Key Points
Successful generation of iPSC lines from FPD patient fibroblasts.
Restoration of megakaryopoiesis after RUNX1 mutation correction.
Abstract
Familial platelet disorder with predisposition to acute myeloid leukemia (FPD/AML) is an autosomal dominant disease of the hematopoietic system that is caused by heterozygous mutations in RUNX1. FPD/AML patients have a bleeding disorder characterized by thrombocytopenia with reduced platelet numbers and functions, and a tendency to develop AML. No suitable animal models exist for FPD/AML, as Runx1+/− mice and zebra fish do not develop bleeding disorders or leukemia. Here we derived induced pluripotent stem cells (iPSCs) from 2 patients in a family with FPD/AML, and found that the FPD iPSCs display defects in megakaryocytic differentiation in vitro. We corrected the RUNX1 mutation in 1 FPD iPSC line through gene targeting, which led to normalization of megakaryopoiesis of the iPSCs in culture. Our results demonstrate successful in vitro modeling of FPD with patient-specific iPSCs and confirm that RUNX1 mutations are responsible for megakaryopoietic defects in FPD patients.
Introduction
Familial platelet disorder with predisposition to acute myeloid leukemia (FPD/AML) is a rare autosomal dominant disease. Patients are characterized by mild to moderate bleeding disorder, thrombocytopenia, and defective platelet functions.1-4 Notably, >35% of patients develop AML during their life. The disease is caused by heterozygous mutations in RUNX1,5 which encodes a transcription factor required for definitive hematopoiesis and is frequently involved in leukemogenesis.6-10 To date, >30 FPD mutations have been identified in RUNX1,11 which can be dominant-negative, haploinsufficient, or hypermorphic.12,13
Existing animal models do not recapitulate all features of FPD/AML, because Runx1+/− animals (both mouse and zebra fish) have no defects in megakaryopoiesis and do not develop leukemia.14,15 On the other hand, induced pluripotent stem cells (iPSCs) have the potential to differentiate into many cell types of the human body so that they may be used to model many human diseases as well as to serve as a valuable resource in the field of regenerative medicine.16-18 If disease-causing mutations can be corrected in patient-specific iPSCs, differentiated to therapeutically relevant cell types, and transplanted back to the patients, the symptoms of numerous diseases might be alleviated and the patients might be cured.
Here, we derived iPSCs from 2 FPD/AML patients and demonstrated that these iPSCs have a megakaryopoietic defect in culture. It is important to note that we were able to rescue the megakaryopoietic defect by correcting the RUNX1 mutation with a gene-targeting strategy enhanced by zinc finger nucleases (ZFNs).
Study design
Patient skin biopsies were performed at the National Institutes of Health Clinical Center, and the fibroblast cultures were established at the Tissue Culture Shared Resource, Georgetown University. Skin biopsies and consent for iPSC derivation were performed under NHGRI IRB-approved protocol (76-HG-0238), in accordance with the Declaration of Helsinki. Human iPSCs were derived using the Yamanaka episomal vector system.19 RUNX1 mutation was corrected with homologous recombination to replace the mutant allele with a targeting vector, facilitated by ZFNs from Sigma. For hematopoietic differentiation in vitro, we followed a published protocol20 with additional modifications. Gene expression profiling was performed using Affymetrix GeneChip Human Gene 2.0 ST Array. The microarray dataset has been deposited to the Gene Expression Omnibus with accession number GSE54295. Transmission electron microscopy (TEM) was performed on cell pellets using standard procedures previously described21 in the Electron Microscopy Laboratory of the National Cancer Institute. Detailed procedures are provided in supplemental Methods (see supplemental Data available on the Blood Web site).
Results and discussion
Generation and characterization of FPD iPSCs
We studied a previously described family consisting of multiple affected members in several generations.1,22 The family carries a C > A transversion (rs121912499) in RUNX1, which causes a Y260X nonsense mutation of the RUNX1 protein. Affected family members have lower platelet counts and platelet aggregation defects; several members have developed AML and myelodysplastic syndromes. We obtained skin biopsies from 2 affected members (III-5 and IV-4 in pedigree 3),22 and we confirmed the presence of the RUNX1 Y260X mutation in the fibroblasts (supplemental Figure 1A). iPSC lines were generated from the fibroblasts using nonintegrating plasmid vectors,19 and their pluripotency was demonstrated through marker staining and teratoma formation (supplemental Figure 1B-C).
