Key Points
Loss of RUNX1 by using genetic knockout or dominant-negative approaches leads to upregulation of its direct target gene Hmga2 in HSPCs.
Expansion of myeloid progenitors caused by the loss of RUNX1 is rescued by loss of Hmga2, suggesting that RUNX1 functions through Hmga2.
Abstract
RUNX1 is a master transcription factor in hematopoiesis and mediates the specification and homeostasis of hematopoietic stem and progenitor cells (HSPCs). Disruptions in RUNX1 are well known to lead to hematologic disease. In this study, we sought to identify and characterize RUNX1 target genes in HSPCs by performing RUNX1 chromatin immunoprecipitation with high-throughput sequencing (ChIP-seq) using a murine HSPC line and complementing this data with our previously described gene expression profiling of primary wild-type and RUNX1-deficient HSPCs (Lineage–/cKit+/Sca1+). From this analysis, we identified and confirmed that Hmga2, a known oncogene, as a direct target of RUNX1. Hmga2 was strongly upregulated in RUNX1-deficient HSPCs, and the promoter of Hmga2 was responsive in a cell-type dependent manner upon coexpression of RUNX1. Conditional Runx1 knockout mice exhibit expansion of their HSPCs and myeloid progenitors as hallmark phenotypes. To further validate and establish that Hmga2 plays a role in inducing HSPC expansion, we generated mouse models of HMGA2 and RUNX1 deficiency. Although mice lacking both factors continued to display higher frequencies of HSPCs, the expansion of myeloid progenitors was effectively rescued. The data presented here establish Hmga2 as a transcriptional target of RUNX1 and a critical regulator of myeloid progenitor expansion.
Introduction
RUNX1, also known as AML1 and CBFα2, is a member of the Runt family of proteins. The major function of RUNX1 is to operate as a DNA-binding transcription factor. Studies over the past 20 years have established RUNX1 as a critical player in hematopoiesis and specification of hematopoietic stem and progenitor cells (HSPCs), because neither process can occur without RUNX1.1-3 The importance of RUNX1 is further validated by its prevalence in a variety of hematologic diseases and malignancies including myelodysplastic syndrome, myeloproliferative neoplasms, and multiple forms for acute myeloid leukemia (AML).4-6 As a transcription factor, RUNX1 binds to DNA regulatory regions to guide the expression of its target genes. Most confirmed RUNX1 target genes are mainly expressed in differentiated blood cells.5,7-9 Direct targets of RUNX1 in HSPCs, however, have largely remained unexplored. The identification of these genes offers an insightful view into how this master regulator influences HSPC biology. To elucidate RUNX1 target genes in HSPCs, we performed genome-occupancy analysis with chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq) using RUNX1 antibodies and a murine HSPC cell line. These data were combined with gene expression signatures from wild-type and RUNX1-deficient HSPCs (Lineage–/cKit+/Sca1+).10 The combination of both sets of data allowed elucidation of Hmga2 as a RUNX1 target gene.
High mobility group AT-hook 2 (HMGA2) is a nonhistone chromatin-binding protein typically associated with enhancers but lacks its own transcriptional activity.11 Its expression is generally highest in stem cells and during embryogenesis.12 Aberrant HMGA2 expression has been associated with a variety of mesenchymal tumors including several examples in the hematopoietic system.13,14 Hence, HMGA2 is considered to be an oncogene. More recently, ectopic expression of Hmga2 in transgenic mouse models or bone marrow transplantation with cells expressing Hmga2 have been demonstrated to result in myeloproliferative disease.15,16 In these models, the HSPC populations are expanded as well as the myeloid progenitors. All of these studies suggest that HMGA2 serves as a critical regulator of proliferation and survival. Conditional deletion of Runx1 in mice has allowed for the study of its role in HSPCs because mice that are null for Runx1 display embryonic lethality.1,2,17,18 Interestingly, one of the most striking phenotypes in Runx1 conditional knockout mice is a marked expansion of their HSPCs and myeloid progenitors.18,19 In this study, we identified Hmga2 as a target gene using RUNX1 ChIP-seq and analyzed the role of RUNX1 in regulating Hmga2 expression. Furthermore, using genetic models of RUNX1 and HMGA2 deficiency, we established that Hmga2 is a crucial regulator of myeloid progenitor expansion.
Methods
Mice
All experimental protocols were approved by the University of California–San Diego (UCSD) Institutional Animal Care and Use Committee and all animals were housed at UCSD. Runx1floxed/floxed and Mx1-Cre mice were kindly provided by Dr Nancy Speck,18 and Hmga2−/− mice were kindly provided by Dr Kiran Chada.20
Chromatin immunoprecipitation and sequencing
The ChIP procedure and antibody were previously described.21 Approximately 5 × 107 EML cells were used to perform 2 replicates. High-throughput sequencing was conducted on a Genome Analyzer II (Illumina, San Diego, CA) resulting in 2.7 × 107 and 2.6 × 107 reads. Sequences were aligned to the reference mouse genome (version mm9). Peak calling was performed using the MACS algorithm.22 Peaks with a false-discovery rate (FDR) of <1% were associated with the closest transcriptional start site (TSS) using PeakAnalyzer.23 De novo motif finding was performed using the MEME software suite (http://meme.nbcr.net).24 Primers are provided in supplemental Table 1, available on the Blood Web site.
Quantitative reverse-transcriptase polymerase chain reaction
RNA was extracted using the TRIzol method (Life Technologies, Carlsbad, CA) or RNeasy Micro Plus kit (Qiagen, Venlo, The Netherlands). cDNA was synthesized using Superscript III (Life Technologies). Primers are provided in supplemental Table 1.
Luciferase assays
Cells were transfected with PEI (Sigma-Aldrich, St. Louis, MO), Nucleofection (Amaxa, Basel, Switzerland), or electroporation (Bio-Rad, Hercules, CA) with 1 to 3 μg luciferase construct, 1 to 1.5 μg pCMV5-empty, or a combination of pCMV5-RUNX125 and pCMV5-CBFβ26 expression constructs, and 50 to 500 ng of Renilla (pRL-null from Promega, Madison, WI) luciferase construct as an internal transfection control for 293T, NIH3T3, K562, and U937 cells. Jurkat cells were transfected by electroporation with these constructs in the same ratios with a total DNA amount of 20 to 25 μg. Cells were collected 24 hours after transfection and lysates were prepared. Manufacturers’ protocols for the Dual-Luciferase Reporter Assay System (Promega) and Monolight 3010 Luminometer (BD Biosciences, San Jose, CA) were followed for ascertaining luciferase activity. Firefly luciferase activity was normalized to Renilla luciferase activity.
