Key Points
Knockdown of the sialyl-transferase, ST3GAL6, in MM inhibits in vivo homing and prolongs survival in xenograft mice.
In MM patients, high expression of ST3GAL6 is associated with inferior overall survival.
Abstract
Glycosylation is a stepwise procedure of covalent attachment of oligosaccharide chains to proteins or lipids, and alterations in this process, especially increased sialylation, have been associated with malignant transformation and metastasis. The role of altered sialylation in multiple myeloma (MM) cell trafficking has not been previously investigated. In the present study we identified high expression of β-galactoside α-2,3-sialyltransferase, ST3GAL6, in MM cell lines and patients. This gene plays a key role in selectin ligand synthesis in humans through the generation of functional sialyl Lewis X. In MRC IX patients, high expression of this gene is associated with inferior overall survival. In this study we demonstrate that knockdown of ST3GAL6 results in a significant reduction in levels of α-2,3–linked sialic acid on the surface of MM cells with an associated significant reduction in adhesion to MM bone marrow stromal cells and fibronectin along with reduced transendothelial migration in vitro. In support of our in vitro findings, we demonstrate significantly reduced homing and engraftment of ST3GAL6 knockdown MM cells to the bone marrow niche in vivo, along with decreased tumor burden and prolonged survival. This study points to the importance of altered glycosylation, particularly sialylation, in MM cell adhesion and migration.
Introduction
Cell trafficking, glycosylation, and sialyltransferases in multiple myeloma
Multiple myeloma (MM) is characterized by the presence of multiple lesions, indicating continuous trafficking of tumor cells to distant bone marrow (BM) niches.1,2 The first step in cellular trafficking is the deceleration of blood cells on target endothelium to below the local blood flow rate, a process known as rolling.3 While rolling, leukocytes are activated by binding to selectins and by chemokines like CXCL12, leading to integrin activation, adhesion, and subsequent migration. Selectin-based adhesion is mediated by the binding of selectins on endothelial cells to sialofucosylated proteins or lipids on selectin ligands. P-selectin glycoprotein ligand 1 (PSGL-1) is highly expressed in MM and can bind to both E- and P-selectin, and the potential therapeutic benefit of selectin inhibition in MM has been demonstrated.4
Functional selectin ligands require posttranslational modification of scaffold proteins by glycosyltransferases and sulfotransferases (Figure 1A). The minimal recognition motif for all selectins is sialyl-Lewisx (sLex) and its isomer sialyl-Lewisa (sLea), which are tetrasaccharides that are synthesized by the combined action of α-1,3-fucosyltransferases, α-2,3-sialyltransferases, 1,4-galactosyltranferases, and N-acetyl-glucosaminyltransferases. Enhanced expression of sLex and sLea structures is frequently associated with cancer, and elevated levels of sialyltransferases have been linked to increased risk of metastatic disease in solid tumors.5
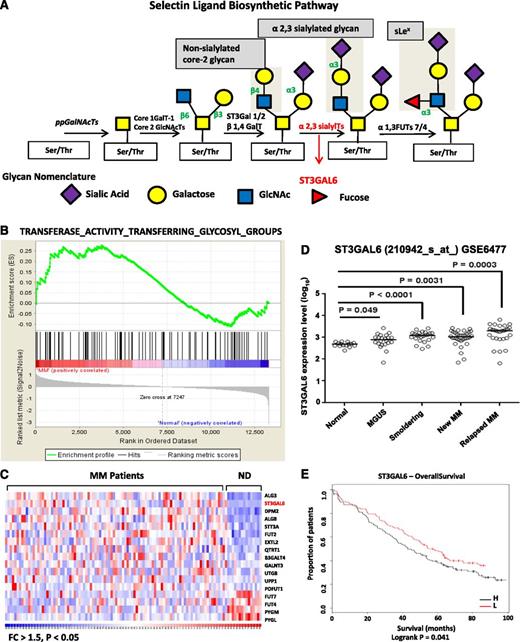
Glycosylation gene expression in MM. (A) Selectin ligand biosynthetic pathway. Sialyltransferases, including ST3GAL6, transfer sialic acid from the activated cytidine 5′-monophospho-N-acetylneuraminic acid to terminal positions on sialylated glycolipids (gangliosides) or to the N- or O-linked sugar chains of glycoproteins. ST3GAL6 contributes to the formation of selectin ligands and sialyl Lewis X, a carbohydrate important in cell-cell recognition. (B) GSEA software analyzed glycosylation-related genes in MM patients from the previously published data set GSE6477. Shown is a graphic enrichment of a publicly available MSigDB gene set (TRANSFERASE_ACTIVITY_TRANSFERRING_GLYCOSYL_GROUPS) in MM patients (left, red) compared with healthy controls (blue, right). False discovery rate (FDR) = 0.24 and normalized enrichment score (NES) = 1.14. (C) Heat map demonstrating significantly altered glycosylation gene expression pattern in MM patients vs normal donors (ND) with fold change (FC) >1.5 and P < .05. ST3GAL6 is highlighted in red as one of the most significantly enriched glycosylation genes in MM patients. (D) Analysis of ST3GAL6 expression in the GSE6477 data set showing significantly increasing expression of ST3GAL6 with disease progression. Comparison of ST3GAL6 expression in healthy donors (Normal) to that in patients with MGUS, smoldering MM, newly diagnosed MM and relapsed MM shows increasing expression levels of this gene (P = .049, < .0001, .0031, and .0003, respectively; see Table 1). (E) Kaplan-Meier survival proportions analysis of patient outcome data from the MRC IX trial demonstrating high levels of ST3GAL6 expression associated with inferior OS: low (L) = 48 months, high (H) = 36 months. Log-rank P = .041. N (H) = 129, N (L) = 130.
Glycosylation gene expression in MM. (A) Selectin ligand biosynthetic pathway. Sialyltransferases, including ST3GAL6, transfer sialic acid from the activated cytidine 5′-monophospho-N-acetylneuraminic acid to terminal positions on sialylated glycolipids (gangliosides) or to the N- or O-linked sugar chains of glycoproteins. ST3GAL6 contributes to the formation of selectin ligands and sialyl Lewis X, a carbohydrate important in cell-cell recognition. (B) GSEA software analyzed glycosylation-related genes in MM patients from the previously published data set GSE6477. Shown is a graphic enrichment of a publicly available MSigDB gene set (TRANSFERASE_ACTIVITY_TRANSFERRING_GLYCOSYL_GROUPS) in MM patients (left, red) compared with healthy controls (blue, right). False discovery rate (FDR) = 0.24 and normalized enrichment score (NES) = 1.14. (C) Heat map demonstrating significantly altered glycosylation gene expression pattern in MM patients vs normal donors (ND) with fold change (FC) >1.5 and P < .05. ST3GAL6 is highlighted in red as one of the most significantly enriched glycosylation genes in MM patients. (D) Analysis of ST3GAL6 expression in the GSE6477 data set showing significantly increasing expression of ST3GAL6 with disease progression. Comparison of ST3GAL6 expression in healthy donors (Normal) to that in patients with MGUS, smoldering MM, newly diagnosed MM and relapsed MM shows increasing expression levels of this gene (P = .049, < .0001, .0031, and .0003, respectively; see Table 1). (E) Kaplan-Meier survival proportions analysis of patient outcome data from the MRC IX trial demonstrating high levels of ST3GAL6 expression associated with inferior OS: low (L) = 48 months, high (H) = 36 months. Log-rank P = .041. N (H) = 129, N (L) = 130.
