | 36 (42) | 5 (25) | 31 (48) | 20 (35) | 16 (57) | ||
| IPI score | |||||||
| 1-2 | 11 (13) | 4 (20) | 7 (11) | .42 | 10 (18) | 1 (4) | .24 |
| 3-5 | 33 (39) | 7 (35) | 26 (40) | 23 (40) | 10 (36) | ||
| Unknown | 41 (48) | 9 (45) | 32 (49) | 24 (42) | 17 (61) | ||
| PIT score | |||||||
| 0-1 | 11 (13) | 5 (25) | 6 (9) | .12 | 10 (18) | 1 (3) | .23 |
| 2-4 | 31 (36) | 6 (30) | 25 (38) | 21 (37) | 10 (36) | ||
| Unknown | 43 (51) | 9 (45) | 34 (52) | 26 (46) | 17 (61) | ||
| Median OS (mo) | 18 | 18 | 25 | .99 | 25 | 17 | .81 |
| 95% confidence interval | (12,51) | (2, 104) | (12, 31) | (11, 51) | (4, NR) | ||
| Variable . | Overall N (%) . | TET2 . | DNMT3A . | ||||
|---|---|---|---|---|---|---|---|
| Unmutated N (%) . | Mutated N (%) . | P value* . | Unmutated N (%) . | Mutated N (%) . | P value* . | ||
| N | 85 | 20 | 65 | 57 | 28 | ||
| Gender | |||||||
| Female | 41 (48) | 9 (45) | 32 (49) | .80 | 31 (54) | 10 (36) | .11 |
| Male | 43 (51) | 11 (55) | 32 (49) | 25 (44) | 18 (64) | ||
| Unknown | 1 (1) | 0 (0) | 1 (1) | 1 (2) | 0 (0) | ||
| Median age at Dx (range) | 69 (30, 89) | 63 (30, 85) | 72 (48, 89) | .060 | 67 (30, 88) | 73 (58, 89) | .037 |
| Stage | |||||||
| II | 1 (1) | 0 (0) | 1 (1) | >.99 | 1 (2) | 0 (0) | .66 |
| III-IV | 28 (33) | 8 (40) | 20 (31) | 19 (33) | 9 (32) | ||
| IV | 23 (27) | 7 (35) | 16 (25) | 18 (32) | 5 (18) | ||
| Unknown | 33 (39) | 5 (25) | 28 (43) | 19 (33) | 14 (50) | ||
| B-Symptoms | |||||||
| Yes | 33 (39) | 7 (35) | 26 (40) | .20 | 23 (40) | 10 (36) | .74 |
| No | 18 (21) | 7 (35) | 11 (17) | 14 (25) | 4 (14) | ||
| Unknown | 34 (40) | 6 (30) | 28 (43) | 20 (35) | 14 (50) | ||
| Elevated LDH | |||||||
| Yes | 31 (36) | 6 (30) | 25 (38) | .038 | 23 (40) | 8 (29) | >.99 |
| No | 21 (25) | 10 (50) | 11 (17) | 15 (26) | 6 (21) | ||
| Unknown | 33 (39) | 4 (20) | 29 (45) | 19 (33) | 14 (50) | ||
| BM involvement | |||||||
| Yes | 10 (12) | 3 (15) | 7 (11) | .18 | 8 (14) | 2 (7) | .45 |
| No | 26 (31) | 9 (45) | 17 (26) | 19 (33) | 7 (25) | ||
| Unknown | 49 (58) | 8 (40) | 41 (63) | 30 (53) | 19 (68) | ||
| ECOG PS | |||||||
| 0-1 | 22 (26) | 6 (30) | 16 (25) | .47 | 17 (30) | 5 (18) | .49 |
| 2-3 | 19 (22) | 3 (15) | 16 (25) | 12 (21) | 7 (25) | ||
| Unknown | 44 (52) | 11 (55) | 33 (51) | 28 (49) | 16 (57) | ||
| Extranodal disease | |||||||
| Yes | 19 (22) | 5 (25) | 14 (22) | .75 | 16 (28) | 3 (11) | .33 |
| No | 30 (35) | 10 (50) | 20 (31) | 21 (37) | 9 (32) | ||
| Unknown | 36 (42) | 5 (25) | 31 (48) | 20 (35) | 16 (57) | ||
| IPI score | |||||||
| 1-2 | 11 (13) | 4 (20) | 7 (11) | .42 | 10 (18) | 1 (4) | .24 |
| 3-5 | 33 (39) | 7 (35) | 26 (40) | 23 (40) | 10 (36) | ||
| Unknown | 41 (48) | 9 (45) | 32 (49) | 24 (42) | 17 (61) | ||
| PIT score | |||||||
| 0-1 | 11 (13) | 5 (25) | 6 (9) | .12 | 10 (18) | 1 (3) | .23 |
| 2-4 | 31 (36) | 6 (30) | 25 (38) | 21 (37) | 10 (36) | ||
| Unknown | 43 (51) | 9 (45) | 34 (52) | 26 (46) | 17 (61) | ||
| Median OS (mo) | 18 | 18 | 25 | .99 | 25 | 17 | .81 |
| 95% confidence interval | (12,51) | (2, 104) | (12, 31) | (11, 51) | (4, NR) | ||
Test includes only known categories. BM, bone marrow; Dx, diagnosis; IPI, International Prognostic Index; NR, not reached; PIT, Prognostic Index for T-cell lymphoma; PS, performance status.
DNMT3A was the second most frequently mutated gene. Strikingly, all 28 (33%) patients with DNMT3A mutations also harbored TET2 mutations (Figure 1; P < .0001). Univariate analysis (Table 1) revealed an association between the DNMT3A mutation and older median age (73 vs 67 years, P = .037). Neither the TET2 nor the DNMT3A mutation significantly affected overall survival (supplemental Figure 3). Seventeen (20%) of the 85 AITLs harbored IDH2 R172 substitutions and 15 co-occurred with TET2 mutations (Figure 1). Although not statistically significant (P = .34), the co-occurrence of IDH2 and TET2 contrasts sharply with the mutually exclusive nature of TET2 and IDH1/2 alterations in acute myeloid leukemia.6,19
Recent reports have suggested that TET2 and DNMT3A mutations among patients with T-cell lymphomas can either exist within CD34+ progenitors or be acquired at later stages of T-cell ontogeny.8,24 In 2 cases, granulocyte colony-stimulating factor–stimulated peripheral blood stem cell (PBSC) products for autologous stem cell transplantation (ASCT) were collected from patients after achieving complete remission. In AITL 21, the only mutation identified was a TET2 InDel present in nearly 50% of reads (supplemental Table 4). We generated myeloid colonies from the PBSC product in the methycellulose culture. In addition, we sorted multiple populations from the PBSC product, including lineage negative (lin−)CD34+CD38−, lin−CD34−CD38+, lin−CD34+CD38+, and lin−CD4+CD10+ cells (similar to the patient’s AITL cells) (supplemental Figure 4). The TET2 InDel was present at ∼50% within the lin−CD34−CD38+ population and absent from the other populations, indicating that cells immunophenotypically distinct from either hematopoietic stem cells or AITL harbored the TET2 mutation and were transplanted back into the patient. Of note, the patient relapsed 11 months after ASCT.
We used the same approach in a separate case with histology that overlapped AITL and peripheral T-cell lymphoma, not otherwise specified. Sequencing of the lymphoma identified DNMT3A R38C, FAS T270I, STAT3 K658N, TET2 R1261H, TP53 S261_splice, and ZFHX3 A438V mutations, but mass spectrometry–based genotyping of myeloid colonies and lin−CD34+CD38− and lin−CD34−CD38+ cells did not detect any of the mutations in any of the samples. This suggests that, in contrast with the TET2 InDel in AITL 21, all of the mutations were acquired within lineage-committed progenitors.
This study defines the first genetic landscape of AITL across >200 genes known to be recurrently mutated in hematologic neoplasms. The complement of genes frequently mutated in AITL (eg, TET2, DNMT3A, and IDH2) resembles myeloid diseases more than other lymphomas, which could help explain the poor outcomes after treatment with regimens developed against B-cell lymphomas. Further studies are needed to determine whether the genetic alterations observed in AITL directly confer malignant phenotypes, whether these phenotypes represent appropriate therapeutic targets, and the extent to which individual mutations are present in non-T cells.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Paul van Hummelen and Aaron Thorner for assistance with sequencing.
This work was supported by the Conquer Cancer Foundation (A.A.L.), Lauri Strauss Leukemia Foundation (A.A.L.), Leukemia and Lymphoma Society (A.A.L.), AIRC 5x1000 (G.O.), Cycle for Survival (S.M.H.), the Stellato Fund (D.M.W.), American Cancer Society (D.M.W.), and a Stand Up To Cancer Innovative Research Grant (D.M.W.).
Authorship
Contribution: O.O. and D.M.W. designed the research and wrote the manuscript; O.W., A.A.L., D.T., M.A.L., N.K., S.K., D.v.B., and S.B., designed and performed the research; J.H.S., J.T.-F., E.H., A.L., D.D., S.J.R., P.P.P., E.J., S.A.P., N.L.H., S.F., G.I., and S.M.H. designed the research and provided vital reagents; and K.S. analyzed data and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for O.W. is Department of Internal Medicine III, Ludwig-Maximilians-University, and Haematologicum, Helmholtz-Center Munich, Munich, Germany.
The current affiliation for J.H.S. is Department of Medicine, University of Arizona Cancer Center, Tucson, AZ.
Correspondence: David Weinstock, Dana-Farber Cancer Institute, 450 Brookline Ave, Dana 510B, Boston, MA 02215; e-mail: dweinstock@partners.org.
References
Author notes
O.O. and O.W. contributed equally to this work.