For additional methods, please see supplemental Methods.
Results
Hmga2 is a direct RUNX1 target gene in HSPCs
To gain further insight into which genes are directly regulated by RUNX1 in HSPCs, we performed RUNX1 ChIP-seq in the murine HSPC-like EML cell line. EML cells rely on stem cell factor for survival and have the capability of differentiating into the erythroid, myeloid, and lymphoid lineages.27 Their multipotency and capacity to be grown in large cultures make the EML cell line an ideal cell system for ChIP-seq studies. Peak calling revealed 6370 peaks with a FDR of <1%. Interestingly, the location of the majority of peaks (73.3%) lay in regions more than 5 kilobases (kb) away from a TSS (Figure 1A). Only a small percentage (10.8%) of peaks were in promoter regions, defined as 3 kb upstream of a TSS (Figure 1B). De novo motif analyses using the MEME suite revealed the most enriched motif to be TGTGGT, which is the known RUNX consensus motif (Figure 1C).28 Other top motifs correspond to the ETS and GATA family of transcription factors, both of which are also important players in hematopoiesis and further confirm that RUNX1 often works with these factors to exert its functions.29-32
RUNX1 genome-wide occupancy in HSPCs as determined by ChIP-seq. (A) The 6370 RUNX1 ChIP-seq peaks with a an FDR <1% are shown in relation to the absolute distance of the TSS of known genes in kb. (B) Distribution of RUNX1 ChIP-seq peaks are grouped based on promoter defined as >3 kb upstream of TSS (10.8%), intron (44.3%), exon (5.7%), and intergenic (39.2%) regions. (C) The 500 peaks with the lowest FDR were submitted to the MEME suite. The top motifs are shown and given with E-values, the associated motif family, and the frequency of the motif found in the 500 peaks.
RUNX1 genome-wide occupancy in HSPCs as determined by ChIP-seq. (A) The 6370 RUNX1 ChIP-seq peaks with a an FDR <1% are shown in relation to the absolute distance of the TSS of known genes in kb. (B) Distribution of RUNX1 ChIP-seq peaks are grouped based on promoter defined as >3 kb upstream of TSS (10.8%), intron (44.3%), exon (5.7%), and intergenic (39.2%) regions. (C) The 500 peaks with the lowest FDR were submitted to the MEME suite. The top motifs are shown and given with E-values, the associated motif family, and the frequency of the motif found in the 500 peaks.
We previously reported the gene expression signatures of HSPCs (Lineage–/cKit+/Sca1+, or LSK cells) from wild-type, Runx1 conditional knockout (Δ/Δ), and RUNX1(41-214)–transplanted mice.10 RUNX1(41-214) (RUNX1SF) is primarily a dominant-negative competitor of endogenous RUNX proteins. Of the 59 genes that were commonly differentially expressed in Runx1Δ/Δ and RUNX1SF HSPCs, 27 have RUNX1 ChIP-seq peaks associated with their loci (Figure 2A and Table 1). We also performed comparisons of our ChIP-seq data with other recently published RUNX1 ChIP-seq data (supplemental Figure 1), which demonstrated further overlap of genes.31,32 The combination of these data and comparisons have revealed a list of high-potential RUNX1 target genes. Within this gene list, we decided first to focus our studies on HMGA2. On the basis of the published information of HMGA2, increased expression of HMGA2 is likely to result in similar cellular phenotypes as observed in RUNX1 deficiency. HMGA2 has been reported to play important roles in regulating cellular proliferation and cancer development.13,33 Our gene expression profiling data obtained from 3 biologically independent sets of RNA prepared from LSK cells indicate that Hmga2 expression is significantly increased (Figure 2B). Using quantitative reverse-transcriptase (polymerase chain reaction (RT-qPCR) studies, we demonstrated that Hmga2 transcript levels were sixfold and 15-fold higher in Runx1Δ/Δ and RUNX1SF HSPCs, respectively, relative to wild-type controls (Figure 2C). In addition, Hmga2 is upregulated in Runx1Δ/Δ granulocyte-macrophage progenitors (GMPs) and common myeloid progenitors (CMPs) relative to wild-type controls (supplemental Figure 2). An increase in Hmga2 transcripts also correlated with increase in HMGA2 protein levels in Runx1Δ/Δ HSPCs, GMPs, and CMPs (Figure 2D-F and supplemental Figure 3).
Hmga2 is a RUNX1 target gene in HSPCs. (A) Venn diagram showing the overlap between the common differentially expressed genes in Runx1Δ/Δ and RUNX1SF HSPCs (n = 59)10 and the genes associated with RUNX1 ChIP-seq peaks. (B) Relative expression of Hmga2 in wild-type, Runx1Δ/Δ, and RUNX1SF HSPCs based on microarray data published in Matsuura et al.10 (C) Validation of upregulation of Hmga2 in microarray results by RT-qPCR using cDNA from wild-type, Runx1Δ/Δ, and RUNX1SF HSPCs. RNA extraction and RT-qPCR were performed from samples of at least 3 independent batches of mice. HMGA2 protein levels by intracellular flow cytometry are shown in (D) LSK, (E) GMP, and (F) CMP cell populations between wild-type and Runx1Δ/Δ mice (n = 5 each). (G) Location of RUNX1 ChIP-seq peaks relative to the Hmga2 gene locus on chromosome 10. IgG control and one RUNX1 ChIP-seq replicate are shown. (H) Confirmation of RUNX1 occupancy regions in relation to the Hmga2 gene locus by ChIP-qPCR. RUNX1 ChIP was compared relative to IgG ChIP and normalized to a negative control region. Data represent 3 replicates of ChIP followed by qPCR.
Hmga2 is a RUNX1 target gene in HSPCs. (A) Venn diagram showing the overlap between the common differentially expressed genes in Runx1Δ/Δ and RUNX1SF HSPCs (n = 59)10 and the genes associated with RUNX1 ChIP-seq peaks. (B) Relative expression of Hmga2 in wild-type, Runx1Δ/Δ, and RUNX1SF HSPCs based on microarray data published in Matsuura et al.10 (C) Validation of upregulation of Hmga2 in microarray results by RT-qPCR using cDNA from wild-type, Runx1Δ/Δ, and RUNX1SF HSPCs. RNA extraction and RT-qPCR were performed from samples of at least 3 independent batches of mice. HMGA2 protein levels by intracellular flow cytometry are shown in (D) LSK, (E) GMP, and (F) CMP cell populations between wild-type and Runx1Δ/Δ mice (n = 5 each). (G) Location of RUNX1 ChIP-seq peaks relative to the Hmga2 gene locus on chromosome 10. IgG control and one RUNX1 ChIP-seq replicate are shown. (H) Confirmation of RUNX1 occupancy regions in relation to the Hmga2 gene locus by ChIP-qPCR. RUNX1 ChIP was compared relative to IgG ChIP and normalized to a negative control region. Data represent 3 replicates of ChIP followed by qPCR.