RUNX1 mutation correction strategy
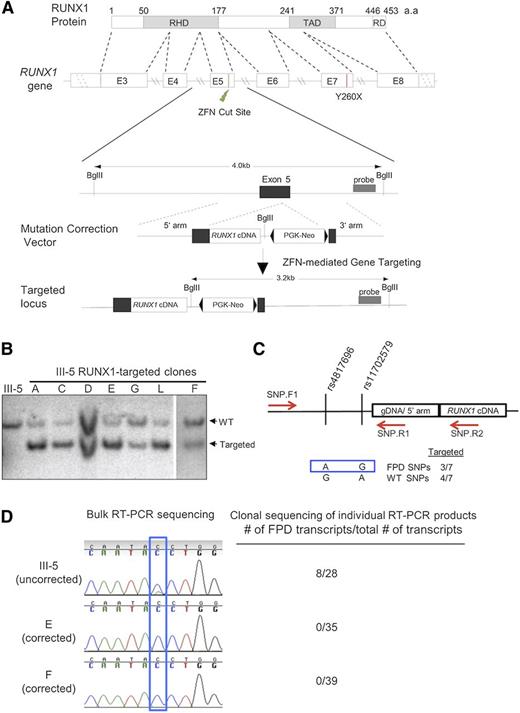
To correct the Y260X mutation in the patient-derived iPSCs and to offer a more universal strategy for correcting other FPD/AML RUNX1 mutations, we designed a donor vector targeting exon 5, which encodes part of the runt-homology domain. The donor vector contains a RUNX1 cDNA fragment containing exons 5 to 8, introns 4 and 5, and a loxP-flanked neo gene. This vector can be used to correct RUNX1 mutations in exons 5 to 8, where more than half of all FPD-associated mutations are located.11 To stimulate gene targeting, we used a pair of RUNX1 ZFNs, which binds and cleaves within exon 5 (Figure 1A).
Targeted gene correction of the mutant RUNX1 allele in FPD/AML iPSCs. (A) Schematic of RUNX1 protein and genomic locus, as well as the gene-targeting strategy to correct the FPD mutation. (Green bolt) ZFN recognition/cleavage site. (Red vertical line) location of the Y260X mutation. Schematic of RUNX1 correction vector is also shown, and proper targeting introduces a new BglII site to the RUNX1 locus. (Gray box) location of the DNA probe for Southern blot hybridization. (B) Screening of selected iPSC clones from gene targeting with Southern blot hybridization using the probe in (A). Clones A to F contain integrations of the targeting vector in the RUNX1 locus. III-5: The FPD iPSC line derived from FPD patient III-5, which was used for targeted mutation correction. “WT” and “Targeted” denote germline and gene-targeted bands (4.0 kb and 3.2 kb) detected by the probe, respectively. (C) Detection of single nucleotide polymorphisms (SNP) by genomic polymerase chain reaction (PCR) to determine which RUNX1 allele was targeted. (Red arrows) locations of primers used in PCR. The combination of AG alleles at the indicated SNPs is associated with the FPD mutant chromosome, and GA allele combination is for the wild-type chromosome. Three of the 7 targeted iPSC lines had insertion of the targeting vector into the FPD chromosome. (D) Sequencing of bulk (sequencing traces) and individual reverse transcription (RT)-PCR products shows only the production of wild-type RUNX1 transcripts (blue peak for C) in corrected clones (clones E and F), whereas the original FPD iPSCs (III-5) produced both mutant (green peak for A) and wild-type (8/28 and 20/28 of the sequenced clones, respectively) RUNX1 transcripts.