Using this background, we examine the altered expression of glycosylation-related genes in MM and demonstrate, using in vitro and in vivo model systems, that knockdown of ST3GAL6 attenuates MM cell adhesion and migration in vitro and reduces the ability of these cells to home to the bone marrow in vivo, resulting in a reduction in tumor burden and prolonged survival of xenograft mice.
This study highlights the important role of sLex-modifying enzymes in alterations of cell trafficking in MM.
Methods
Gene expression analysis
We performed gene set enrichment analysis (GSEA) to determine whether the glycosylation-related gene sets were enriched in MM patients (GSE64776 ). GSEA was performed following the developer’s protocol (http://www.broad.mit.edu/gsea/).7 Analysis of publically available gene expression data (GSE 6477) and data that were provided to us by collaborators (MRC IX trial data8 ) was carried out to examine the expression of ST3GAL6 in MM patients and healthy controls. When available, the association between high levels of expression of ST3GAL6 and patient outcomes were examined. Survival data were generated using Kaplan-Meier survival analysis and log-rank test (GraphPad Software, La Jolla, CA).
Cell lines and primary cells
The RPMI-8226 and MM1S cell lines were purchased from ATCC (Manassas, VA) and were authenticated by Molecular Diagnostics Laboratory Research Services at Dana-Farber Cancer Institute. Primary MM cells were obtained from BM aspirates of individual MM patients using CD138+ microbead selection (Miltenyi Biotech, Auburn, CA) as previously described.9 Bone marrow stromal cells (BMSCs) were obtained from healthy donors and MM patient fresh BM samples and were isolated as described.10 Human umbilical vein endothelial cells (HUVECs) were purchased from Lonza. All human MM cell lines and BMSCs were cultured as in previous studies.4,9 Green-fluorescent-protein and luciferase-positive (GFP-Luc+) MM1S cells were generated using lentivirus infection, as previously described.11 Calcein-AM cells were obtained from Invitrogen (Carlsbad, CA). For hypoxia experiments, cells were cultured in 1% O2 for 24 hours. Informed consent was obtained from all patients in accordance with the Declaration of Helsinki. These studies were approved by the Dana-Farber Cancer Institute Institutional Review Board and the Research Ethics Committee, Galway University Hospital.
DNA synthesis assay and cell cycle analysis
The proliferation rate of MM1S cells (scrambled control and shST3GAL6 cells) was measured by cell counting using trypan blue exclusion and DNA synthesis using [3H]-thymidine uptake (Perkin Elmer, Waltham, MA) as previously described.12 For cell cycle analysis, MM cells were stained with propidium iodide (PI; Sigma-Aldrich, St. Louis, MO) and cell cycle was determined using flow cytometry as previously described.12 Cell survival was measured using MTT assays.
Flow cytometry
Expression of α-2,3 sialic acid at the cell surface was analyzed by flow cytometry using biotinylated Maackia amurensis (MAA) lectin (Vector Labs, Burlingame, CA) conjugated to streptavadin APC (BD Biosciences, Franklin Lakes, NJ). Expression of HECA-452 was analyzed by flow cytometry using Cutaneous Lymphocyte Antigen (BD Biosciences).
shRNA-mediated ST3GAL6 gene silencing
To determine the role of ST3GAL6 in MM biology, we established ST3GAL6 knockout RPMI-8226 and MM1S-GFP-Luc cell lines using a lentiviral system as previously described.13 The efficiency of ST3GAL6 knockdown was assessed by real-time quantitative polymerase chain reaction (qRT-PCR) using the following primers—forward primer: TTG CCT CTC TGC TGA GGT TT; reverse primer: CCT CCA TTA CCA ACC ACC AC.14 The resultant stable ST3GAL6 knockdown (shST3GAL6) cell line was compared with the scrambled control cell line in all subsequent functional assays. The sense and antisense oligonucleotide sequence for construction of ST3GAL6 shRNA were as follows: clone no 10402; (NM_006100.2-1332s1c1, target sequence CCTTTGCACTACTATGGGAAT [clone A2], NM_006100.2-1110s1c1 target sequence CCAGCCTTAAACCTGATTTAT [clone A3]).
Adhesion assays
Assays for adhesion of MM cells to HUVECs and MM BMSCs were performed using 96-well plates. HUVECs or MM BMSCs (5 × 103 cells/well) were cultured to confluence overnight in 96-well plates to establish a monolayer. MM cells were serum-starved for 4 hours, prelabeled with calcein AM, added to the HUVECs or BMSCs, and allowed to adhere for 2 hours at 37°C. Nonadherent cells were aspirated off, HUVECs or BMSCs were washed in phosphate-buffered saline, and fluorescence intensity was measured using a fluorescent-plate reader (Ex/Em _ 485/520 nm). Fibronectin adhesion assays were performed using an in vitro adhesion assay plate coated with fibronectin following the manufacturer’s recommendations (EMD Biosciences, San Diego, CA) as previously described.15
Transendothelial migration assays
Migration assays were performed as previously described.4 Briefly, HUVECs (5 × 103 cells/well) were incubated overnight in the upper chambers of transwell migration assay plates (pore size 0.8 µm; Corning Life Sciences, Acton, MA). The migration ability of MM scrambled control cells vs shST3GAL6 cells was assessed after 4 hours of serum starvation. MM cells were prelabeled with calcein AM and then placed in the upper migration chambers. 500 µL of BMSC-conditioned DMEM media was added to the lower chambers. After 4 hours at 37°C, cells that migrated to the lower chambers were counted on a fluorescent plate reader (Molecular Devices, Sunnyvale, CA).
Rolling assays
Rolling assays were performed using an in vitro flow chamber assay system (Cellix, Dublin, UK). In brief, MM cells (2 × 106/mL, 40 µL/channel) were pipetted into individual channels of P-selectin–coated chips (15 µg/mL) and shear stress of 1 dyne.cm2 was applied with live-frame capture every 0.5 seconds for a minimum of 30 frames per repeat.
Immunoblotting
Immunoblotting was performed as previously described.16 Briefly, whole-cell lysates were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinyldene fluoride (PVDF) membranes (Bio-Rad Laboratories, Hercules, CA). The antibodies used for immunoblotting included anti-ST3GAL6 antibody (Sigma-Aldrich), anti-phospho-Src (P-Src; Tyr 416), anti-phospho-cofilin (P-cofilin), and anti-phospho-paxilin (P-paxilin) with anti-actin or anti-α-tubulin (Cell Signaling Technology, Danvers, MA) used as protein-loading controls.