Table of 27 common genes differentially expressed in Runx1Δ/Δ and RUNX1SF HSPCs that overlap with RUNX1 genome occupancy
| Symbol . | Description . | Runx1Δ/Δ array . | RUNX1SF array . | Peak location . |
|---|---|---|---|---|
| Adamtsl4 | ADAMTS-like 4 | 2.42 | 3.37 | chr3:95511225-95512034 |
| Alcam | activated leukocyte cell adhesion molecule | −2.05 | −4.28 | chr16:54016417-54016980 |
| Atp8a2 | ATPase, aminophospholipid transporter, class I, type 8A, member 2 | 2.05 | 2.84 | chr14:60691216-60691929 |
| chr14:60748188-60748880 | ||||
| Bdh1 | 3-hydroxybutyrate dehydrogenase, type 1 | −2.35 | −5.59 | chr16:30967909-30969214 |
| chr16:30971738-30972608 | ||||
| chr16:31080812-31081818 | ||||
| chr16:31209726-31210547 | ||||
| Cd72 | CD72 antigen | −2.08 | −2.32 | chr4:43469362-43470612 |
| Cpa3 | carboxypeptidase A3 | 8.36 | 13.89 | chr3:20141473-20142494 |
| chr3:20143698-20144216 | ||||
| Csf2rb | colony stimulating factor 2 receptor, β, low-affinity (granulocyte-macrophage) | 2.41 | 2.74 | chr15:78097680-78098791 |
| chr15:78150945-78151767 | ||||
| Cyb561 | cytochrome b561 | −3.78 | −3.34 | chr11:105807879-105808525 |
| chr11:105820316-105821213 | ||||
| chr11:105918137-105919089 | ||||
| Deptor | DEP domain containing MTOR-interacting protein | 2.72 | 3.15 | chr15:54963581-54964309 |
| chr15:55022569-55023361 | ||||
| chr15:55049202-55050131 | ||||
| Fhdc1 | FH2 domain containing 1 | 3.48 | 4.5 | chr3:84264390-84265426 |
| chr3:84295898-84296552 | ||||
| chr3:84307981-84308734 | ||||
| Fscn1 | fascin homolog 1, actin bundling protein (Strongylocentrotus purpuratus) | −3.17 | −32.73 | chr5:143581961-143584992 |
| chr5:143656547-143657461 | ||||
| chr5:143715033-143716206 | ||||
| Gzmb | granzyme B | −3.94 | −4.51 | chr14:56880645-56881698 |
| chr14:56884808-56885570 | ||||
| chr14:56914779-56915975 | ||||
| chr14:56933192-56933620 | ||||
| chr14:57190343-57191097 | ||||
| Hmga2 | high mobility group AT-hook 2 | 4.75 | 4.34 | chr10:119760560-119761484 |
| chr10:119883733-119884448 | ||||
| chr10:120053435-120054404 | ||||
| Igf2r | insulin-like growth factor 2 receptor | −2.37 | −10.81 | chr17:12947269-12947943 |
| Itga2b | integrin α 2b | 2.5 | 4.19 | chr11:102330569-102332184 |
| Itga9 | integrin α 9 | 3.32 | 2.83 | chr9:118541479-118542316 |
| chr9:118558410-118559222 | ||||
| chr9:118625078-118626008 | ||||
| chr9:118659770-118660359 | ||||
| chr9:118792266-118792827 | ||||
| Jam3 | junction adhesion molecule 3 | 3.49 | 4.41 | chr9:26906891-26907613 |
| Krt80 | keratin 80 | 7.36 | 7.9 | chr15:101218519-101219226 |
| Lcp2 | lymphocyte cytosolic protein 2 | 2.12 | 2.16 | chr11:33950589-33951437 |
| chr11:33963605-33964645 | ||||
| chr11:33867349-338683 | ||||
| chr11:33900230-33901036 | ||||
| chr11:33909353-33911207 | ||||
| Map3k6 | mitogen-activated protein kinase kinase kinase 6 | 2.01 | 2.39 | chr4:132793901-132794220 |
| Pcdh7 | protocadherin 7 | 5.41 | 9.62 | chr5:58108350-58109816 |
| chr5:58126695-58127555 | ||||
| chr5:58232582-58233087 | ||||
| chr5:58248528-58249096 | ||||
| chr5:58422191-58422711 | ||||
| Podxl | podocalyxin-like | 2.81 | 4.19 | chr6:32107983-32109068 |
| Slc24a3 | solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 | 4.95 | 7.09 | chr2:145050905-145052230 |
| chr2:145087407-145088337 | ||||
| Spp1 | secreted phosphoprotein 1 | −2.41 | −3.6 | chr5:104863923-104864409 |
| Stx3 | syntaxin 3 | 2.66 | 3.32 | chr19:11881628-11882355 |
| Tjp1 | tight junction protein 1 | 6.45 | 6.76 | chr7:72518004-72518826 |
| chr7:73323785-73326321 | ||||
| chr7:73436915-73437490 | ||||
| chr7:73448143-73449672 | ||||
| Zcchc18 | zinc finger, CCHC domain containing 18 | −2.5 | −10.84 | chrX:133520640-133521165 |
| Symbol . | Description . | Runx1Δ/Δ array . | RUNX1SF array . | Peak location . |
|---|---|---|---|---|
| Adamtsl4 | ADAMTS-like 4 | 2.42 | 3.37 | chr3:95511225-95512034 |
| Alcam | activated leukocyte cell adhesion molecule | −2.05 | −4.28 | chr16:54016417-54016980 |
| Atp8a2 | ATPase, aminophospholipid transporter, class I, type 8A, member 2 | 2.05 | 2.84 | chr14:60691216-60691929 |
| chr14:60748188-60748880 | ||||
| Bdh1 | 3-hydroxybutyrate dehydrogenase, type 1 | −2.35 | −5.59 | chr16:30967909-30969214 |
| chr16:30971738-30972608 | ||||
| chr16:31080812-31081818 | ||||
| chr16:31209726-31210547 | ||||
| Cd72 | CD72 antigen | −2.08 | −2.32 | chr4:43469362-43470612 |
| Cpa3 | carboxypeptidase A3 | 8.36 | 13.89 | chr3:20141473-20142494 |
| chr3:20143698-20144216 | ||||
| Csf2rb | colony stimulating factor 2 receptor, β, low-affinity (granulocyte-macrophage) | 2.41 | 2.74 | chr15:78097680-78098791 |
| chr15:78150945-78151767 | ||||
| Cyb561 | cytochrome b561 | −3.78 | −3.34 | chr11:105807879-105808525 |
| chr11:105820316-105821213 | ||||
| chr11:105918137-105919089 | ||||
| Deptor | DEP domain containing MTOR-interacting protein | 2.72 | 3.15 | chr15:54963581-54964309 |
| chr15:55022569-55023361 | ||||
| chr15:55049202-55050131 | ||||
| Fhdc1 | FH2 domain containing 1 | 3.48 | 4.5 | chr3:84264390-84265426 |