Targeted gene correction of the mutant RUNX1 allele in FPD/AML iPSCs. (A) Schematic of RUNX1 protein and genomic locus, as well as the gene-targeting strategy to correct the FPD mutation. (Green bolt) ZFN recognition/cleavage site. (Red vertical line) location of the Y260X mutation. Schematic of RUNX1 correction vector is also shown, and proper targeting introduces a new BglII site to the RUNX1 locus. (Gray box) location of the DNA probe for Southern blot hybridization. (B) Screening of selected iPSC clones from gene targeting with Southern blot hybridization using the probe in (A). Clones A to F contain integrations of the targeting vector in the RUNX1 locus. III-5: The FPD iPSC line derived from FPD patient III-5, which was used for targeted mutation correction. “WT” and “Targeted” denote germline and gene-targeted bands (4.0 kb and 3.2 kb) detected by the probe, respectively. (C) Detection of single nucleotide polymorphisms (SNP) by genomic polymerase chain reaction (PCR) to determine which RUNX1 allele was targeted. (Red arrows) locations of primers used in PCR. The combination of AG alleles at the indicated SNPs is associated with the FPD mutant chromosome, and GA allele combination is for the wild-type chromosome. Three of the 7 targeted iPSC lines had insertion of the targeting vector into the FPD chromosome. (D) Sequencing of bulk (sequencing traces) and individual reverse transcription (RT)-PCR products shows only the production of wild-type RUNX1 transcripts (blue peak for C) in corrected clones (clones E and F), whereas the original FPD iPSCs (III-5) produced both mutant (green peak for A) and wild-type (8/28 and 20/28 of the sequenced clones, respectively) RUNX1 transcripts.
Targeted RUNX1 mutation correction in FPD/AML iPSCs
The RUNX1 ZFNs and the donor vector were transfected into FPD iPSC line III-5 using nucleofection.23 G418+ clones were screened with Southern blot hybridization. All 7 of the screened clones were correctly targeted at the RUNX1 locus (Figure 1B). We determined the chromosome with vector integration in each clone by identifying a unique haplotype associated with the mutant allele (Figure 1C), and we found that 3 of the 7 clones were targeted in the mutated chromosome. By RT-PCR and sequencing, we found 2 corrected iPSC lines, E and F, produced only wild-type transcripts, whereas the original patient iPSC line produced both wild-type and mutant RUNX1 transcripts (Figure 1D).
In vitro hematopoietic differentiation of patient and mutation-corrected iPSC lines
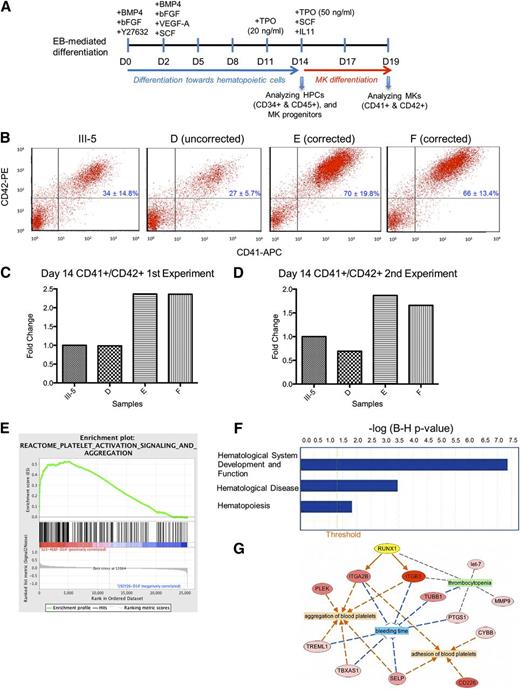
We next assessed the megakaryopoietic potential of the patient (III-5) and targeted iPSCs using a published differentiation protocol20,23 with improvements for megakaryopoiesis (Figure 2A, and supplemental Methods). The targeted iPSC lines consisted of 2 mutation-corrected clones (E and F) and an iPSC clone that was targeted at the wild-type allele (clone D). The patient and the 2 gene-targeted iPSC clones produced similar percentages of CD34+CD45+ HPCs on days 14 and 19 (supplemental Figure 2). Clone D showed reduced percentages of CD34+CD45+ HPCs on day 14, but showed numbers comparable to other iPSC lines on day 19. Significantly, the 2 mutation-corrected iPSC lines produced ∼40% to 60% more CD41+CD42+ MKs than the patient and the wild-type allele-targeted iPSC lines at both time points (Figure 2B-D and supplemental Table 5). This observation is consistent with previous work showing that MK production from CD34+ HPCs in FPD patients is ∼60% lower.11 Moreover, TEM analysis showed that the MKs, differentiated from the FPD patient iPSCs, contained many vacuoles, and the differentiating platelets had a reduced number of α granules and lacked dense granules (supplemental Figure 3A). These findings are similar to those reported earlier.11 It is important to observe that these defects disappeared in the MKs differentiated from the mutation-corrected iPSCs (supplemental Figure 3B). Together, our data demonstrate that a heterozygous RUNX1 mutation causes defective megakaryopoiesis, and targeted mutation correction in patient iPSCs rescues the defect.
Hematopoietic differentiation and differential gene expression analysis of FPD and mutation-corrected iPSC lines. (A) Schematic of an improved hematopoietic differentiation method starting with embryoid body (EB) formation and ending with megakaryocyte (MK) formation. HPCs, hematopoietic progenitor cells. (B) Day 14 fluorescence-activated cell sorter analysis of differentiated cells demonstrating rescue of megakaryopoietic defects in the mutation-corrected iPSC lines (clones E and F) compared with the FPD patient iPSC line (III-5). The iPSC clone D was targeted in the wild-type RUNX1 allele, therefore the Y260X mutation was still present. (C-D) (Bar graphs) fold changes of the percentages of CD41+CD42+ MK cells in culture on day 14 for each targeted clone as compared with the patient iPSC line, III-5, in 2 independent experiments (panels C and D). (E) Gene set enrichment analysis plot distribution of the Reactome Platelet Activation Signaling and Aggregation gene set across the ranked microarray data for day 14 mutation-corrected clones E and F vs patient clone III-5. The normalized enrichment score = 1.75, the nominal P < .01, and the FDR q = .0115. A full list of genes within this gene set is presented in supplemental Table 2. (F) Selected physiological system development and functions that are significantly enriched in differentially expressed genes (DEGs) from mutation-corrected iPSC clones E and F on day 14, as identified by ingenuity pathway analysis (IPA). (Orange vertical line) cutoff threshold of the Benjamini-Hochberg multiple testing correction (B-H p-value) = .05. “Hematological System Development and Function” is the most significantly enriched pathway, “Hematological Disease” ranks sixth, and “Hematopoiesis” ranks 47th among 55 enriched pathways/categories. A full list of significantly enriched physiological system developments and functions can be found in supplemental Table 3. (G) Network representation of processes that are related to disease (green), or predicted to be either activated (orange), or inhibited (blue), because of differential expression of genes between mutation-corrected iPSC lines (clones E and F) and the FPD iPSC line III-5 from day 14. The network was constructed using IPA. A selected list of FPD-related categories and diseases are listed in supplemental Table 4. RUNX1 (highlighted in yellow), upregulated genes (shades of red), and most upregulated gene (dark red). (Arrows) indicate predicted activation (orange), inhibition (blue), or no prediction (gray).
Hematopoietic differentiation and differential gene expression analysis of FPD and mutation-corrected iPSC lines. (A) Schematic of an improved hematopoietic differentiation method starting with embryoid body (EB) formation and ending with megakaryocyte (MK) formation. HPCs, hematopoietic progenitor cells. (B) Day 14 fluorescence-activated cell sorter analysis of differentiated cells demonstrating rescue of megakaryopoietic defects in the mutation-corrected iPSC lines (clones E and F) compared with the FPD patient iPSC line (III-5). The iPSC clone D was targeted in the wild-type RUNX1 allele, therefore the Y260X mutation was still present. (C-D) (Bar graphs) fold changes of the percentages of CD41+CD42+ MK cells in culture on day 14 for each targeted clone as compared with the patient iPSC line, III-5, in 2 independent experiments (panels C and D). (E) Gene set enrichment analysis plot distribution of the Reactome Platelet Activation Signaling and Aggregation gene set across the ranked microarray data for day 14 mutation-corrected clones E and F vs patient clone III-5. The normalized enrichment score = 1.75, the nominal P < .01, and the FDR q = .0115. A full list of genes within this gene set is presented in supplemental Table 2. (F) Selected physiological system development and functions that are significantly enriched in differentially expressed genes (DEGs) from mutation-corrected iPSC clones E and F on day 14, as identified by ingenuity pathway analysis (IPA). (Orange vertical line) cutoff threshold of the Benjamini-Hochberg multiple testing correction (B-H p-value) = .05. “Hematological System Development and Function” is the most significantly enriched pathway, “Hematological Disease” ranks sixth, and “Hematopoiesis” ranks 47th among 55 enriched pathways/categories. A full list of significantly enriched physiological system developments and functions can be found in supplemental Table 3. (G) Network representation of processes that are related to disease (green), or predicted to be either activated (orange), or inhibited (blue), because of differential expression of genes between mutation-corrected iPSC lines (clones E and F) and the FPD iPSC line III-5 from day 14. The network was constructed using IPA. A selected list of FPD-related categories and diseases are listed in supplemental Table 4. RUNX1 (highlighted in yellow), upregulated genes (shades of red), and most upregulated gene (dark red). (Arrows) indicate predicted activation (orange), inhibition (blue), or no prediction (gray).