Immunohistochemistry
Bone marrow biopsies were obtained from 6 MM patients, 3 monoclonal gammopathy of undetermined significance (MGUS) patients, and 3 healthy subjects. Consecutive serial sections cut from each FFPET block were used for immunohistochemistry (IHC). Four-μm sections were cut onto polysine slides, allowed to dry overnight at 40°C, dewaxed in xylene, and rehydrated through graded alcohol to water. Rabbit polyclonal anti-ST3GAL6 antibody was obtained from ATLAS antibodies (Stockholm, Sweden). Full details of the IHC protocol are available in the supplemental Methods. Slides were dual-stained with mouse monoclonal anti-human-CD138 (BD Biosciences, Dublin, UK) and polyclonal rabbit anti-human ST3GAL6 antibody. For detection of HECA-452 expression, BM biopsies were dual-stained with rat IgM, κ anti-human HECA-452 (BD Pharmingen, San Diego, CA), and CD138.
Animals
Approval for animal studies was obtained from the Dana-Farber Cancer Institute and Massachusetts General Hospital Institutional Animal Care and Use Committees. Female 6- to 7-week-old, severe combined immunedeficient beige (SCID-Bg) mice were obtained from Taconic Laboratories. Anesthesia was performed by intraperitoneal injection of ketamine/xylazine (Lloyd Laboratories, Shenandoah, IA) at 80 mg/kg per 12 mg/kg body weight before in vivo imaging sessions. At the study end points, the mice were sacrificed by inhalation of CO2. For survival studies, mice were followed until the development of hind-limb paralysis, at which time they were sacrificed according to our institutional animal protocol.
Xenograft models
Tumors were established in mice using MM1s-GFP-Luc+ cells (5 × 106/ mouse), which were injected into the tail vein of SCID-Bg mice (n = 3/group for homing, n = 12 for tumor burden of which 3 per group were also assessed for engraftment, n = 14 for survival).
Bioluminescent imaging
To detect tumor burden on a weekly basis, mice were anesthetized and injected with 75 mg/kg luciferin (Xenogen, Hopkinton, MA) and imaged for bioluminescence 5 minutes after the injection.
In vivo confocal microscopy
Using the SCID-Bg xenograft mice, homing and engraftment of MM cells to distant bone marrow niches was tracked in vivo, by using in vivo confocal microscopy.4,17,18 Briefly, MM cells that homed to the BM 18 hours after intravenous (IV) injection, or that were engrafted 4 weeks after injection, were imaged in vivo using a Zeiss 710 confocal system (Carl Zeiss Microimaging, Jena, Germany) on an upright examiner stand with a custom stage. A skin flap was made in the scalp of the mice to expose the underlying dorsal skull surface. High-resolution images with cellular detail were obtained through the intact mouse skull at depths of up to 250 μm from the surface of the skull using a 10 × 0.45 NA Plan-Apo objective (Carl Zeiss Microimaging). GFP was excited with the 488-nm line on an Argon laser. Blood vessels were imaged using Evans Blue (100 µL IV) (Sigma-Aldrich) excited with a 633-nm laser. Emission signals were collected by the Zeiss internal confocal Quasar detectors.
Statistical analysis
Statistical differences between experimental groups were analyzed using GraphPad Prism (GraphPad Software). Student t tests were used to compare means (2-tailed; α = 0.05). P values < .05 were considered statistically significant. Kaplan-Meier survival probability curves were constructed to compare groups graphically, and log-rank analysis was used to test for significant differences in survival.
Results
GSEA confirms overexpression of glycosylation-related signatures in MM
Enrichment of glycosylation-related signatures in MM was demonstrated by GSEA7 using an MSigDB gene set and GSE6477 (Figure 1B, heat map of the entire gene set in supplemental Figure 1). The false-discovery rate (FDR) q-value was 0.24, and normalized enrichment score was 1.14, with FDR values <0.25 considered significant.7 ST3GAL6 emerged from this analysis as being one of the most significantly increased genes in MM patients compared with healthy donors, as shown in the heat map in Figure 1C. Moreover, further analysis of GSE6477 demonstrates significantly higher levels of ST3GAL6 expression as the disease progresses from MGUS through smoldering disease to newly diagnosed MM (Figure 1D and Table 1).
ST3GAL6 expression with disease progression
| . | P value . | 95% Confidence interval . |
|---|---|---|
| Healthy vs MGUS | .049 | 0.0008498-0.3576 |
| Healthy vs smoldering | <.0001 | 0.2308-0.4988 |
| Healthy vs new MM | .0031 | 0.1059-0.4901 |
| Healthy vs relapsed MM | .0003 | 0.2388-0.7260 |
| . | P value . | 95% Confidence interval . |
|---|---|---|
| Healthy vs MGUS | .049 | 0.0008498-0.3576 |
| Healthy vs smoldering | <.0001 | 0.2308-0.4988 |
| Healthy vs new MM | .0031 | 0.1059-0.4901 |
| Healthy vs relapsed MM | .0003 | 0.2388-0.7260 |
High expression of ST3GAL6 is associated with inferior survival in myeloma patients
To further validate the influence of this gene in MM patients, we assessed the effect of expression on survival of patients using GEP data from the MRC IX trial (Figure 1E). Patients expressing higher levels of ST3GAL6 had a significantly reduced overall survival (OS) compared with those with lower expression levels (48 vs 36 months, respectively; log-rank P = .041). On multivariate analysis, ST3GAL6 is an independent predictor of reduced OS after adjustment for known prognostic variables (hazard ratio [HR] = 1.47) (Table 2). Subset analysis revealed that the adverse effect on survival of high ST3GAL6 expression was restricted to patients who did not receive either high-dose therapy or thalidomide (P = .0071 and P = .0173, respectively) (supplemental Figure 2).
ST3GAL6 as a prognostic variable in MM
| . | HR . | HR 95% confidence interval . | P value . |
|---|---|---|---|
| High ST3GAL6 | 1.47 | 1.01-2.13 | .04 |
| ISS stage | 1.5 | 1.20-1.87 | <.001 |
| Adverse IgH | 1.5 | 1.0-2.37 | .05 |
| Gain 1q | 1.5 | 1.05-2.28 | .03 |
| . | HR . | HR 95% confidence interval . | P value . |
|---|---|---|---|
| High ST3GAL6 | 1.47 | 1.01-2.13 | .04 |
| ISS stage | 1.5 | 1.20-1.87 | <.001 |
| Adverse IgH | 1.5 | 1.0-2.37 | .05 |
| Gain 1q | 1.5 | 1.05-2.28 | .03 |
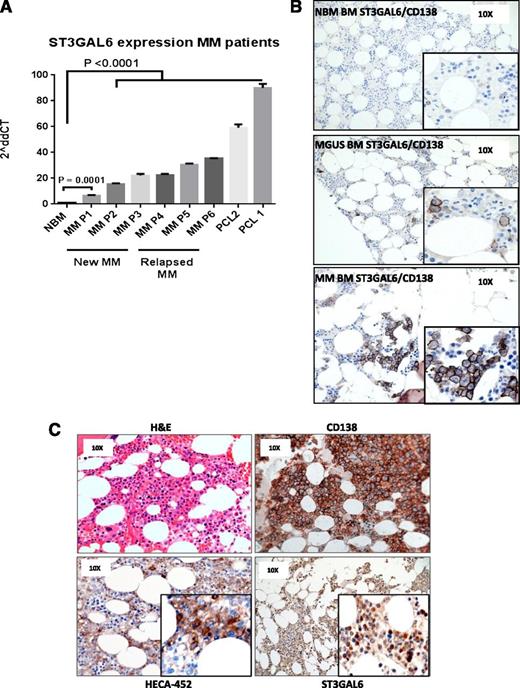
ST3GAL6 is expressed at high levels in CD138+ cells from MM patients
Comparing the expression of ST3GAL6 in primary CD138+ cells from MM patients compared with healthy donors, we noted significantly higher levels of ST3GAL6 in newly diagnosed (n = 3, P = .0001) and relapsed disease (n = 3, P < .0001), with the highest levels of expression seen in patients with secondary plasma cell leukemia (n = 2) (Figure 2A; P < .0001). Immunohistochemistry staining for ST3GAL6 protein was carried out comparing healthy donors with MGUS (n = 3) and MM (n = 6) patients (Figure 2B). This revealed ST3GAL6 expression in MGUS and MM cases that was not apparent in healthy donors. In some cases of MM showing high levels of ST3GAL6, there were corresponding high levels of staining with the monoclonal antibody HECA-452, which binds to sLex and reacts with both cutaneous lymphocyte antigen (CLA) and hematopoietic cell E-/L-selectin ligand (HCELL) (Figure 2C).19
ST3GAL6 expression MM patients. (A) qRT-PCR demonstrating expression levels of ST3GAL6 mRNA isolated from CD138+ cells of MM patients; newly diagnosed (New MM) n = 3, relapsed MM n = 3, or plasma cell leukemia patients (PCL) n = 2 compared with CD138+ cells isolated from normal healthy bone marrow (NBM). Newly diagnosed patients had a significantly higher level of ST3GAL6 mRNA, and patients with relapsed MM and PCL demonstrated increasingly higher levels with disease progression: NBM vs P1 P = .0001, NBM vs all other patients P < .0001. (B) Immunohistochemistry (IHC): dual staining for CD138 and ST3GAL6 demonstrating minimal expression of ST3GAL6/CD138 in healthy donor bone marrow (NBM, top panel, n = 3). Positive expression of ST3GAL6 was noted in patients with MGUS (middle panel, n = 3) and in MM patients (bottom panel, n = 3). ST3GAL6 cytoplasmic brown staining, CD138 membranous dark brown/black staining, blue counterstain. (C) IHC, 4 panels from MM patient (n = 6) demonstrating: hematoxylin and eosin (top left); CD138 (top right); HECA-452 (bottom left), which recognizes sialofucosylated glycans and sialyl Lewis X (sLex); and ST3GAL6 (bottom right), demonstrating in this patient a high ST3GAL6 expression with a corresponding high expression of HECA-452.
ST3GAL6 expression MM patients. (A) qRT-PCR demonstrating expression levels of ST3GAL6 mRNA isolated from CD138+ cells of MM patients; newly diagnosed (New MM) n = 3, relapsed MM n = 3, or plasma cell leukemia patients (PCL) n = 2 compared with CD138+ cells isolated from normal healthy bone marrow (NBM). Newly diagnosed patients had a significantly higher level of ST3GAL6 mRNA, and patients with relapsed MM and PCL demonstrated increasingly higher levels with disease progression: NBM vs P1 P = .0001, NBM vs all other patients P < .0001. (B) Immunohistochemistry (IHC): dual staining for CD138 and ST3GAL6 demonstrating minimal expression of ST3GAL6/CD138 in healthy donor bone marrow (NBM, top panel, n = 3). Positive expression of ST3GAL6 was noted in patients with MGUS (middle panel, n = 3) and in MM patients (bottom panel, n = 3). ST3GAL6 cytoplasmic brown staining, CD138 membranous dark brown/black staining, blue counterstain. (C) IHC, 4 panels from MM patient (n = 6) demonstrating: hematoxylin and eosin (top left); CD138 (top right); HECA-452 (bottom left), which recognizes sialofucosylated glycans and sialyl Lewis X (sLex); and ST3GAL6 (bottom right), demonstrating in this patient a high ST3GAL6 expression with a corresponding high expression of HECA-452.
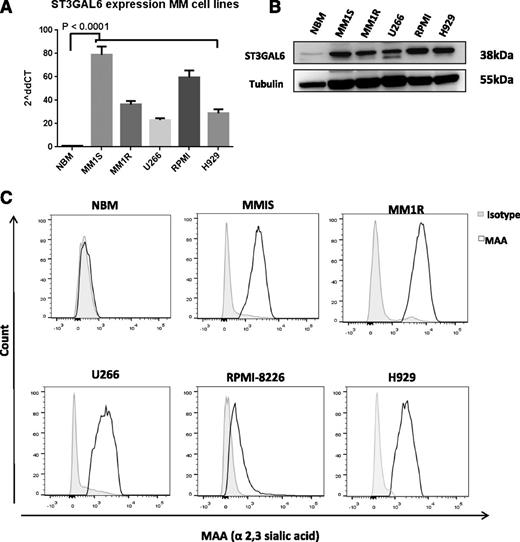
ST3GAL6 is expressed at high levels in MM cell lines
Screening of 5 MM cell lines for ST3GAL6 expression (MM1S, MM1R, U266, RPMI-8226, and H929) demonstrated significantly higher levels of ST3GAL6 mRNA and protein compared with healthy CD138+ cells (Figure 3A-B). Expression of α-2,3 sialic acid, the synthesis of which is contributed to by ST3GAL6, was also demonstrated to be higher at the surface of these MM cell lines compared with CD138+ cells from healthy donor BM using flow cytometry for MAA lectin (Figure 3C). Hypoxia is known to induce the expression of sialyltransferases in cancer; to test whether this may be occurring in MM, we assessed gene expression after 24 hours of hypoxic culture, and this demonstrated induction of ST3GAL6 in MM cell lines (supplemental Figure 3F).
ST3GAL6 expression MM cell lines. (A) ST3GAL6 expression MM cell lines; qRT-PCR for ST3GAL6 gene expression in MM cell lines MM1S, MM1R, U266, RPMI-8226 (RPMI), and H929 compared with CD138 cells isolated from normal healthy donor bone marrow (NBM) P < .0001. (B) Western blot analysis demonstrating the expression level of ST3GAL6 protein in CD138+ cells isolated from the bone marrow of healthy controls (NBM) compared with MM cell lines MM1S, MM1R, U266, RPMI8226, and H929. (C) Flow cytometry demonstrating surface expression of MAA lectin, which binds to α-2,3 sialylated glycans, at the surface of MM cell lines MM1S, MM1R, U266, RPMI8226, and H929 compared with CD138+ cells isolated from healthy donor bone marrow (NBM). MAA is synthesized by ST3GAL6, and the presence of this lectin at the cell surface is indicative of ST3GAL6 activity in these cells.
ST3GAL6 expression MM cell lines. (A) ST3GAL6 expression MM cell lines; qRT-PCR for ST3GAL6 gene expression in MM cell lines MM1S, MM1R, U266, RPMI-8226 (RPMI), and H929 compared with CD138 cells isolated from normal healthy donor bone marrow (NBM) P < .0001. (B) Western blot analysis demonstrating the expression level of ST3GAL6 protein in CD138+ cells isolated from the bone marrow of healthy controls (NBM) compared with MM cell lines MM1S, MM1R, U266, RPMI8226, and H929. (C) Flow cytometry demonstrating surface expression of MAA lectin, which binds to α-2,3 sialylated glycans, at the surface of MM cell lines MM1S, MM1R, U266, RPMI8226, and H929 compared with CD138+ cells isolated from healthy donor bone marrow (NBM). MAA is synthesized by ST3GAL6, and the presence of this lectin at the cell surface is indicative of ST3GAL6 activity in these cells.
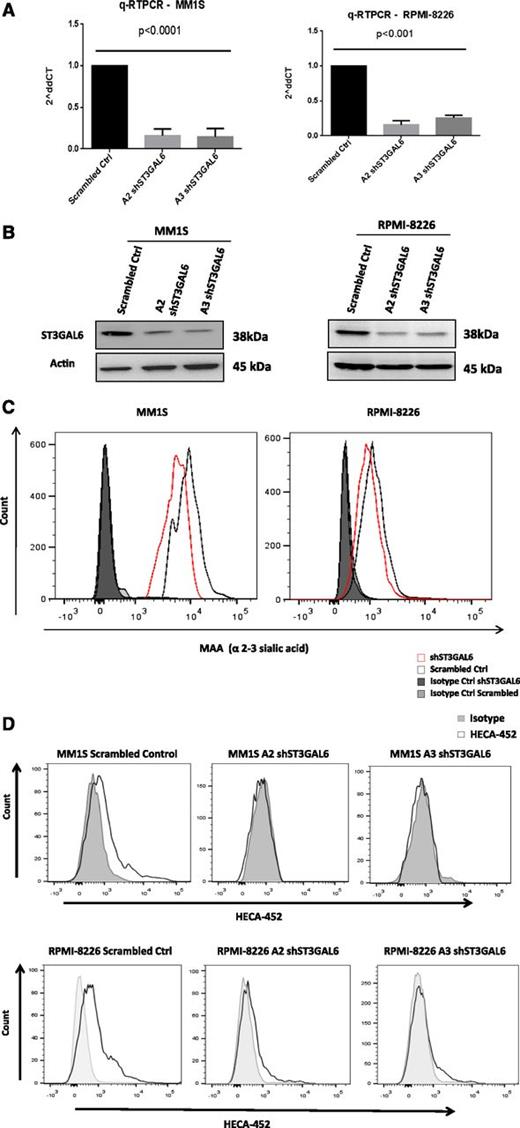
Knockdown of ST3GAL6 in MM cell lines
To investigate the functional role of ST3GAL6 in MM, we performed lentiviral-mediated knockdown in 2 MM cell lines, MM1S and RPMI-8226. The efficiency of this knockdown is demonstrated using qRT-PCR (Figure 4A) and immunoblotting (Figure 4B). Knockdown of ST3GAL6 in MM cell lines resulted in a significant reduction in the amount of α-2,3 sialic acid at the surface of the cells (Figure 4C; MM1S scrambled vs MM1S A2 clone, P = .0006; RPMI-8226 scrambled vs RPMI-8226 A2 clone, P = .001), indicating that in these cells ST3GAL6 contributes to the synthesis of this glycan, which is a component of selectin ligands. A proportion of MM cells from the MM1S and RPMI-8226 cell lines demonstrate positive staining for HECA-452; this was absent in the ST3GAL6 knockdown cells (comparison of mean fluorescent intensity (MFI)s: MM1S scrambled vs MM1S clone A2, P = .014; scrambled vs MM1S clone A3, P = .0059; RPMI scrambled vs RPMI-8226 clone A2, P = .005; scrambled vs RPMI-8226 clone A3, P = .001.); importantly, this indicates reduced reactivity with CLA and HCELL, which are sialofucosylated glycoforms of PSGL-1 and CD44, respectively, which can bind E-selectin (Figure 4D). Knockdown of ST3GAL6 did not affect MM cell proliferation, as assessed by thymidine uptake or cell-counting methods (supplemental Figure 3A-B), and there was no effect on cell cycle or apoptosis (supplemental Figure 3D-E).
ST3GAL6 Knockdown MM cell lines. (A) qRT-PCR demonstrating efficiency of lentiviral-mediated knockdown of ST3GAL6 in MM1S (left) and RPMI-8226 (right) cell lines vs corresponding scrambled controls. shST3GAL6 MM1S cells vs scrambled control, P < .0001; shST3GAL6 RPMI-8226 cells vs scrambled controls, P < .001. (B) Western blot analysis for ST3GAL6 protein expression in MM1S and RPMI-8226 cell lines compared with scrambled controls, demonstrating significant reduction of protein expression in knockdown cell lines. (C) Flow cytometry demonstrating reduced expression of MAA lectin on MM1S and RPMI-8226 cells after the knockdown of ST3GAL6 in these cell lines. Comparison of MFIs: MM1S scrambled vs MM1S A2 clone, P = .0006; RPMI-8226 scrambled vs RPMI-8226 A2 clone, P = .001. (D) Flow cytometry demonstrating reduced expression of HECA452 on MM1S and RPMI-8226 shST3GAL6 cells compared with scrambled control cells. Comparison of MFIs: MM1S scrambled vs MM1S clone A2, P = .014; scrambled vs MM1S clone A3, P = .0059. RPMI scrambled vs RPMI-8226 clone A2, P = .005; scrambled vs RPMI-8226 clone A3, P = .001.
ST3GAL6 Knockdown MM cell lines. (A) qRT-PCR demonstrating efficiency of lentiviral-mediated knockdown of ST3GAL6 in MM1S (left) and RPMI-8226 (right) cell lines vs corresponding scrambled controls. shST3GAL6 MM1S cells vs scrambled control, P < .0001; shST3GAL6 RPMI-8226 cells vs scrambled controls, P < .001. (B) Western blot analysis for ST3GAL6 protein expression in MM1S and RPMI-8226 cell lines compared with scrambled controls, demonstrating significant reduction of protein expression in knockdown cell lines. (C) Flow cytometry demonstrating reduced expression of MAA lectin on MM1S and RPMI-8226 cells after the knockdown of ST3GAL6 in these cell lines. Comparison of MFIs: MM1S scrambled vs MM1S A2 clone, P = .0006; RPMI-8226 scrambled vs RPMI-8226 A2 clone, P = .001. (D) Flow cytometry demonstrating reduced expression of HECA452 on MM1S and RPMI-8226 shST3GAL6 cells compared with scrambled control cells. Comparison of MFIs: MM1S scrambled vs MM1S clone A2, P = .014; scrambled vs MM1S clone A3, P = .0059. RPMI scrambled vs RPMI-8226 clone A2, P = .005; scrambled vs RPMI-8226 clone A3, P = .001.
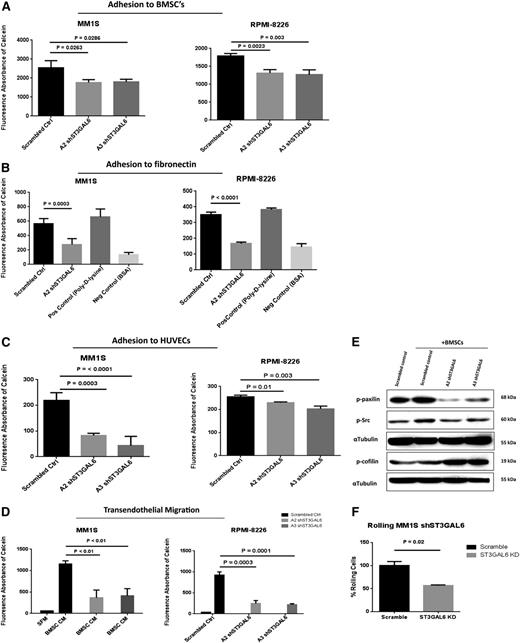
Knockdown of ST3GAL6 in MM cell lines reduces adhesion to MM BMSCs, HUVECs, and fibronectin
Given the potential for knockdown of ST3GAL6 to result in an alteration of carbohydrate-mediated interactions with endothelial cells, and components of the BM microenvironment, we performed in vitro adhesion assays. A significant reduction in adhesion to BMSCs was demonstrated for MM1S and RPMI-8226 shST3GAL6 clones A2 and A3 (Figure 5A; P = .0263, .0286, .0023, and .003, respectively). A reduction in adhesion to fibronectin-coated plates was also seen in shST3GAL6 cells (Figure 5B) (MM1S, P = .0003; RPMI-8226, P < .0001). We also assessed the ability of shST3GAL6 cells to adhere to endothelial cells. Knockdown of ST3GAL6 in both MM1S and RPMI-8225 cells resulted in reduced MM cell adhesion to HUVECs in vitro compared with scrambled control cells (Figure 5C) (MM1S, P = .0003 and < .0001 for clones A2 and A3, respectively; RPMI-8226, P = .01 and .003 for clones A2 and A3, respectively).
In vitro functional effect of ST3GAL6 knockdown in MM cell lines. (A) Effect of ST3GAL6 knockdown on the ability of MM1S and RPMI-8226 cells to adhere to BMSCs isolated from the BM of MM patients. MM1S clone A2, P = .0263; clone A3 P = .0286. RPMI-8226 clone A2, P = .0023, clone A3 P = .003. (B) Adhesion of ST3GAL6 knockdown cells to fibronectin in vitro compared with corresponding scrambled control cells. MM1S A2 clone, P = .0003; RPMI-8226, P < .0001. Similar results were obtained for MM1S and RPMI-8226 A3 clones (data not shown). (C) Adhesion of MM1S and RPMI-8226 shST3GAL6 cells to HUVECs compared with scrambled control cells. Both shST3GAL6 knockdown cells had a significantly reduced ability to adhere to HUVECs in vitro. MM1S clone A2, P = .0003; clone A3 P < .0001. RPMI-8226 clone A2, P = .01; clone A3 P = .003. (D) Migration of ST3GAL6 knockdown cells to BMSC-conditioned media (BMSC CM) in a transendothelial migration assay. The ability of ST3GAL6 knockdown cells to achieve transendothelial migration was significantly reduced compared with scrambled control cells. MM1S clone A2, P < .01; clone A3, P < .01. RPMI-8226 clone A2, P = .0003; A3 P = .0001. (E) Western blot analysis demonstrating reduced levels of adhesion-related proteins in MM1S shST3GAL6 cells that were co-cultured with the BMSCs from MM patients for 24 hours. Phosphorylated paxilin was reduced in shST3GAL6 compared with scrambled control cells cultured in the presence of BMSCs. Phosphorylation of cofilin was greater in the MM cells co-cultured with BMSCs. We also noted an induction of P-Src in scrambled control cells upon co-culture with BMSCs, which was not apparent in the shST3GAL6 cells. α-tubulin was used as a loading control. (F) Rolling of shST3GAL6 cells on P-selectin compared with scrambled control cells demonstrates a reduced ability of these cells to roll on P-selectin (P = .02).
In vitro functional effect of ST3GAL6 knockdown in MM cell lines. (A) Effect of ST3GAL6 knockdown on the ability of MM1S and RPMI-8226 cells to adhere to BMSCs isolated from the BM of MM patients. MM1S clone A2, P = .0263; clone A3 P = .0286. RPMI-8226 clone A2, P = .0023, clone A3 P = .003. (B) Adhesion of ST3GAL6 knockdown cells to fibronectin in vitro compared with corresponding scrambled control cells. MM1S A2 clone, P = .0003; RPMI-8226, P < .0001. Similar results were obtained for MM1S and RPMI-8226 A3 clones (data not shown). (C) Adhesion of MM1S and RPMI-8226 shST3GAL6 cells to HUVECs compared with scrambled control cells. Both shST3GAL6 knockdown cells had a significantly reduced ability to adhere to HUVECs in vitro. MM1S clone A2, P = .0003; clone A3 P < .0001. RPMI-8226 clone A2, P = .01; clone A3 P = .003. (D) Migration of ST3GAL6 knockdown cells to BMSC-conditioned media (BMSC CM) in a transendothelial migration assay. The ability of ST3GAL6 knockdown cells to achieve transendothelial migration was significantly reduced compared with scrambled control cells. MM1S clone A2, P < .01; clone A3, P < .01. RPMI-8226 clone A2, P = .0003; A3 P = .0001. (E) Western blot analysis demonstrating reduced levels of adhesion-related proteins in MM1S shST3GAL6 cells that were co-cultured with the BMSCs from MM patients for 24 hours. Phosphorylated paxilin was reduced in shST3GAL6 compared with scrambled control cells cultured in the presence of BMSCs. Phosphorylation of cofilin was greater in the MM cells co-cultured with BMSCs. We also noted an induction of P-Src in scrambled control cells upon co-culture with BMSCs, which was not apparent in the shST3GAL6 cells. α-tubulin was used as a loading control. (F) Rolling of shST3GAL6 cells on P-selectin compared with scrambled control cells demonstrates a reduced ability of these cells to roll on P-selectin (P = .02).
ST3GAL6 influences MM cell transendothelial migration
Migratory ability was also affected in shST3GAL6 MM cells compared with scrambled controls, as shown by a reduced ability of these cells to migrate to BMSC-conditioned media in a transendothelial cell migration assay incorporating HUVECs (Figure 5D). Compared with scrambled control cells, MM1S and RPMI-8226 shST3GAL6 cells had a reduced ability to migrate (MM1S clone A2 vs scrambled control P < .01, A3 P < .01; RPMI-8226 clone A2 P = .0003, A3 P = .0001).
ST3GAL6 knockdown attenuates Src activation in MM cells
To assess the effect of ST3GAL6 knockdown on adhesion and migration-related proteins, we performed Western blot analyses after co-culture of shST3GAL6 MM1S cells or scrambled controls with primary BMSCs from MM patients. ST3GAL6 knockdown attenuated Src and paxilin activation with a corresponding increased phoshorylation and inactivation of cofilin (Figure 5E).
Ability of MM cells to roll on P-selectin is affected by ST3GAL6 knockdown
Given the potential impact of a reduction in the availability of carbohydrate epitopes on selectin binding, we assessed the ability of ST3GAL6 knockdown cells to roll on P-selectin in vitro. There was a modest but significant reduction in the ability of shST3GAL6 cells to roll on P-selectin compared with scrambled control cells (Figure 5F; P = .02).
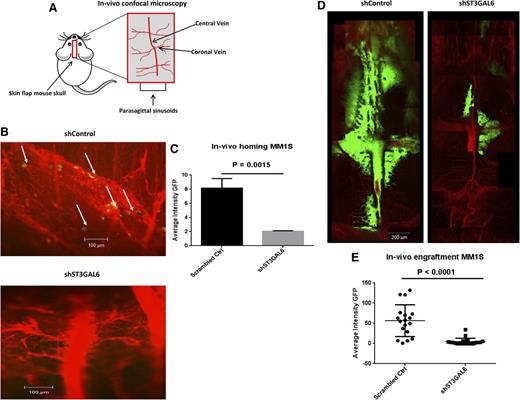
Knockdown of ST3GAL6 reduces MM cell homing and engraftment in vivo
Given the potential for knockdown of ST3GAL6 to affect functions of selectin ligands, and based on our in vitro data showing reduced adhesive and migratory properties of these cells, we assessed the ability of shST3GAL6 cells to home to the BM in vivo and to engraft. Mice were assessed for the presence of MM cells in the BM of the skull 18 hours after injection of MM1S shST3GAL6 GFP+ cells or scrambled control cells (5 × 106 cells/mouse) using in vivo confocal microscopy (Figure 6A). We noted a reduction in homing of shST3GAL6 GFP+ cells in the skull BM of mice 18 hours after IV injection compared with mice that had received scrambled control cells; representative images are shown in Figure 6B (n = 3/group). This reduction in homing was statistically significant for shST3GAL6 cells (Figure 6C; P = .0015). Mice were again evaluated after 4 weeks to demonstrate sustained attenuation of colonization and engraftment of shST3GAL6 cells at this site. We noted a marked reduction in the presence of shST3GAL6 GFP+ cells compared with scrambled control cells indicating that the initial reduction in homing translated to fewer cells reaching the BM and a reduced tumor burden over time. Representative images are shown in Figure 6B (n = 3/group). This reduction in engraftment was statistically significant (Figure 6E; P < .0001).
In vivo ST3GAL6 knockdown. (A) Graphic representation of the anatomic region of the mouse skull examined using confocal microscopy. (B) Representative in vivo confocal microscopy images of scrambled control MM1S-GFP+ cells demonstrating homing to the BM of SCID-Bg mice 18 hours after tail vein injection of 1 × 106 cells (left panel). In these mice, GFP+ cells were visible within the vessels and in the surrounding BM niche, indicating their homing ability. The right panel demonstrates the corresponding vessels in mice that received shST3GAL6 cells; in these mice, there was no evidence of GFP+ cells arriving to the BM, indicating an impaired ability of these cells to home. Images are representative of n = 3 per group. (C) Quantification of in vivo homing of shST3GAL6 or scrambled control GFP+ cells to the BM in vivo. shST3GAL6 cells demonstrated a reduced homing ability, P = .0015. (D) Representative in vivo confocal microscopic images demonstrating a reduction in engraftment and tumor growth of ST3GAL6 knockdown cells in the BM niche of the skull 4 weeks after IV injection of 5 × 106 shST3GAL6 or scrambled control cells (n = 3/group). (E) Quantification of in vivo engraftment of shST3GAL6 cells vs scrambled controls. shST3GAL6 cells demonstrated significantly reduced engraftment, P < .0001.
In vivo ST3GAL6 knockdown. (A) Graphic representation of the anatomic region of the mouse skull examined using confocal microscopy. (B) Representative in vivo confocal microscopy images of scrambled control MM1S-GFP+ cells demonstrating homing to the BM of SCID-Bg mice 18 hours after tail vein injection of 1 × 106 cells (left panel). In these mice, GFP+ cells were visible within the vessels and in the surrounding BM niche, indicating their homing ability. The right panel demonstrates the corresponding vessels in mice that received shST3GAL6 cells; in these mice, there was no evidence of GFP+ cells arriving to the BM, indicating an impaired ability of these cells to home. Images are representative of n = 3 per group. (C) Quantification of in vivo homing of shST3GAL6 or scrambled control GFP+ cells to the BM in vivo. shST3GAL6 cells demonstrated a reduced homing ability, P = .0015. (D) Representative in vivo confocal microscopic images demonstrating a reduction in engraftment and tumor growth of ST3GAL6 knockdown cells in the BM niche of the skull 4 weeks after IV injection of 5 × 106 shST3GAL6 or scrambled control cells (n = 3/group). (E) Quantification of in vivo engraftment of shST3GAL6 cells vs scrambled controls. shST3GAL6 cells demonstrated significantly reduced engraftment, P < .0001.
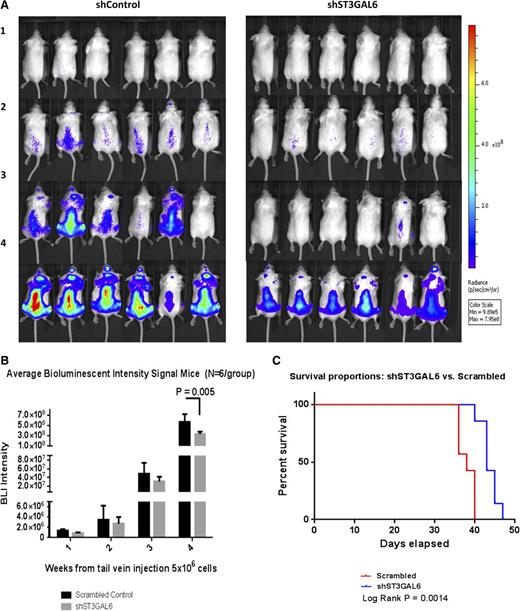
Knockdown of ST3GAL6 decreases MM tumor burden and increases survival in vivo
To examine the effect of ST3GAL6 knockdown on tumor progression in vivo SCID-Bg mice (n = 6/group) were injected with MM1S shST3GAL6 Luc+ cells or scrambled control cells (5 × 106 cells/mouse) via tail vein and underwent weekly bioluminescent imaging (BLI) to assess tumor growth. Mice receiving shST3GAL6 cells demonstrated a statistically significant reduction in tumor burden (Figure 7A-B; P = .005). After completion of the in vivo study qRT-PCR of RNA extracted from MM cells isolated from the BM of the mice confirmed preservation of knockdown of ST3GAL6 (supplemental Figure 3G). A separate study was carried out to evaluate the effect of ST3GAL6 knockdown on the survival of xenograft mice (n = 7/group), and Kaplan-Meier analysis was performed (Figure 7C) demonstrating prolonged survival for mice receiving shST3GAL6 cells vs scrambled control cells (log-rank P = .0014).
Effect of ST3GAL6 knockdown on tumor burden and survival in mice. (A) Tumor burden as assessed by BLI: shown are weeks 1, 2, and 3, and final imaging at week 4 post-injection of 5 × 106 cells (n = 6/group). A reduction in tumor burden was noted in mice that received shST3GAL6 cells compared with scrambled controls. (B) Average BLI signal was quantified over the course of the 4-week study. This demonstrated a statistically significant reduction in tumor burden for mice that received shST3GAL6 cells compared with scrambled controls cells; P = .005. (C) Kaplan-Meier survival probability curve. The effect of knockdown of ST3GAL6 in MM cells on survival of xenograft mice was evaluated in a separate survival study. Mice injected IV with 5 × 106 shST3GAL6 cells had a superior survival to mice injected with scrambled control cells (n = 7/group), log-rank P = .0014.
Effect of ST3GAL6 knockdown on tumor burden and survival in mice. (A) Tumor burden as assessed by BLI: shown are weeks 1, 2, and 3, and final imaging at week 4 post-injection of 5 × 106 cells (n = 6/group). A reduction in tumor burden was noted in mice that received shST3GAL6 cells compared with scrambled controls. (B) Average BLI signal was quantified over the course of the 4-week study. This demonstrated a statistically significant reduction in tumor burden for mice that received shST3GAL6 cells compared with scrambled controls cells; P = .005. (C) Kaplan-Meier survival probability curve. The effect of knockdown of ST3GAL6 in MM cells on survival of xenograft mice was evaluated in a separate survival study. Mice injected IV with 5 × 106 shST3GAL6 cells had a superior survival to mice injected with scrambled control cells (n = 7/group), log-rank P = .0014.
Discussion
Until recently, there has been little focus on the role of glycosylation in the biology and disease progression of MM. However, as has been seen in several solid malignancies,20-23 there is accruing evidence that altered glycosylation is important in MM.24-26 We were particularly interested in identifying alterations in glycosylation with the potential to impact cell trafficking in MM, given the crucial role that this process is known to play in cell-cell interactions and hematogenous metastasis of cancer cells.27 We report enrichment of glycosylation genes in MM, and we identified the sialyltransferase ST3GAL6 as being highly expressed. Knockdown of ST3GAL6 in our study resulted in a reduction in the adhesion and migratory abilities of MM cells in vitro, which was supported by a reduced ability of these cells to home to the bone marrow in vivo.
The sialyltransferases are enzymes that transfer sialic acid from the activated cytidine 5′-monophospho-N-acetylneuraminic acid (CMP-NeuAc) to terminal positions on sialylated glycolipids (gangliosides) or to the N- or O-linked sugar chains of glycoproteins (Figure 1A) Studies performed in knockout mice lacking either ST3GAL4 or ST3GAL6 have uncovered a critical role for ST3GAL6 in the generation of functional selectin ligands in mice.28 Indeed, in murine neutrophils, deletion of ST3GAL6 results in a modest reduction in MAA binding only, along with a moderate reduction in both E- and P-selectin ligand expression. Our data suggest that ST3GAL6 may play a more critical role in the generation of E-selectin ligands in human myeloma cells. That we only see a modest reduction in MAA binding is likely a result of ongoing production of α-2,3 sialic acid by ST3GAL4, which may be incorporated into selectin ligands along with deposition of α-2,3 sialic acid on other glycoproteins and glycolipids by other α-2,3-sialyltransferases.
Selectin ligands, such as PSGL-1, have recently shown to play an important role in the pathobiology of MM.4,21,22 Studies of selectins and their ligands have highlighted the importance of posttranslational glycosylation in the process of leukocyte tethering, rolling, and transendothelial migration, which are central to cellular trafficking.29-31 PSGL-1 is the predominant physiologic ligand for P-selectin and L-selectin, but when modified by HECA-452 reactive glycans (CLA), it can also serve as an E-selectin ligand.24 Posttranslational modifications of PSGL-1 are important for optimal selectin binding; to bind to P-selectin, PSGL-1 requires an α-2,3-sialylated and α-1,3-fucosylated core 2 O-glycan; and to bind to E-selectin, PSGL-1 requires core 2 α-1,3-fucosylated and α-2,3-sialylated O-glycans, indicating a crucial role for ST3GAL6 in the function of PSGL-1 as both an E- and P-selectin ligand.32,33
Therefore, our in vivo findings of reduced tumor burden and prolonged survival of mice receiving shST3GAL6 cells may be related to a reduced ability of these cells to traffic to the BM as a result of a reduction in the availability of functional selectin ligands in these cells. In addition to the potential effect on P-selectin binding, the reduced homing and engraftment of shST3GAL6 cells in the BM in vivo may be influenced by the loss of E-selectin binding, because CLA can serve as an E-selectin ligand, and E-selectin is constitutively expressed by the BM microvasculature.34 Loss of PSGL-1 in mice leads to moderate reduction in rolling on E-selectin in vivo, whereas PSGL-1–deficient hematopoietic progenitors have reduced E-selectin–dependent BM homing; thus E-selectin could conceivably play a dominant role in trafficking of sLex-expressing MM progenitors in the BM niche.35,36
The mechanisms leading to increased ST3GAL6 gene expression in MM are unclear. It has previously been demonstrated in colon cancer that hypoxia induces expression of glycosylation genes, resulting in an increase in selectin ligand expression and adhesion to endothelial selectin.37 In keeping with this finding, we have noted an induction of ST3GAL6 expression in MM cells after hypoxic culture. It appears from our in vitro data that silencing ST3GAL6 primarily mediates changes in migration and adhesion: in support of this, we noted reduced phosphorylation of various proteins involved in adhesion and migration, including P-Src and P-Paxilin, agreeing with previous studies in which sialyltransferase inhibition was shown to reduce the activation of adhesion-related signaling in lung and breast cancers.38
ST3GAL6 overexpression may have potentially important clinical implications. In our analysis of the MRC-IX GEP data, we noted a reduced OS for patients with high levels of ST3GAL6 expression in the nonintensive treatment arm, and for patients who did not receive thalidomide, this could potentially indicate that treatment intensification or the use of thalidomide or other immunomodulatory drugs may be beneficial in patients with high levels of ST3GAL6. However, further studies will be needed to address this question. Finally, patients with high levels of ST3GAL6, and/or other sialyltransferases involved in selectin ligand synthesis, may benefit from potential future therapeutic strategies such as the use of selectin or sialyltransferase inhibitors, with the ultimate aim of preventing dissemination and achieving superior outcomes for MM patients.4,38-40
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by the Health Research Board (HRB) NSAFP, and HRB CSA 2012, Ireland; and in part by the National Institutes of Health, National Cancer Institute grant (RO1 CA154648).
Authorship
Contribution: S.V.G., L.J., A.N., S.M., Yong Z., A.S., M.R.R., A.M.R., I.M.G., and M.E.O. designed and performed research, analyzed data, and prepared the manuscript; L.S.M. performed immunohistochemistry; P.W., M.M., Y.M., I.S., Yu Z., and W.Z. analyzed data; and G.M. contributed MRC-IX gene expression data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael E. O’Dwyer, Room G011, Bioscience Research Building, National University of Ireland, Galway, Ireland; e-mail: michael.odwyer@nuigalway.ie.
References
Author notes
I.M.G. and M.E.O. are cosenior authors.