| chr3:84295898-84296552 | ||||
| chr3:84307981-84308734 | ||||
| Fscn1 | fascin homolog 1, actin bundling protein (Strongylocentrotus purpuratus) | −3.17 | −32.73 | chr5:143581961-143584992 |
| chr5:143656547-143657461 | ||||
| chr5:143715033-143716206 | ||||
| Gzmb | granzyme B | −3.94 | −4.51 | chr14:56880645-56881698 |
| chr14:56884808-56885570 | ||||
| chr14:56914779-56915975 | ||||
| chr14:56933192-56933620 | ||||
| chr14:57190343-57191097 | ||||
| Hmga2 | high mobility group AT-hook 2 | 4.75 | 4.34 | chr10:119760560-119761484 |
| chr10:119883733-119884448 | ||||
| chr10:120053435-120054404 | ||||
| Igf2r | insulin-like growth factor 2 receptor | −2.37 | −10.81 | chr17:12947269-12947943 |
| Itga2b | integrin α 2b | 2.5 | 4.19 | chr11:102330569-102332184 |
| Itga9 | integrin α 9 | 3.32 | 2.83 | chr9:118541479-118542316 |
| chr9:118558410-118559222 | ||||
| chr9:118625078-118626008 | ||||
| chr9:118659770-118660359 | ||||
| chr9:118792266-118792827 | ||||
| Jam3 | junction adhesion molecule 3 | 3.49 | 4.41 | chr9:26906891-26907613 |
| Krt80 | keratin 80 | 7.36 | 7.9 | chr15:101218519-101219226 |
| Lcp2 | lymphocyte cytosolic protein 2 | 2.12 | 2.16 | chr11:33950589-33951437 |
| chr11:33963605-33964645 | ||||
| chr11:33867349-338683 | ||||
| chr11:33900230-33901036 | ||||
| chr11:33909353-33911207 | ||||
| Map3k6 | mitogen-activated protein kinase kinase kinase 6 | 2.01 | 2.39 | chr4:132793901-132794220 |
| Pcdh7 | protocadherin 7 | 5.41 | 9.62 | chr5:58108350-58109816 |
| chr5:58126695-58127555 | ||||
| chr5:58232582-58233087 | ||||
| chr5:58248528-58249096 | ||||
| chr5:58422191-58422711 | ||||
| Podxl | podocalyxin-like | 2.81 | 4.19 | chr6:32107983-32109068 |
| Slc24a3 | solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 | 4.95 | 7.09 | chr2:145050905-145052230 |
| chr2:145087407-145088337 | ||||
| Spp1 | secreted phosphoprotein 1 | −2.41 | −3.6 | chr5:104863923-104864409 |
| Stx3 | syntaxin 3 | 2.66 | 3.32 | chr19:11881628-11882355 |
| Tjp1 | tight junction protein 1 | 6.45 | 6.76 | chr7:72518004-72518826 |
| chr7:73323785-73326321 | ||||
| chr7:73436915-73437490 | ||||
| chr7:73448143-73449672 | ||||
| Zcchc18 | zinc finger, CCHC domain containing 18 | −2.5 | −10.84 | chrX:133520640-133521165 |
Upon examination of the Hmga2 gene locus, ChIP-seq revealed RUNX1-binding regions in the promoter, upstream, downstream, and third intron regions (Figure 2G). All except the promoter region were determined to be significant by MACS.22 These 3 significant RUNX1-binding regions are in agreement with recently published RUNX1 ChIP-seq data using murine HSPC lines.31 The binding of RUNX1 to these regions was confirmed using ChIP-qPCR (Figure 2H). The locations of these regions relative to the Hmga2 TSS suggest that they may serve as enhancers and/or silencers for the regulation of Hmga2 expression. Together, our combined gene expression and ChIP analyses show that RUNX1 directly binds to the Hmga2 locus and influences its expression.
The Hmga2 promoter is regulated by RUNX1 in a cell type–dependent manner and is independent of canonical RUNX binding sites
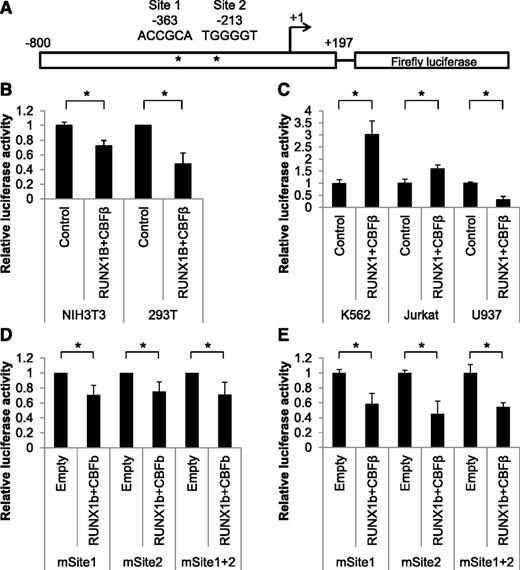
ChIP studies indicate that RUNX1 binds to the previously reported Hmga2 promoter region (base pairs [bp] −800 to +197 relative to the TSS).34 To study how RUNX1 regulates Hmga2 expression, a DNA fragment containing this region was inserted into the multiple cloning site of the pGLX promoterless luciferase reporter vector. The pGLX vector is a modified version of pGL2, which does not contain any RUNX consensus binding sites.26 The cloned Hmga2 promoter DNA fragment has 2 RUNX consensus sites at bp −363 and −213 and a polypyrimidine/polypurine tract spanning bp −84 to −25, which gives the promoter much of its activity (Figure 3A).34 To examine whether RUNX1 affects Hmga2 promoter activity, we conducted transient transfection assays in nonhematopoietic cell lines (NIH3T3 and 293T) and hematopoietic cell lines (K562, Jurkat, and U937). In NIH3T3 and 293T cells, coexpression of the full-length promoter with RUNX1 and its partner CBFβ reduces luciferase activity to 72% and 47%, respectively (Figure 3B). These results suggest that RUNX1 acts as a transcriptional repressor on the Hmga2 promoter in these 2 adherent cell lines. Interestingly, in the K562 and Jurkat leukemia cell lines, RUNX1 increases luciferase activity by threefold and 1.6-fold, respectively (Figure 3C). Because these cell lines represent erythromyeloid and lymphoid lineages, respectively, we also conducted experiments using U937 cells, which more closely represent monocyte/myeloid cells (Figure 3C). In this myeloid representative cell line, RUNX1 acts to suppress luciferase activity to 31%. Thus, RUNX1 regulates Hmga2 promoter activity in all of the cell lines tested but acts on the promoter in a positive or negative manner depending on the cell and/or tissue type. Most importantly, RUNX1 acts as a transcriptional repressor of Hmga2 in myeloid cell types.