Gene expression profiling of differentiated hematopoietic cells
We performed microarray analysis of patient (III-5) and mutation-corrected (clones E and F) iPSC lines collected on day 14. DEGs with fold change values >2 and Benjamini-Hochberg adjusted P values < .05 were identified via Partek. We observed a total of 92 DEGs in clones E and F as compared with III-5 (supplemental Table 1). It is interesting to note that previously recognized genes involved in megakaryopoiesis, such as platelet glycoprotein IIb (ITGA2B; also known as CD41)24 and integrin β3 (ITGB3; also known as CD61),25 were significantly upregulated in the corrected iPSCs (supplemental Table 1). In fact, there is good correlation between the gene expression pattern in the differentiated mutation-corrected iPSCs and a gene signature associated with platelets (Figure 2E and supplemental Table 2). In addition, other pathways that are relevant to the pathogenesis of FPD, such as “Hematological System Development and Function,” Hematological Disease,” and “Hematopoiesis,” were identified as significantly enriched in the differentiated mutation-corrected iPSCs (Figure 2F, supplemental Tables 3 and 4). Network reconstruction identified RUNX1 as the gene positioned in the network to account for key differences observed between the patient and corrected samples (Figure 2G).
Taken together, these data support the hypothesis that mutation correction rescues the function of RUNX1 in megakaryopoiesis. These isogenic iPSCs will facilitate research to understand better the function of RUNX1 and to screen small molecules that regulate or modulate its actions. If these patient-specific and mutation-corrected iPSCs could be differentiated to functional hematopoietic stem cells, they could potentially be used to treat FPD/AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the FPD patients for their participation and support of our research, Dionyssia Clagett from Georgetown University for establishing fibroblast lines from the FPD patients, Shinya Yamanaka and Keisuke Okita from the Center for iPS Cell Research and Application, Kyoto University, for episomal reprogramming vectors, Lisa Garrett, Gene Elliott, and Michael Eckhaus from National Institutes of Health for teratoma formation assay and analysis, Kunio Nagashima from National Institutes of Health for electron microscopy, Marjan Huizing from National Institutes of Health for the review of TEM images, and Zack Z Wang, Zhaohui Ye, and Siddharth Shah from Johns Hopkins University for discussions and assistance with MK differentiation of human iPSCs.
This work was supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health, National Institutes of Health grants U01-HL107446 and 2R01-HL073781 (L.C.) and 2R01-DK-78340 (M.S.H.).
Authorship
Contribution: J.P.C., E.M.K., Y.G., and A.G.E. designed and performed the experiments and analyzed the data; N.S.T. and E.M.K. analyzed the microarray data; M.S.H. contributed the patient materials; L.C. and P.P.L. designed and organized the experiments and analyzed the data; and J.P.C., E.M.K., L.C., and P.P.L. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for J.P.C. is Genome Engineering Center, Washington University, St. Louis, MO.
Correspondence: P. Paul Liu, 49 Convent Dr, Building 49, Room 3A26, Bethesda, MD 20892; e-mail: pliu@mail.nih.gov.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal