Key Points
AML patients with isolated trisomy 13 have a very poor clinical outcome
Isolated trisomy 13 in AML is associated with a high frequency of mutations in SRSF2 (81%) and RUNX1 (75%)
Abstract
In acute myeloid leukemia (AML), isolated trisomy 13 (AML+13) is a rare chromosomal abnormality whose prognostic relevance is poorly characterized. We analyzed the clinical course of 34 AML+13 patients enrolled in the German AMLCG-1999 and SAL trials and performed exome sequencing, targeted candidate gene sequencing and gene expression profiling. Relapse-free (RFS) and overall survival (OS) of AML+13 patients were inferior compared to other ELN Intermediate-II patients (n=855) (median RFS, 7.8 vs 14.1 months, P = .006; median OS 9.3 vs. 14.8 months, P = .004). Besides the known high frequency of RUNX1 mutations (75%), we identified mutations in spliceosome components in 88%, including SRSF2 codon 95 mutations in 81%. Recurring mutations were detected in ASXL1 (44%) and BCOR (25%). Two patients carried mutations in CEBPZ, suggesting that CEBPZ is a novel recurrently mutated gene in AML. Gene expression analysis revealed a homogeneous expression profile including upregulation of FOXO1 and FLT3 and downregulation of SPRY2. This is the most comprehensive clinical and biological characterization of AML+13 to date, and reveals a striking clustering of lesions in a few genes, defining AML+13 as a genetically homogeneous subgroup with alterations in a few critical cellular pathways. Clinicaltrials.gov identifiers: AMLCG-1999: NCT00266136; AML96: NCT00180115; AML2003: NCT00180102; and AML60+: NCT00893373
Introduction
Acquired isolated trisomy 13 (+13) is a rare cytogenetic alteration in acute myeloid leukemia (AML). In a retrospective study of 22 856 AML patients from the Mayo Clinic, its incidence was 0.7%.1 So far, the prognostic relevance of AML+13 has not been extensively studied, but assumed to be unfavorable based on small or heterogeneous patient cohorts.2-4 However, according to the European LeukemiaNet (ELN) classification, AML+13 is currently classified in the Intermediate-II genetic group.5 AML+13 is frequently associated with FAB M0 morphology and shows a high frequency (80% to 100%) of RUNX1 mutations.6,7 Overexpression of FLT3 (located in band q12 on chromosome 13) due to a gene dosage effect was proposed as a potential mechanism of leukemogenesis in AML+13.6,7 The possibility that AML+13 might be a marker for treatment response to lenalidomide has recently been raised.8
Constitutional aneuploidy is linked to increased cancer risk.9 For example, Down syndrome (trisomy 21) predisposes to megakaryoblastic leukemia with a high frequency of acquired GATA1 mutations.10 Trisomy 13 (Patau syndrome) is a severe congenital disorder with cerebral, cardiac, and renal malformations.11 An association of Patau syndrome and solid neoplasms including neuroblastoma and nephroblastoma was reported.12 In the literature, we found a single case report of Patau syndrome with congenital myeloid leukemia.13 Considering that the vast majority of infants with Patau syndrome die before 1 year of age,11 it remains unclear whether constitutional trisomy 13 predisposes to myeloid neoplasia.
We set out to characterize the clinical course of AML+13 patients and to elucidate the underlying spectrum of molecular genetic changes by exome sequencing, targeted sequencing, and gene expression profiling.
Materials and methods
Patients
In this analysis, a subgroup of patients enrolled in the German AML Cooperative Group (AMLCG) (NCT00266136) multicenter AMLCG-1999 trial, and the AML96, AML2003, and AML60+ trials of the Study Alliance Leukemia (SAL) was studied (for details, see supplemental Figure 1A-B on the Blood Web site).14-17 All patients received intensive induction chemotherapy as described elsewhere.14-17 The AMLCG and SAL clinical trials were approved by the local institutional review boards of all participating centers and informed consent was obtained from all patients in accordance with the Declaration of Helsinki.
Exome sequencing
To perform exome sequencing, genomic DNA of available paired diagnostic and remission samples was extracted from archived bone marrow (BM) samples and fragmented for library preparation as described previously.18,19 Protein-coding regions were enriched using the SureSelect Human All Exon V4 Kit (Agilent), followed by multiplexed 80 bp paired-end sequencing on an Illumina Genome Analyzer IIx. In total, at least 3.2 Gb of raw sequence data were generated per sample (mean 3.5 Gb; quality metrics are summarized in supplemental Table 1). Raw sequence reads were filtered by Illumina’s chastity filter and mapped to the NCBI human hg19 RefSeq reference genome using BWA mapper with default parameters.20 Insufficiently mapped sequence reads (cutoff Q13, according to 95% confidence of correct mapping) and polymerase chain reaction (PCR) duplicate reads were removed using SAMtools21 ; realignment of mapped reads was performed using the Genome Analysis Toolkit to reduce false-positive single nucleotide variant calls.22 Candidates for somatically acquired mutations were detected using VarScan with the following parameters: coverage ≥ 10×, variant allele frequency ≥ 20%, variant base calling quality ≥ Q13, and variant reads ≥ 3.23 Positions with evidence for a variant in the corresponding remission sample or annotated polymorphism (as listed in dbSNP v135) were excluded.
Targeted amplicon sequencing
A selection of genes identified by exome sequencing (n = 9) and a panel of genes recurringly mutated in AML (n = 42) were studied by targeted amplicon sequencing (Haloplex; Agilent) in all AMLCG AML+13 patients with available material (16 of 23). The resulting libraries were sequenced in a single run on a MiSeq instrument. Sequence data were aligned to the human reference genome (version hg19) using BWA.20 Single nucleotide variants and short insertions or deletions were called using VarScan 2 and Pindel, respectively.24,25
In addition, Sanger sequencing of genomic DNA was performed for additional validation of selected mutations. Primer sequences and PCR conditions (for SRSF2) are shown in supplemental Tables 2 and 3). PCR products were purified using NucleoFast 96 PCR Clean-up Kit (Macherey Nagel, Düren, Germany) and bi-directional sequencing was performed on an ABI 3500xL Genetic Analyzer using the BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). Sequences were aligned and compared with the reference sequences (NCBI accession numbers: NC_000002.11 [CEBPZ], NG_027868.1 [ASXL1], and NG_032905.1 [SRSF2]) using the Sequencher software (Gene Codes Corporation, Ann Arbor, MI)
Gene expression analysis
To further characterize the AML+13 subgroup, we compared gene expression profiles of 9 patients with AML+13 to 509 AML patients with various genetic abnormalities (except for numerical alterations affecting chromosome 13). The gene expression data set was published previously and is publicly available through the Gene Expression Omnibus Web site (GSE37642).26 Eight of 9 patients were also included in the genetic analysis. Details of sample preparation, hybridization, and image acquisition were described previously.26 For probe-to-probe set summarization, we used custom chip definition files based on GeneAnnot version 2.0 (available at http://www.xlab.unimo.it/GA_CDF/) as reported before.18 Only the 17 389 probe sets present on both the Affymetrix HG-U133A and B chips, and the HG-U133 plus 2.0 chips were included in the analysis. To eliminate the batch effect resulting from the use of different chip designs, we applied an empirical Bayesian method as described previously.27
Gene set enrichment analysis (GSEA) was performed with GSEA software (MIT) using the “c5_all” collection consisting of 1454 gene sets derived from the controlled vocabulary of the Gene Ontology project.28
Statistical analyses
All statistical analyses were performed using the R 2.12.2 and 3.0.1 software31 and routines from the biostatistics software repository Bioconductor, and SPSS version 21.0 (SPSS Inc., Chicago, IL). Two-sided Fisher’s exact test was used to compare categorical variables, while Wilcoxon Mann-Whitney U test was applied for continuous variables. Adjustment for multiple hypothesis testing was performed using the Benjamini-Hochberg procedure.32 Complete remission (CR) was defined as hematologic recovery with at least 1000 neutrophils per μL and at least 100 000 platelets per μL, and < 5% BM blasts in at least one measurement.33 Relapse-free survival (RFS) was defined as time from the date of CR until relapse, or death. Overall survival (OS) was defined as time from study entry until death from any cause. Patients alive without an event were censored at the time of their last follow-up. The prognostic impact of AML+13 was evaluated according to the Kaplan-Meier method and the log-rank test. To adjust for other potential prognostic variables, we derived multivariate Cox models for RFS and OS. The following variables were included in the models, based on their role as potential confounders and availability of data: age (as a continuous parameter), sex, BM blasts at initial diagnosis and on day 16, Eastern Cooperative Oncology Group (ECOG) performance status, white blood cell (WBC) count, platelet count, hemoglobin, serum lactate dehydrogenase (LDH) level, de novo vs secondary AML, and presence of AML+13. No variable selection technique was applied, and all variables were retained in the final models. P ≤ .05 was considered significant.
Results
Isolated trisomy 13 is associated with poor prognosis
We evaluated the cytogenetic reports of 6836 AML patients with available follow up data treated within the multicenter AMLCG-1999 and SAL trials for aneuploidy of chromosome 13. A total of 264 patients (3.9%) lacked sufficient cytogenetic data. Additional copies of chromosome 13 were reported in 99 of 6572 patients (incidence, 1.5%). Our analyses focused on patients with isolated trisomy (n = 33) or tetrasomy 13 (n = 1) (incidence, 0.5%). Patients with additional numerical alterations of the sex chromosomes (n = 2) were included. These 34 patients (AML+13) were categorized into the Intermediate-II genetic category according to the ELN recommendations.5 The remaining 65 patients had heterogeneous additional cytogenetic aberrations (aAML+13), frequently in the context of a complex karyotype, and were mostly classified as “adverse” according to ELN criteria (Favorable, n = 1; Intermediate-II, n = 20; Adverse, n = 44). AML+13 patients (n = 34 [AMLCG, n = 23; SAL, n = 11]) were compared with 850 ELN Intermediate-II genetic group patients without +13 enrolled in the same clinical trials. Detailed patient characteristics are given in Table 1 (and separated for the AMLCG and SAL subgroups in supplemental Table 4A-B). The study design is summarized in supplemental Figure 1A-B. In the combined data set, AML+13 patients were significantly older (P = .004) and had higher initial BM blast counts (P = .02), but significantly lower LDH levels (P = .009) than other patients in the ELN Intermediate-II genetic group. AML+13 and aAML+13 patients had similar baseline characteristics, except for significantly lower LDH levels and a higher CR rate in AML+13 and lower platelet counts than aAML+13 (supplemental Table 4C).
Patient characteristics
| Variable . | AML+13* . | Control Group* . | P . |
|---|---|---|---|
| No. of patients | 34 | 850 | |
| Median age, years (range) | 64 (43-80) | 59 (17-84) | .004 |
| Male sex, no. (%) | 24 (70) | 465 (55) | .08 |
| WBC count, G/l, median (range) | 10 (1-318) | 11 (0.1-365) | .64 |
| Hemoglobin, g/dl, median (range) | 8.9 (4.6-12.8) | 9.2 (2.9-17.2) | .2 |
| Platelet count, G/l, median (range) | 77 (1-399) | 54 (1-1760) | .23 |
| LDH (U/l), median (range) | 269 (155-1011) | 414 (115-11140) | .009 |
| BM blasts, %, median (range) | 80 (11-100) | 68 (11-100) | .02 |
| BM blasts at day 16, %, median (range) | 5 (0-85) | 9 (0-100) | .78 |
| Performance status (ECOG) ≥ 2 (%) | 8 (26) | 263 (34) | .44 |
| de novo AML (%) | 26 (76) | 646 (76) | 1.0 |
| Allogeneic transplantation, no. (%) | 6 (18) | 180 (21) | .83 |
| CR, no. (%) | 21 (62) | 471 (55) | .49 |
| Relapse, no. (%) | 18 (86) | 327 (69) | .14 |
| Deceased, no. (%) | 31 (91) | 644 (76) | .04 |
| Variable . | AML+13* . | Control Group* . | P . |
|---|---|---|---|
| No. of patients | 34 | 850 | |
| Median age, years (range) | 64 (43-80) | 59 (17-84) | .004 |
| Male sex, no. (%) | 24 (70) | 465 (55) | .08 |
| WBC count, G/l, median (range) | 10 (1-318) | 11 (0.1-365) | .64 |
| Hemoglobin, g/dl, median (range) | 8.9 (4.6-12.8) | 9.2 (2.9-17.2) | .2 |
| Platelet count, G/l, median (range) | 77 (1-399) | 54 (1-1760) | .23 |
| LDH (U/l), median (range) | 269 (155-1011) | 414 (115-11140) | .009 |
| BM blasts, %, median (range) | 80 (11-100) | 68 (11-100) | .02 |
| BM blasts at day 16, %, median (range) | 5 (0-85) | 9 (0-100) | .78 |
| Performance status (ECOG) ≥ 2 (%) | 8 (26) | 263 (34) | .44 |
| de novo AML (%) | 26 (76) | 646 (76) | 1.0 |
| Allogeneic transplantation, no. (%) | 6 (18) | 180 (21) | .83 |
| CR, no. (%) | 21 (62) | 471 (55) | .49 |
| Relapse, no. (%) | 18 (86) | 327 (69) | .14 |
| Deceased, no. (%) | 31 (91) | 644 (76) | .04 |
Significant P values are indicated in bold.
All patients were enrolled in the AMLCG-99 or SAL trials and received intensive induction treatment. All patients are classified as ELN Intermediate-II; AML+13: patients with isolated tri- or tetrasomy 13, additional aberrations of the sex chromosomes are allowed.
Twenty-one AML+13 patients (62%, 95% confidence interval [CI]: 44% to 77%) reached CR, compared with 471 (55%, 95% CI: 52% to 59%) of ELN Intermediate-II patients without +13 (P = .49). However, 18 of these 21 patients (86%, 95% CI: 63% to 96%) relapsed.
In the AMLCG trial, AML+13 was associated with inferior RFS and OS (median RFS = 8.7 vs 14.1 months, P = .02; median OS = 7 vs 13.9 months, P = .01; Figure 1A-B), whereas in the SAL cohort, the differences between AML+13 and other ELN Intermediate-II patients did not reach significance (RFS, P = .12; OS, P = .29; supplemental Figure 2A), possibly due to the small number of AML+13 cases (n = 11). RFS and OS in the combined SAL and AMLCG cohort were inferior for the AML+13 group compared with other ELN Intermediate-II patients (median RFS = 7.8 vs 14.1 months, P = .006; median OS = 9.3 vs 14.8 months, P = .004; Figure 1C-D).
RFS and OS in AML patients. (A-B) AMLCG cohort. (C-D) Combined AMLCG and SAL cohort. Kaplan–Meier estimates of RFS and OS are significantly reduced for the AML+13 subgroup within the ELN Intermediate-II genetic group.
RFS and OS in AML patients. (A-B) AMLCG cohort. (C-D) Combined AMLCG and SAL cohort. Kaplan–Meier estimates of RFS and OS are significantly reduced for the AML+13 subgroup within the ELN Intermediate-II genetic group.
In a multivariate analysis in the combined AMLCG and SAL cohorts that adjusted for other known prognostic markers, AML+13 remained a significant variable within the ELN Intermediate-II genetic group for OS, but not for RFS (Table 2).
Multivariate analysis
| Variable‡ . | RFS* . | OS† . | ||
|---|---|---|---|---|
| HR (95% CI) . | P . | HR (95% CI) . | P . | |
| Age (10 y increase) | 1.33 (1.21-1.46) | <.001 | 1.38 (1.27-1.5) | <.001 |
| BM blasts on day 16 (10% increase) | 1.04 (0.97-1.09) | .08 | 1.02 (1.02-1.09) | .002 |
| WBC (10 G/l increase) | 1.02 (0.99-1.05) | .15 | 1.02 (1-1.05) | .04 |
| de novo vs secondary AML | 1.02 (0.75-1.4) | .89 | 1.26 (1-1.59) | .05 |
| AML+13 | 1.47 (0.82-2.62) | .2 | 1.65 (1.03-2.63) | .04 |
| Variable‡ . | RFS* . | OS† . | ||
|---|---|---|---|---|
| HR (95% CI) . | P . | HR (95% CI) . | P . | |
| Age (10 y increase) | 1.33 (1.21-1.46) | <.001 | 1.38 (1.27-1.5) | <.001 |
| BM blasts on day 16 (10% increase) | 1.04 (0.97-1.09) | .08 | 1.02 (1.02-1.09) | .002 |
| WBC (10 G/l increase) | 1.02 (0.99-1.05) | .15 | 1.02 (1-1.05) | .04 |
| de novo vs secondary AML | 1.02 (0.75-1.4) | .89 | 1.26 (1-1.59) | .05 |
| AML+13 | 1.47 (0.82-2.62) | .2 | 1.65 (1.03-2.63) | .04 |
Significant P values are indicated in bold.
n = 378, number of events = 275 (114 patients excluded due to missing covariables).
n = 549, number of events = 410 (335 patients excluded due to missing covariables).
Only variables with P ≤ .05 in either model are shown. The following variables were included in both models: sex, age (continuous variable), BM blasts at initial diagnosis and day 16, ECOG performance status, WBC count, platelet count, hemoglobin, serum LDH level, de novo vs secondary AML, and AML+13 status.
There was no significant difference in RFS (P = .74) or OS (P = .82) between the AML+13 and aAML+13 subgroups, despite the high frequency of adverse cytogenetic alterations in the aAML+13 group (supplemental Figure 2B). We also compared the AMLCG AML+13 group (n = 23) to 463 patients treated on the AMLCG-1999 trial who had adverse cytogenetics. Baseline characteristics for these cohorts are shown in supplemental Table 4D. There was no significant difference regarding RFS (P = .78) or OS (P = .98) between both groups (supplemental Figure 2C).
High frequency of mutations affecting SRSF2, RUNX1, ASXL1, and BCOR in AML+13
To systematically identify somatic mutations associated with AML+13, we performed exome sequencing of paired diagnostic and remission samples from 2 patients with AML+13 (patients no. 8 and 11). We identified nonsynonymous leukemia-specific mutations affecting 36 genes, including RUNX1, ASXL1, BCOR, ZRSR2, NUP188, and CEBPZ. No recurring mutations were observed between the 2 patients. Nonsynonymous mutations in protein-coding transcripts are summarized in supplemental Table 5.
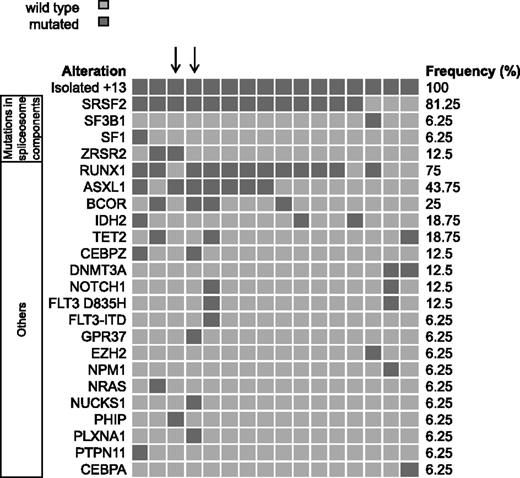
Targeted amplicon sequencing was performed on 16 AML+13 patient samples. Consistent with previous reports,6,7 we found a high frequency of RUNX1 mutations (n = 12, 75%). In addition, we detected mutations in spliceosome components in 14 AML+13 patients (88%), including SRSF2 codon 95 mutations in 13 patients (81%) and an SF3B1 mutation in 1 patient. The association of spliceosome component mutations (SRSF2, SF3B1, SF1, and ZRSR2) with RUNX1 mutations was significant (P = .05). Additional recurring mutations affected ASXL1 (n = 7, 44%) and BCOR (n = 4, 25%), and occurred with RUNX1 and SRSF2 mutations but these associations did not reach statistical significance (ASXL1-SRSF2, P = .21; ASXL1-RUNX1, P = .34; BCOR-SRSF2, P = .53; and BCOR-RUNX1, P = .53). The 2 patients without mutations in the splicing machinery had DNMT3A mutations, which were also mutually exclusive with mutations in RUNX1 or ASXL1. Two patients carried mutations in CEBPZ, thus establishing CEBPZ as a novel recurrently mutated gene in AML. Details of all detected nonsynonymous variants are shown in Figure 2 and supplemental Table 6.
Frequency distribution of recurrently mutated genes in AML+13. Distribution of mutated genes in 16 patients with AML+13. Patients show a high frequency of mutations in spliceosome components and in RUNX1, ASXL1, and BCOR. Arrows highlight the 2 patients who were exome-sequenced.
Frequency distribution of recurrently mutated genes in AML+13. Distribution of mutated genes in 16 patients with AML+13. Patients show a high frequency of mutations in spliceosome components and in RUNX1, ASXL1, and BCOR. Arrows highlight the 2 patients who were exome-sequenced.
The mutations in SRSF2 and CEBPZ were confirmed by Sanger sequencing (results summarized in supplemental Table 6). The correlation of the results from Sanger sequencing and targeted high throughput sequencing was 100% (for details, see supplemental Figure 3). In one of the patients with a CEBPZ mutation and an available remission sample, we could confirm the somatic nature of the mutation (supplemental Figure 3).
Both patients characterized by exome sequencing carried SRSF2 mutations at codon 95, as identified by amplicon sequencing. However, these mutations were not detected by exome sequencing due to low coverage of this region in both samples. These results show that our targeted sequencing approach detects mutations in AML candidate genes with high sensitivity and specificity, including mutations in regions not covered by exome sequencing.
To further explore the association between RUNX1 and SRSF2 mutations, we analyzed the SRSF2 gene in a cohort of 14 patients with a known RUNX1 mutation and normal karyotype AML (CN-AML).34 We found mutations in SRSF2 in 3 of the 14 patients (21%).
Distinct gene expression pattern of AML+13
We identified 678 probe sets as significantly (P ≤ .05 after adjustment for multiple testing) deregulated (upregulated, 492; downregulated, 186) in AML+13 patients (n = 9), when compared to AML patients with various other cytogenetic abnormalities (n = 509). Detailed patient characteristics are given in supplemental Table 7. Only 59 (8.7%) of these probe sets were localized on chromosome 13, but of those, 55 were upregulated and only 4 were downregulated. Upregulated probe sets on chromosome 13 included FOXO1, FLT3, (Figure 3A) and RB1. The strongest downregulated probe set on chromosome 13 belonged to the tumor suppressor gene SPRY2 (Figure 3B), which is a negative regulator of receptor tyrosine kinases. As described before, FLT3 is significantly upregulated in AML+13, compared with all other AML samples in our gene expression data set (P = .04). However, as shown in Figure 3A, FLT3 expression in AML shows a complex pattern with a wide range of expression levels, and AML+13 is not the only entity associated with high FLT3 levels.
Gene expression profile of AML+13. (A-B) FLT3 and SPRY2 expression in AML subgroups. Boxplot showing FLT3 (A) and SPRY2 (B) expression levels in various cytogenetic AML subgroups. The boxes indicate the upper and lower quartiles. The band within the boxes represents the median. Outliers are plotted as individual points. FLT3 expression is significantly higher in AML+13 compared with all other samples (P = .04). However, in several individual samples of various cytogenetic subgroups, FLT3 was expressed at higher levels compared with AML+13. SPRY2 expression is significantly lower in AML+13 (P < .001). (C) Clustering of AML+13 using 21 probe sets. Heatmap visualizing hierarchical clustering of AML+13 samples according to the 21 most differentially expressed probe sets (log-fold change ≥ 2 or ≤ −2 and adjusted P-value < .001) compared with AML with various other cytogenetic aberrations except for +13. All AML+13 samples cluster closely together, indicating a highly homogenous expression profile of this subgroup. (D) Regional gene expression on chromosome 13 in AML+13. Expression levels of probe sets located on chromosome 13 displayed by MACAT analysis in AML+13 patients (n = 9) compared with AML with various other cytogenetic abnormalities (except +13, n = 519). Scores for probe sets are shown as black dots. The sliding average of the 0.025 and 0.975 quantiles of the permuted scores are visualized as gray lines. The sliding average permuted scores (red line), and highlighted regions (yellow-dotted), where the score exceeds the quantiles, are plotted along chromosome 13. Despite the majority of probe sets showing elevated expression levels as expected, some regions were characterized by significantly lower expression levels.
Gene expression profile of AML+13. (A-B) FLT3 and SPRY2 expression in AML subgroups. Boxplot showing FLT3 (A) and SPRY2 (B) expression levels in various cytogenetic AML subgroups. The boxes indicate the upper and lower quartiles. The band within the boxes represents the median. Outliers are plotted as individual points. FLT3 expression is significantly higher in AML+13 compared with all other samples (P = .04). However, in several individual samples of various cytogenetic subgroups, FLT3 was expressed at higher levels compared with AML+13. SPRY2 expression is significantly lower in AML+13 (P < .001). (C) Clustering of AML+13 using 21 probe sets. Heatmap visualizing hierarchical clustering of AML+13 samples according to the 21 most differentially expressed probe sets (log-fold change ≥ 2 or ≤ −2 and adjusted P-value < .001) compared with AML with various other cytogenetic aberrations except for +13. All AML+13 samples cluster closely together, indicating a highly homogenous expression profile of this subgroup. (D) Regional gene expression on chromosome 13 in AML+13. Expression levels of probe sets located on chromosome 13 displayed by MACAT analysis in AML+13 patients (n = 9) compared with AML with various other cytogenetic abnormalities (except +13, n = 519). Scores for probe sets are shown as black dots. The sliding average of the 0.025 and 0.975 quantiles of the permuted scores are visualized as gray lines. The sliding average permuted scores (red line), and highlighted regions (yellow-dotted), where the score exceeds the quantiles, are plotted along chromosome 13. Despite the majority of probe sets showing elevated expression levels as expected, some regions were characterized by significantly lower expression levels.
A total of 21 probe sets showed highly significant deregulation (log-fold change ≥ 2 or ≤ −2 and adjusted P-value < .001) and were therefore used for clustering (supplemental Table 8). The result of the clustering is shown in Figure 3C. Consistent with the results from our genetic analysis, AML+13 shows a homogenous gene expression profile that is distinct from other AML subsets.
Surprisingly, some genes located on chromosome 13 showed significantly lower expression in AML+13 compared with patients with two copies of chromosome 13. The differential regional gene expression of AML+13 patient samples across chromosome 13 is visualized in Figure 3D (for details, see supplemental Table 9A-B). Despite the additional copy of chromosome 13, we identified several regions on chromosome 13 with significantly reduced gene expression levels compared with patients with two copies of chromosome 13.
By using GSEA, we see a potential deregulation of gene sets associated with cytoplasmatic and nuclear transport and the regulation of transcription. Details are given in supplemental Table 10. We could also observe that the expression levels of the transcription factor FOXO1 correlated with higher expression levels of a predefined gene set consisting of target genes of this transcription factor (nominal P-value: .02; false discovery rate: .23). In summary, our gene expression studies reveal a complex picture of deregulated genes in AML+13 patients with a potential role in leukemogenesis. Some of these genes, such as SPRY2 (Figure 3B) are downregulated despite their location on chromosome 13.
Finally, we compared the results of our gene expression analysis with data derived from the comparison of RUNX1-mutated and wild type AML with CN-AML.34 This 85 gene RUNX1 signature showed an overlap of 28 genes (33%) with differentially expressed genes in AML+13 (supplemental Table 11).
Discussion
Our study is the first to show that AML+13 patients have a significantly inferior RFS and OS compared with patients with other intermediate-risk cytogenetic abnormalities in a homogeneously treated cohort. Based on these findings, AML+13 should be considered as a subgroup associated with an extremely poor outcome. Furthermore, we provide evidence that AML+13 leukemia is genetically homogenous, not only on the cytogenetic but also on the molecular level. AML+13 is not only associated with a high frequency of RUNX1 mutations, but also with mutations in SRSF2, ASXL1, and BCOR. To our knowledge, the incidence of mutations in SRSF2 in AML+13 is the highest of any AML or myelodysplastic syndrome (MDS) subgroup reported so far.35,36 An association between SRSF2 and RUNX1 mutations was already reported in patients with MDS.35 We provide first evidence that an association between these mutations could also be observed in AML with RUNX1 mutations. However, larger studies are necessary to verify this observation.
It is intriguing to speculate about functional interactions between mutations in these two genes and trisomy 13. It remains unclear whether mutations targeting SRSF2 and RUNX1, and trisomy 13, affect a common pathway or different but complementary pathways on the way to leukemia. Although one of these lesions likely represents a near compulsory additional hit required by the initial event, the order of these events remains elusive. In light of the high prevalence of acquired GATA1 mutations in AML of Down syndrome patients,10 it is very likely that the chromosomal aneuploidy is the first event and determines the subsequent acquisition of mutations in precisely defined genes.
There is some, but limited overlap of recurrently mutated genes in AML and MDS. However, the high incidence of spliceosome gene mutations in both MDS and AML+13 is striking. A case report of 2 AML+13 patients who achieved sustained complete morphologic and cytogenetic remission while treated with high-dose, single-agent lenalidomide suggests a potential role of spliceosome gene mutations in the response to lenalidomide, which is also used in MDS therapy.8 Otrock et al recently reported an association of lenalidome response with distinct mutation patterns.37
Of note, only one SRSF2 mutation was found in 200 AML patients studied by whole exome or whole genome sequencing.38 This SRSF2-mutated patient also had a RUNX1 mutation. The study included a total of 19 RUNX1-mutated patients.38 As is obvious from our study, it is likely that some SRSF2 mutations in this study might have gone undetected, since exome sequencing may miss these mutations due to inefficient target enrichment.
It was proposed that overexpression of FLT3, which localizes to chromosome 13, could play a crucial role in AML+13.6,7 Our study confirms an elevated expression level of FLT3 in the AML+13 subgroup. However, the levels are similar to other cytogenetic AML subgroups without additional chromosome 13, showing that high FLT3 expression levels are not a defining feature of AML+13. Nevertheless, these findings do not rule out that high FLT3 expression levels are an important leukemic driver in AML+13. High FLT3 expression levels might be achieved by other mechanisms than an additional copy of chromosome 13 in other leukemias. Our gene expression analysis suggests several possible alternative or additional consequences of trisomy 13. FOXO1 is overexpressed in AML+13, and GSEA revealed upregulated sets of FOXO1 target genes. Recurrent mutations in FOXO1 associated with poor survival were recently discovered in diffuse large B-cell lymphoma.39 Furthermore, activation of FOXO1 was observed in ∼40% of AML patients.40 Inhibition of FOXO1 leads to reduced leukemic cell growth.40 The tumor suppressor gene SPRY2, a negative regulator of receptor tyrosine kinases, had strikingly low expression levels even though it is located on chromosome 13 (Figure 3B). Downregulation of SPRY2 was previously reported in a variety of solid tumors.41-44 It is challenging to explain the underlying mechanism for this apparently contradictory result (ie, the downregulation despite an additional gene copy). Potential mechanisms for low SPRY2 expression include epigenetic inactivation, submicroscopic deletions of SPRY2, or mutations in upstream regulators of SPRY2. These results again demonstrate the complexity of gene regulation and indicate that the concept of gene dosage is inadequate to explain all effects of an additional chromosome 13. Our gene expression data show a distinct gene expression profile of AML+13 partially overlapping with RUNX1- mutated CN-AML.
The striking association of mutations affecting only a few distinct genes in AML+13 suggests a strong synergism of these lesions during leukemogenesis. The fact that mutations in RUNX1, ASXL1, and upregulation of FLT3 were previously reported as markers of poor prognosis in AML clearly suggests that the combination of these lesions is responsible for the extremely poor outcome of AML+13.
In summary, we discovered the highest incidence of SRSF2 mutations in a specific AML subgroup reported so far. This rare, but genetically extremely homogenous group of AML+13 leukemia is characterized by concurrent mutations of SRSF2 and RUNX1, as well as a specific gene expression profile. Consistent with other studies, our findings suggest a connection between mutations of RUNX1 and SRSF2 in myeloid leukemogenesis. AML+13 is associated with inferior survival despite intensive treatment. Therefore, new treatment strategies are highly warranted.
The discovery of rare, genetically homogenous AML subgroups indicates that the genetic complexity of AML is extremely high but mutations do not occur randomly. Despite the increasing number of comprehensively characterized AML cases, the understanding of oncogenic collaboration poses a challenge ahead.
Presented in abstract form at the 55th annual meeting of the American Society of Hematology, New Orleans, LA, December 7-10, 2013.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all participants and recruiting centers of the AMLCG and SAL trials.
This work was supported by a grant from the German Cancer Aid (109031) to P.A.G. and S.K.B., and start-up funding from the Ludwig-Maximilians-Universität to T.H. and K.H.M. (FöFoLe 798/774 and 783). S.K.B., K.S., and P.A.G. acknowledge support from the German Research Council (DFG) (Collaborative Research Center 684 Molecular Mechanisms of Normal and Malignant Hematopoesis, projects A6, A12, and start-up funding 2011).
Authorship
Contribution: T.H., K.H.M., and. P.A.G. conceived and designed the experiments; T.H., K.H.M., L.H., E.Z., B.K., and S.K. performed experiments; T.H., K.H.M., S.V., M.H., and V.J. analyzed data; S.V. and A.G. provided bioinformatics support; H.B. managed the Genome Analyzer IIx platform; B.K., A.D., E.Z., Z.P., P.M.K., S.S., S.K.B., and K.S. characterized patient samples; M.C.S., W.E.B., T.B., B.J.W., and W.H. coordinated the AMLCG clinical trial; P.A.G., U.M., K.S., and S.K.B. supervised the project; T.H., K.H.M., S.K.B., and P.A.G. wrote the manuscript; and C.R., F.S., M.B., and G.E. coordinated the SAL clinical trials, selected, contributed, and analyzed SAL data.
Conflict-of-interest disclosure: P.A.G. and S.K. received honoraria from Illumina. The remaining authors declare no competing financial interests.
Correspondence: Philipp A. Greif, University Hospital Grosshadern, Ludwig-Maximilians Universität, Marchioninistr. 15, 81377 Munich, Germany; e-mail: pgreif@med.uni-muenchen.de and p.greif@dkfz-heidelberg.de.