RUNX1 controls transcription of Hmga2 via its promoter in a tissue-specific manner. (A) Diagram of promoter-luciferase construct showing RUNX consensus Sites 1 and 2(*) at bp −363 and −213, respectively. (B) NIH3T3 and 293T cell lines were transfected with the full-length Hmga2 promoter-luciferase construct, and luciferase assays were performed 24 hours after transfection. Coexpression was conducted with an empty expression vector (Control) or RUNX1 and CBFβ expression vectors. Error bars indicate standard deviation and are from at least 3 replicates. (C) K562, Jurkat, and U937 cell lines were transfected with the full-length Hmga2 promoter-luciferase construct, and luciferase assay was performed 24 hours after transfection. Coexpression was conducted with an empty expression vector (Control) or RUNX1 and CBFβ expression vectors. Error bars indicate standard deviation and are from at least 3 replicates. (D) NIH3T3 cells were transfected with promoter-luciferase constructs with Site1, Site2, or both sites mutated. Luciferase assays were performed 24 hours after transfection. Coexpression was conducted with an empty expression vector (Control) or RUNX1 and CBFβ expression vectors. Error bars indicate standard deviation and are from at least 3 replicates. (E) U937 cells were transfected with the constructs indicated in (D) and luciferase assays were performed 24 hours after transfection. Coexpression was conducted with an empty expression vector (Control) or RUNX1 and CBFβ expression vectors. Error bars indicate standard deviation from at least 3 replicates. *P < .05.
RUNX1 controls transcription of Hmga2 via its promoter in a tissue-specific manner. (A) Diagram of promoter-luciferase construct showing RUNX consensus Sites 1 and 2(*) at bp −363 and −213, respectively. (B) NIH3T3 and 293T cell lines were transfected with the full-length Hmga2 promoter-luciferase construct, and luciferase assays were performed 24 hours after transfection. Coexpression was conducted with an empty expression vector (Control) or RUNX1 and CBFβ expression vectors. Error bars indicate standard deviation and are from at least 3 replicates. (C) K562, Jurkat, and U937 cell lines were transfected with the full-length Hmga2 promoter-luciferase construct, and luciferase assay was performed 24 hours after transfection. Coexpression was conducted with an empty expression vector (Control) or RUNX1 and CBFβ expression vectors. Error bars indicate standard deviation and are from at least 3 replicates. (D) NIH3T3 cells were transfected with promoter-luciferase constructs with Site1, Site2, or both sites mutated. Luciferase assays were performed 24 hours after transfection. Coexpression was conducted with an empty expression vector (Control) or RUNX1 and CBFβ expression vectors. Error bars indicate standard deviation and are from at least 3 replicates. (E) U937 cells were transfected with the constructs indicated in (D) and luciferase assays were performed 24 hours after transfection. Coexpression was conducted with an empty expression vector (Control) or RUNX1 and CBFβ expression vectors. Error bars indicate standard deviation from at least 3 replicates. *P < .05.
Because the Hmga2 promoter contains 2 RUNX consensus sites, we sought to ascertain whether these sites mediate RUNX1 interaction with the promoter by using promoter constructs with these regions truncated or mutated. In NIH3T3 cells, mutation of either or both of the presumptive RUNX1-binding sites in the promoter does not abrogate the repression of luciferase upon co-expression of RUNX1 and CBFβ (Figure 3D). Constructs harboring truncations of the promoter at the 5′ or 3′ end also failed to affect RUNX1-mediated repression (supplemental Figure 4). The same mutation constructs were used in U937 cells. The coexpression of RUNX1 and CBFβ was still able to repress luciferase activity with the 3 mutation constructs (Figure 3E). These results suggest that RUNX1 does not regulate the Hmga2 promoter through these 2 sites and likely exerts its effect by interacting with other intermediary factors that bind to the promoter. Given the ability of RUNX1 to interact with a variety of other transcriptional regulators,5 this possibility seems likely regarding the regulation of Hmga2.
Runx1 intronic and intergenic binding regions mediate Hmga2 transcription
Significant RUNX1-binding regions were discovered in intronic and intergenic regions of the Hmga2 locus (Figure 2H and supplemental Figure 5). We sought to determine how these regions affect the transcription of Hmga2 and whether they contribute to the effect of RUNX1 on the Hmga2 promoter. The 3 regions were cloned downstream of the luciferase gene in the Hmga2 promoter-luciferase construct (Figure 4A). The upstream binding element adds significant basal luciferase activity relative to the full-length promoter alone in all cell lines tested (Figure 4B-C). Interestingly, in NIH3T3 cells, addition of the upstream element induces enough transcriptional activation to counteract the repressive effect caused by coexpression of RUNX1 and CBFβ (Figure 4B). For example, we no longer observe significant repression in the Promoter + Upstream case compared with Promoter alone. The same trends are observed using the same constructs in U937 cells (Figure 4C). Accordingly, the upstream element continues to result in heightened transcriptional activity in K562 cells (supplemental Figure 6). Physical interaction between the upstream element and the promoter was confirmed by 3C-qPCR assay (Figure 4D). The increase in transcriptional activity induced by the upstream element, and confirmation of physical interaction between the upstream element and the promoter suggest that it may potentially act as an enhancer element.
RUNX1 distal binding regions exert differential effects on Hmga2 expression. (A) Schematic of distal element-promoter luciferase constructs. The distal elements being tested are the RUNX1-binding regions that are downstream, upstream, and in the intron of the Hmga2 locus. Distal element sequences were cloned downstream of the Hmga2 promoter in the promoter-luciferase constructs. (B) These constructs were cotransfected with empty vector (Control) or with RUNX1 and CBFβ (designated as RUNX1) expression constructs into NIH3T3 cells. Luciferase activity was performed 24 hours after transfection, and error bars indicate standard deviation from 4 biological replicates. (C) The same experiment as described in (B) was performed using U937 cells. Luciferase activity was performed 24 hours after transfection, and error bars indicate standard deviation from 3 biological replicates. For (B) and (C), Promoter alone is designated as “P,” Promoter + Upstream is “P + U,” Promoter + Downstream is “P + D,” and Promoter + Intron is “P + I.” (D) Diagram and chart showing enrichment of interaction between the promoter and upstream element (located at approximately −139 kb upstream) as demonstrated by 3C-qPCR assay, which was performed using EML cells. Data shown are average of 2 independent 3C assays and error bars indicate standard deviation. *P < .05.
RUNX1 distal binding regions exert differential effects on Hmga2 expression. (A) Schematic of distal element-promoter luciferase constructs. The distal elements being tested are the RUNX1-binding regions that are downstream, upstream, and in the intron of the Hmga2 locus. Distal element sequences were cloned downstream of the Hmga2 promoter in the promoter-luciferase constructs. (B) These constructs were cotransfected with empty vector (Control) or with RUNX1 and CBFβ (designated as RUNX1) expression constructs into NIH3T3 cells. Luciferase activity was performed 24 hours after transfection, and error bars indicate standard deviation from 4 biological replicates. (C) The same experiment as described in (B) was performed using U937 cells. Luciferase activity was performed 24 hours after transfection, and error bars indicate standard deviation from 3 biological replicates. For (B) and (C), Promoter alone is designated as “P,” Promoter + Upstream is “P + U,” Promoter + Downstream is “P + D,” and Promoter + Intron is “P + I.” (D) Diagram and chart showing enrichment of interaction between the promoter and upstream element (located at approximately −139 kb upstream) as demonstrated by 3C-qPCR assay, which was performed using EML cells. Data shown are average of 2 independent 3C assays and error bars indicate standard deviation. *P < .05.
The downstream element was also tested in NIH3T3 and U937 cells. In both cell lines, the effect of coexpression of RUNX1 and CBFβ remained intact and did not differ significantly from the promoter element by itself. In U937 cells, the downstream element also added significantly more luciferase activity compared with promoter alone.
Finally, the constructs containing the intron element with the Hmga2 promoter were used for promoter-luciferase assays. In contrast to the upstream element, the intron element significantly represses basal luciferase activity relative to the full-length promoter in all of the cell lines tested (Figure 4B-C). In NIH3T3 and U937 cells, the promoter and intron construct continues to result in transcriptional repression when coexpressed with RUNX1 and CBFβ (Figure 4B). In K562 cells, addition of the intron element induces enough transcriptional repression to counteract the activation caused by co-expression of RUNX1 and CBFβ (supplemental Figure 6). Similar to the case of the Promoter + Upstream element in NIH3T3 cells, where the trend of repression is still observed but no longer significant, in K562 cells the activation caused by coexpression of RUNX1 and CBFβ is still observed but no longer significant. These results suggest that the upstream and intron regions are enhancer and silencer elements, respectively.
HMGA2 does not modulate the effect of RUNX1 deficiency on HSPC expansion
Our current studies indicate that Hmga2 expression is directly regulated by RUNX1 and that Hmga2 expression is significantly increased in HSPCs in the absence of RUNX1 or in the presence of RUNX1SF (Figure 2). One of the most dramatic phenotypes of Runx1Δ/Δ mice and RUNX1SF mice is expansion of their HSPC populations.10,17,18 HMGA2 is generally thought to be a proliferation-inducing factor. Transgenic mice expressing Hmga2 and overexpression of Hmga2 via retroviral transduction and bone marrow transplantation have both been recently shown to induce myeloproliferative disease in mice.15,16 Because Hmga2 is a target gene of RUNX1, upregulation of Hmga2 in RUNX1-deficient HSPCs may contribute to their expansion. To examine the role of HMGA2 in HSPC expansion caused by loss of RUNX1 function, we generated littermates of wild type, Hmga2−/−, Runx1Δ/Δ, and Hmga2 and Runx1Δ/Δ double-knockout (Double KO) mice for further studies. The bone marrow compartments of these 4 genotypes of mice were analyzed by staining with cell-surface markers followed by flow cytometry. As expected, RUNX1 deficiency led to a significant expansion of LSK, LT-HSC, and ST-HSC populations (Figure 5A-D). However, lack of HMGA2 did not have any effects on HSPC frequency. The Double KO mice also displayed HSPC expansion, suggesting that loss of HMGA2 does not affect HSPC expansion as a result of the deficiency of RUNX1.
HMGA2 does not modulate the effect of RUNX1 deficiency on HSPC expansion. Percentages of hematopoietic stem cell populations analyzed by flow cytometry, including (A) LSK cells, (B) LT-HSCs, and (C) ST-HSCs. For LSK: wild-type (Hmga2+/+ or Hmga2+/−, Runx1(fl/fl), Mx1Cre–, n = 15), Hmga2−/− (Hmga2−/−, Runx1(fl/fl), Mx1Cre–, n = 9), Runx1Δ/Δ (Hmga2+/+ or Hmga2+/−, Runx1(fl/fl), Mx1Cre+, n = 21), Double KO (Hmga2−/−, Runx1(fl/fl), Mx1Cre+, n = 8). For LT-HSCs and ST-HSCs: wild-type (n = 10), Hmga2−/− (n = 4), Runx1Δ/Δ (n = 10), Double KO (n = 4). (D) Representative flow cytometry gating of LSK and SLAM populations for each of the 4 genotypes of mice and their averages are shown. (E) Percentages of CLP populations were analyzed by flow cytometry. For CLPs: wild-type (n = 5), Hmga2−/− (n = 5), Runx1Δ/Δ (n = 10), Double KO (n = 4). (F) Averages of frequencies of Annexin V–positive/7AAD-negative cells from the LSK cells of each genotype (n = 4 each) are shown. *P < .05.
HMGA2 does not modulate the effect of RUNX1 deficiency on HSPC expansion. Percentages of hematopoietic stem cell populations analyzed by flow cytometry, including (A) LSK cells, (B) LT-HSCs, and (C) ST-HSCs. For LSK: wild-type (Hmga2+/+ or Hmga2+/−, Runx1(fl/fl), Mx1Cre–, n = 15), Hmga2−/− (Hmga2−/−, Runx1(fl/fl), Mx1Cre–, n = 9), Runx1Δ/Δ (Hmga2+/+ or Hmga2+/−, Runx1(fl/fl), Mx1Cre+, n = 21), Double KO (Hmga2−/−, Runx1(fl/fl), Mx1Cre+, n = 8). For LT-HSCs and ST-HSCs: wild-type (n = 10), Hmga2−/− (n = 4), Runx1Δ/Δ (n = 10), Double KO (n = 4). (D) Representative flow cytometry gating of LSK and SLAM populations for each of the 4 genotypes of mice and their averages are shown. (E) Percentages of CLP populations were analyzed by flow cytometry. For CLPs: wild-type (n = 5), Hmga2−/− (n = 5), Runx1Δ/Δ (n = 10), Double KO (n = 4). (F) Averages of frequencies of Annexin V–positive/7AAD-negative cells from the LSK cells of each genotype (n = 4 each) are shown. *P < .05.
In addition, the common lymphoid progenitor (CLP) populations were assessed. Although we observed a trend toward higher frequencies of CLPs in the absence of RUNX1, this difference was not significant (Figure 5E). In addition, neither Hmga2−/− nor Double KO mice exhibited any significant differences in CLP frequency.
The HSPCs in Runx1Δ/Δ mice have been described to have lower levels of apoptosis overall.35 In addition, ectopic expression of Hmga2 has also been demonstrated to lead to lower levels of apoptosis.36 To ascertain whether Hmga2 has any role in mediating apoptosis in the context of RUNX1 loss-of-function, we examined the frequency of apoptosis in LSK cells by using Annexin V staining (Figure 5F). Hmga2−/− and Double KO mice exhibited higher levels, whereas Runx1Δ/Δ mice generally exhibited lower levels of Annexin V–positive cells, demonstrating that Hmga2 plays an important role in regulating apoptosis in this population and that it is necessary for the decreased apoptosis associated with loss of RUNX1.
Hmga2 contributes to myeloid progenitor expansion caused by the loss of RUNX1
Another major phenotype of Runx1Δ/Δ mice is their expansion of the myeloid progenitors, specifically the GMP and CMP populations.18,19 Intriguingly, the role of HMGA2 in inducing proliferation in the hematopoietic system has primarily been biased toward the myeloid lineage.15,16 To test the involvement of HMGA2 in regulating myeloid progenitors in RUNX1-deficient mice, we analyzed the bone marrow compartments of wild-type, Hmga2−/−, Runx1Δ/Δ, and Double KO mice and focused on their myeloid progenitor cells (Figure 6A). As expected, Runx1Δ/Δ mice displayed an expansion of the GMP population compared with wild-type (1.41% of total bone marrow vs 0.59%, P < .001). Hmga2−/− mice, on the other hand, had significantly fewer GMPs compared with wild-type (0.25%, P < .001). In Double KO mice, the expansion of the GMPs is significantly rescued compared with Runx1Δ/Δ mice (0.57%, P < .001). The CMP populations displayed a similar trend, whereas megakaryocyte-erythrocyte progenitors (MEPs) were not affected (Figure 6B-C). Representative flow cytometry charts and averages for each genotype are shown (Figure 6D). Thus the loss of Hmga2 in a RUNX1-deficient genetic background brings the GMP and CMP frequency back down to wild-type levels. Together these results suggest that Hmga2 contributes to the myeloid progenitor expansion caused by the loss of RUNX1 function.
Lack of HMGA2 rescues myeloid progenitor expansion caused by loss of RUNX1. Percentages of hematopoietic stem cell populations analyzed by flow cytometry, including (A) GMPs, (B) CMPs, and (C) MEPs. For myeloid progenitor staining, mice used were: wild-type (Hmga2+/+ or Hmga2+/−, Runx1(fl/fl), Mx1Cre–, n = 9), Hmga2−/− (Hmga2−/−, Runx1(fl/fl), Mx1Cre–, n = 9), Runx1Δ/Δ (Hmga2+/+ or Hmga2+/−, Runx1(fl/fl), Mx1Cre+, n = 14), and Double KO (Hmga2−/−, Runx1(fl/fl), Mx1Cre+, n = 8). (D) Representative flow cytometry gating of GMP, CMP, and MEP populations on each of the 4 genotypes of mice and their averages are shown. (E-G) Averages of frequencies of Annexin V–positive/7AAD-negative cells from the (E) GMP, (F) CMP, and (G) MEP gates of each genotype (n = 4 each) are shown. *P < .05.
Lack of HMGA2 rescues myeloid progenitor expansion caused by loss of RUNX1. Percentages of hematopoietic stem cell populations analyzed by flow cytometry, including (A) GMPs, (B) CMPs, and (C) MEPs. For myeloid progenitor staining, mice used were: wild-type (Hmga2+/+ or Hmga2+/−, Runx1(fl/fl), Mx1Cre–, n = 9), Hmga2−/− (Hmga2−/−, Runx1(fl/fl), Mx1Cre–, n = 9), Runx1Δ/Δ (Hmga2+/+ or Hmga2+/−, Runx1(fl/fl), Mx1Cre+, n = 14), and Double KO (Hmga2−/−, Runx1(fl/fl), Mx1Cre+, n = 8). (D) Representative flow cytometry gating of GMP, CMP, and MEP populations on each of the 4 genotypes of mice and their averages are shown. (E-G) Averages of frequencies of Annexin V–positive/7AAD-negative cells from the (E) GMP, (F) CMP, and (G) MEP gates of each genotype (n = 4 each) are shown. *P < .05.
In addition, Hmga2 regulates the decreased apoptosis observed in the myeloid progenitors of Runx1Δ/Δ mice because Double KO myeloid progenitors continue to show increased frequencies of Annexin V–positive cells (Figure 6E-G).
Discussion
RUNX1 has been implicated in a variety of blood-related diseases and neoplasms.5 Many of these diseases originate from mutations in RUNX1 that occur at the stage of the HSPC and result in the loss of normal RUNX1 function. As a transcription factor, the primary biological activity of RUNX1 is to control the expression of its target genes. The goal of this study was to determine which genes RUNX1 directly targets at the level of the HSPC and whether those genes have any functions in regulating HSPCs. To this end, we used a combination of differential gene expression analyses and genome-wide transcription factor occupancy to identify prospective RUNX1 direct targets. For gene expression analyses, 2 models of RUNX1 deficiency were used—an inducible conditional knockout model for RUNX1 and a bone marrow transplantation model using RUNX1SF as a dominant-negative regulator of endogenous RUNX proteins. To elucidate DNA occupancy of RUNX1 on a genome-wide scale, ChIP-seq was conducted on an HSPC-like cell line using RUNX1 antibody. The result was a condensed list of high-potential direct target genes that are regulated by RUNX1 at the level of the HSPC (Table 1). The prospect of modulating the activity of these genes provides potential candidates for creating therapies against diseases caused by the loss of RUNX1 function and offers an intriguing look into the mechanisms of how perturbed RUNX1 function may result in disease.
All of these genes have diverse roles in mediating various functions both inside and outside of the hematologic system. Briefly, Csf2rb codes for the common β-chain subunit found in the granulocyte macrophage colony-stimulating factor (GM-CSF), IL-3, and IL-5 receptors and is important in regulating specific cytokine responses.37 Gzmb and Igf2r have both been implicated in regulating cytotoxic T cell–mediated apoptosis.38 Lcp2 is necessary for T-cell development.39 Another subset of genes are involved in cell-to-cell interactions in blood cells, like Alcam40 and Jam3,41 and may help to mediate interactions between HSPCs and the stem cell niche. These are just a few examples of genes that have blood-specific functions. Others such as Krt8042 and Tjp143 have described functions primarily in nonhematopoietic cells. Some genes such as Fhdc1 and Zcchc18 have biological roles that are relatively unstudied or unknown. The diversity of functions in this list further confirms that RUNX1 is a master transcription factor and that disrupting RUNX1 function can have a variety of consequences resulting from disruption of its target genes. Ultimately, our analysis exposed the oncogene Hmga2 for further study. HMGA2 has known roles in mediating cellular proliferation, and a recent study described the role of HMGA2 in both fetal and adult HSPCs, but interactions between RUNX1 and HMGA2 have remained largely unexplored.13,36 The connection of these 2 genes in regulating various hematopoietic processes is a highly interesting avenue for further investigation.
Promoter-luciferase assays demonstrated that Hmga2 is directly regulated by RUNX1, but the regulation is cell type–specific, as shown by the contrasting results in a variety of cell lines. Cell lines like K562 and Jurkat cells express different sets of transcriptional regulators and cofactors that may not be present in adherent cell lines like NIH3T3 and 293T cells, or more myeloid-like lines like U937 cells. Various members of the ETS and GATA families offer just a few examples of transcription factors that primarily function in the hematopoietic system. These hematopoietic-specific transcription factors may collaborate with RUNX1 to result in cell-specific regulation of Hmga2. Furthermore, the factors cooperating with RUNX1 in regulating Hmga2 expression can differ in HSPCs, differentiated hematopoietic cells, or leukemia cells. Identification of which partner factor(s) RUNX1 interacts with in these various contexts is a topic of ongoing study.
For the first time, distal regulatory regions in and around the Hmga2 locus were demonstrated to have effects on Hmga2 expression. Interestingly, the effects exerted by the upstream and intron regions were constant and did not depend on the cell type in which they were tested. When these regions were added to the promoter-luciferase assays, the upstream and intron regions were associated with transcriptional activation and repression, respectively, but did not affect RUNX1-mediated transcription specific to each cell line tested. Hence, the upstream and intron regions most likely serve as enhancer and silencers of Hmga2, respectively. Another possibility is that RUNX1 binding to these regulatory elements contributes to the regulation of endogenous Hmga2 gene expression in HSPCs. Under conditions of transient transfection, however, the relatively higher levels of exogenous RUNX1 expression is sufficient for binding with its collaborating factors at the Hmga2 proximal promoter region, which may overwhelm its effects from the binding to these other regulatory regions.
In addition to demonstrating that Hmga2 is a transcriptional target of RUNX1, we showed that it contributes to GMP and CMP expansion resulting from loss of RUNX1 function. Importantly, the GMP population has often been associated with harboring leukemia stem cells in a variety of different leukemias.44-46 A recent study focused specifically on the RUNX1-ETO (RE) fusion protein described the ability of RE to induce expansion of the GMP population and that the GMP RE-expressing population can induce a leukemia-like state in an in vivo mouse model.46 Loss of RUNX1 in various conditional knockout models has also been shown to expand both the GMP and CMP populations and induce altered hematopoietic states.18,19 Hence, loss of RUNX1 function either through mutation or by involvement in chromosomal translocation leads to an increase in cells capable of eventually transitioning to leukemia. Without RUNX1 acting as a tumor suppressor, Hmga2 is allowed to become upregulated to induce a premalignant state. Limiting the expansion of GMPs by decreasing or regulating the amounts of HMGA2 may provide another method of controlling leukemia progression through keeping the number of leukemia stem cells in check.
Various cases of HMGA2 disruption associated with hematologic disease have been described and are typically associated with perturbing the myeloid lineages.47-49 Notably, some mutations result in truncation of the 3′ untranslated region of HMGA2. This region of HMGA2 contains binding sites for the let-7 family of miRNAs, which target a variety of cellular mediators and is a major regulator of HMGA2 levels.50,51 Let-7 family members have been reported to be lower in RE-positive leukemia patients and mouse models of RE,52,53 but various large gene expression studies in RE-positive leukemia patients have reported contrasting levels of HMGA2 when compared with non-RE–positive leukemia patients (The Cancer Genome Atlas). No comprehensive study has been conducted comparing the expression of let-7 family members or HMGA2 in myelodysplastic syndrome or myeloproliferative neoplasm patients based on RUNX1 mutation status, but such an investigation offers an interesting future direction to further establish the role HMGA2 in the context of RUNX1 loss-of-function. In summary, our study establishes Hmga2 as a target gene of RUNX1 and that Hmga2 mediates myeloid progenitor expansion caused by loss of RUNX1 function.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Nancy Speck for providing the Runx1 conditional knockout mice and Dr Kiran Chada for providing Hmga2 knockout mice, Feng Yue and Uli Wagner from Dr Bing Ren’s laboratory for help with high throughput sequencing and analyses, and everyone in the Zhang Laboratory and Dr Daniel Tenen for their helpful and enlightening discussions.
The work was supported by funding from the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (DK080665, DK098808) and a National Research Service Award (HL103106) (K.L.).
Authorship
Contribution: K.L. designed, performed, and analyzed experiments and wrote the manuscript; A.M., R.D., Y.H., A.G.S., M.Y., S.M., and S.W. performed or helped with experiments; H.H. analyzed data; and D.-E.Z. designed and analyzed experiments and supervised manuscript preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for Y.H. and H.H. is Department of Hematology, Juntendo University School of Medicine, Tokyo, Japan.
The current affiliation for S.M. is Department of Medicine, Boston University School of Medicine, Boston, MA.
Correspondence: Dong-Er Zhang, Moores UCSD Cancer Center, University of California–San Diego, 3855 Health Sciences Dr, La Jolla, CA 92093; e-mail: d7zhang@ucsd.edu.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal