Key Points
EBV infection leads to PRMT5 overexpression and global epigenetic changes that are essential to drive B-lymphocyte transformation.
Highly selective PRMT5 inhibitors represent a novel, first-in-class drug that restores critical regulatory checkpoints in lymphoma cells.
Abstract
Epigenetic events that are essential drivers of lymphocyte transformation remain incompletely characterized. We used models of Epstein-Barr virus (EBV)–induced B-cell transformation to document the relevance of protein arginine methyltransferase 5 (PRMT5) to regulation of epigenetic-repressive marks during lymphomagenesis. EBV+ lymphomas and transformed cell lines exhibited abundant expression of PRMT5, a type II PRMT enzyme that promotes transcriptional silencing of target genes by methylating arginine residues on histone tails. PRMT5 expression was limited to EBV-transformed cells, not resting or activated B lymphocytes, validating it as an ideal therapeutic target. We developed a first-in-class, small-molecule PRMT5 inhibitor that blocked EBV-driven B-lymphocyte transformation and survival while leaving normal B cells unaffected. Inhibition of PRMT5 led to lost recruitment of a PRMT5/p65/HDAC3-repressive complex on the miR96 promoter, restored miR96 expression, and PRMT5 downregulation. RNA-sequencing and chromatin immunoprecipitation experiments identified several tumor suppressor genes, including the protein tyrosine phosphatase gene PTPROt, which became silenced during EBV-driven B-cell transformation. Enhanced PTPROt expression following PRMT5 inhibition led to dephosphorylation of kinases that regulate B-cell receptor signaling. We conclude that PRMT5 is critical to EBV-driven B-cell transformation and maintenance of the malignant phenotype, and that PRMT5 inhibition shows promise as a novel therapeutic approach for B-cell lymphomas.
Introduction
Genomic instability and acquired genetic mutations have been well documented as relevant drivers of the multistep oncogenic process.1 Although much progress has been made in understanding the complex interplay between genetic, inflammatory, and microenvironmental variables in cancer etiology, the contribution of aberrant epigenetic regulation in driving initiation and maintenance of malignancy remains incompletely characterized. A major limiting factor in documenting the progression of aberrant epigenetic activity that occurs during cellular immortalization and transformation has been the lack of spontaneous, reproducible models.
The Epstein-Barr virus (EBV) is a human, B-lymphotropic, γ-herpesvirus associated with the development of B-cell lymphomas.2 Here, we address the significance of the dysregulation of an epigenetic modifier, the protein arginine methyltransferase 5 (PRMT5), during the transformation process using EBV as a tool to drive B-lymphocyte immortalization and maintenance of the malignant phenotype.
The human genome encodes 11 protein arginine methyltransferases (PRMTs) that covalently modify arginine residues in histone and nonhistone proteins contributing to a wide variety of cellular regulatory networks.3-6 Both type I and II PRMTs catalyze monomethylation at the ω-NH2 of arginine, however, they differ in their ability to add the second methyl group, either asymmetrically (type I) or symmetrically (type II).3,4,6 PRMT5 is a type II arginine methyltransferase that catalyzes symmetric dimethylation of histone proteins H3 (S2Me-H3R8) and H4 (S2Me-H4R3) altering chromatin structure to promote transcriptional target gene silencing.7-16 A growing number of nonhistone proteins involved in the control of multiple regulatory networks, including the EBV-encoded gene products EBNA1 and EBNA2, have also been identified to interact with PRMT5.4,17-21
PRMT5 overexpression is involved in the proliferation and survival of mantle cell lymphoma (MCL)15 and diffuse large B-cell lymphoma (DLBCL) cells.22 Although these studies demonstrated direct association between PRMT5 and malignant B-cell survival and proliferation, the overall relevance of PRMT5 overexpression during the transformation process remains unclear.
Here, we demonstrate PRMT5 overexpression to be dependent on a latent membrane protein 1 (LMP1)–driven nuclear factor-κB (NF-κB)–repressive complex recruited to the microRNA96 (miR96) promoter which silences this regulatory miR and promotes PRMT5 overexpression, its translocation to the B lymphoblast nucleus and transcriptional silencing of critical tumor suppressor genes. We target this oncogenic pathway with a first-in-class, highly selective, small-molecule inhibitor of PRMT5 that allowed us to study the epigenetic events occurring during B-cell transformation. Interfering with PRMT5 activity prevents establishment of B-cell immortalization and maintenance of malignant phenotype, in part, by affecting differential NF-κB/p65 subunit association with transcriptional activation complexes that restore expression of tumor suppressors. This work provides insight into the essential contribution of PRMT5 to B-cell lymphomagenesis, and provides rationale for targeting this enzyme in B-cell non-Hodgkin lymphomas (NHLs).
Methods
In vivo development of spontaneous EBV lymphomas and establishment of transformed cell lines
The severe combined immune-deficient mouse (hu-PBL-SCID) model of EBV-driven lymphomagenesis is described in detail in the supplemental Methods (available on the Blood Web site). Fully transformed lymphoblastoid cell lines (LCLs) were derived from the processing of tissues involved with tumor.
B-cell isolation and activation are described in the supplemental Methods.
EBV-induced immortalization of B lymphocytes
LCLs (“immortalized LCLs”) were obtained 30 to 45 days following in vitro infection of normal B lymphocytes with EBV-containing supernatant from the B95.8 cell line following standard protocols.23,24 The immortalized cell lines D-5, D-9, D-22, D-25, D-27, D-28, D-32, and D-33 were each obtained from different donors. Further details are provided in the supplemental Methods.
Antibodies and reagents are listed in the supplemental Methods.
Patient primary tumor samples and immunohistochemistry studies
Formalin-fixed samples were obtained from 3 patients with reactive lymph nodes (EBV−) and from 23 patients with EBV+ malignant lymphoproliferative disorders (LPDs) (8 Burkitt lymphomas, 3 plasmablastic lymphomas, 3 EBV+ DLBCLs of the elderly, 7 posttransplant DLBCLs, 1 posttransplant peripheral T-cell lymphoma, and 1 polymorphic posttransplant LPD). Additionally, tumor morphology (hematoxylin and eosin), EBV status (EBV-encoded RNA), and PRMT5 localization was assessed in primary EBV+ tumors (n = 3) that spontaneously developed in the hu-PBL-SCID mouse model of EBV-LPD. Further details are presented in the supplemental Methods.
Comparative modeling of hPRMT5 enzyme, structure-based in silico screen for hPRMT5 inhibitors, and histone methyltransferase assay
A detailed description of human PRMT5 (hPRMT5) model development and discovery of the hPRMT5 small-molecule inhibitor is provided in “Results” and the supplemental Methods.
Small interference RNA (siRNA) transfection and short hairpin RNA (shRNA) infection are described in the supplemental Methods.
Apoptosis analysis, immunofluorescence, confocal microscopy, western blot, and immunoprecipitation (IP) experiments were performed using standard techniques and are described in the supplemental Methods.
Proliferation assay
Proliferation of B cells infected with EBV ± PRMT5 shRNA lentiviral treatment was determined by tritiated thymidine incorporation as detailed in the supplemental Methods.
Transcriptome sequencing
RNAseq was performed using standard protocols including RNA integrity check, poly-A selection, and Truseq library preparation. Further details are provided in the supplemental Methods.
Quantitative real-time polymerase chain reaction (qRT-PCR) and chromatin IP (ChIP) assay were performed using standard techniques as detailed in the supplemental Methods.
Statistical analysis
To statistically validate data generated using multiple samples within different groups, analysis of variance was used to calculate the P values. To identify differentially expressed genes or recruitment of various chromatin remodelers between 2 groups, paired t tests were used to calculate the P values. In all cases, GraphPad Prism4 software was used to generate figures and P values. All the experiments were performed in triplicate unless otherwise specified.
Results
PRMT5 is overexpressed in EBV+ primary lymphomas, EBV-transformed and EBV-immortalized B cells
Immunohistochemistry of 23 primary human EBV+ lymphomas showed that both PRMT5 and associated epigenetic marks, S2Me-H4R3 and S2Me-H3R8, were markedly overexpressed with a nuclear/cytoplasmic pattern in all cases of EBV+ lymphomas and EBV-LPD as compared with normal or reactive lymph nodes (Table 1; Figure 1A).
PRMT5 expression profile in lymphomas
| Histologic subtype . | PRMT5 . | H4R3 . | H3R8 . |
|---|---|---|---|
| Burkitt lymphoma | 8/8 | 8/8 | 8/8 |
| Plasmablastic lymphoma | 3/3 | 3/3 | 3/3 |
| EBV+ DLBCL of the elderly | 3/3 | 3/3 | 3/3 |
| Monomorphic PTLD, DLBCL | 7/7 | 7/7 | 7/7 |
| Monomorphic PTLD, PTCL/NOS | 1/1 | 1/1 | 1/1 |
| Polymorphic PTLD | 1/1 | 1/1 | 1/1 |
| Total | 23/23 | 23/23 | 23/23 |
| Histologic subtype . | PRMT5 . | H4R3 . | H3R8 . |
|---|---|---|---|
| Burkitt lymphoma | 8/8 | 8/8 | 8/8 |
| Plasmablastic lymphoma | 3/3 | 3/3 | 3/3 |
| EBV+ DLBCL of the elderly | 3/3 | 3/3 | 3/3 |
| Monomorphic PTLD, DLBCL | 7/7 | 7/7 | 7/7 |
| Monomorphic PTLD, PTCL/NOS | 1/1 | 1/1 | 1/1 |
| Polymorphic PTLD | 1/1 | 1/1 | 1/1 |
| Total | 23/23 | 23/23 | 23/23 |
NOS, not otherwise specified; PTCL, peripheral T-cell lymphoma; PTLD, posttransplant lymphoproliferative disorder.
PRMT5 is overexpressed in EBV+ primary lymphomas, EBV-transformed and EBV-immortalized B cells. (A) Immunohistochemical nuclear/cytoplasmic expression of both PRMT5 and its epigenetic marks, S2Me-H3R8 and S2Me-H4R3, in Burkitt lymphoma (BL) (left column: ×400) and plasmablastic lymphoma (PL) (second column, from left; ×400); examples of nuclear dark red positivity are highlighted by black arrows and in the insets. The pattern of PRMT5, S2Me-H3R8, and S2Me-H4R3 expression in germinal centers (GCs) and mantle zone (MZ) of reactive lymph nodes is illustrated (third column, from left: ×400). Images of negative controls are shown (×400). (B) Western blot of PRMT5 expression in resting (RB), activated B cells (AB), and transformed LCLs obtained from hu-PBL-SCID mouse lymphomas generated from 4 separate donors (60A, C7M3, 100, 147). (C) Western blot of PRMT5 expression in RB, AB cells, and fully immortalized LCLs generated by infecting with EBV normal B cells from 6 separate healthy donors (C7M3, D-9, D-22, D-27, D-28, D-32). (D) Western blot of PRMT5 expression at various time points following in vitro infection of normal B cells with EBV. LCL is a fully immortalized cell line; Jurkat was used as positive control. Actin used as loading control. (E-G) Confocal microscopy of PRMT5 (E), LMP1 and EBNA2 (F), and PRMT5 epigenetic marks, S2Me-H4R3 and S2Me-H3R8 (G), at various time points following in vitro EBV infection of normal B cells. LCL is a fully immortalized cell line (G); C7M3 is a fully transformed cell line used as positive control (E).
PRMT5 is overexpressed in EBV+ primary lymphomas, EBV-transformed and EBV-immortalized B cells. (A) Immunohistochemical nuclear/cytoplasmic expression of both PRMT5 and its epigenetic marks, S2Me-H3R8 and S2Me-H4R3, in Burkitt lymphoma (BL) (left column: ×400) and plasmablastic lymphoma (PL) (second column, from left; ×400); examples of nuclear dark red positivity are highlighted by black arrows and in the insets. The pattern of PRMT5, S2Me-H3R8, and S2Me-H4R3 expression in germinal centers (GCs) and mantle zone (MZ) of reactive lymph nodes is illustrated (third column, from left: ×400). Images of negative controls are shown (×400). (B) Western blot of PRMT5 expression in resting (RB), activated B cells (AB), and transformed LCLs obtained from hu-PBL-SCID mouse lymphomas generated from 4 separate donors (60A, C7M3, 100, 147). (C) Western blot of PRMT5 expression in RB, AB cells, and fully immortalized LCLs generated by infecting with EBV normal B cells from 6 separate healthy donors (C7M3, D-9, D-22, D-27, D-28, D-32). (D) Western blot of PRMT5 expression at various time points following in vitro infection of normal B cells with EBV. LCL is a fully immortalized cell line; Jurkat was used as positive control. Actin used as loading control. (E-G) Confocal microscopy of PRMT5 (E), LMP1 and EBNA2 (F), and PRMT5 epigenetic marks, S2Me-H4R3 and S2Me-H3R8 (G), at various time points following in vitro EBV infection of normal B cells. LCL is a fully immortalized cell line (G); C7M3 is a fully transformed cell line used as positive control (E).
Primary tumors from the hu-PBL-SCID mouse model of EBV lymphoma (n = 3) also assessed by immunohistochemistry showed abundant levels of cytoplasmic and nuclear PRMT5 (supplemental Figure 1). Western blot analysis of transformed LCLs (60A, C7M3, 100, 147) confirmed PRMT5 overexpression whereas resting or activated B-lymphocyte, CD4+ T-cell, CD8+ T-cell, and monocyte preparations did not, suggesting that PRMT5 expression is associated with a malignant phenotype rather than proliferation of physiologically activated B lymphocytes (Figure 1B; supplemental Figure 2).
Western blot analysis of normal B cells either physiologically activated or infected with EBV (B95.8 strain or control nontransforming EBV strain) showed that (in contrast to resting, activated B cells or B cells infected with the nontransforming strain of EBV) immortalized LCLs (D-9, D-22, D-27, D-28, D-32) expressed abundant PRMT5 levels (Figure 1C) equivalent to that seen in transformed LCLs (Figure 1B). PRMT5 expression was consistently seen ∼8 days following infection of B cells with EBV by western blot (Figure 1D). Confocal microscopy studies documented detection of PRMT5 as early as 4 days postinfection (Figure 1E), a time that coincided with expression of the LMP1 viral oncoprotein (Figure 1F), translocation of PRMT5 from cytoplasm to nucleus as early as day 8 postinfection, and continued increase of nuclear expression intensity during the immortalization process, eventually becoming equivalent to that seen in transformed LCLs (Figure 1E). Expression of high levels of nuclear PRMT5 coincided with acquisition of global PRMT5-catalyzed histone epigenetic marks (S2Me-H4R3 and S2Me-H3R8) (Figure 1G) with relative loss of the type I PRMT1 asymmetric histone mark A2Me2-H4R3 (supplemental Figure 3). These observations led us to hypothesize that PRMT5 overexpression occurred soon after EBV infection rather than being acquired after establishment of the transformed state.
PRMT5 is a therapeutic target for B-cell NHLs
Pilot experiments evaluating the effects of shRNA knockdown of PRMT5 in LCLs led to decreased viability (supplemental Figure 4A; data not shown). EBV immortalization assays with B lymphocytes transfected with lentiviral preparations designed to express a PRMT5 complementary DNA showed enhanced viability and proliferation 5 and 12 days following infection, whereas cells transfected with an shRNA designed to knockdown PRMT5 showed reduced viability (supplemental Figure 4B-C) (P < .05). These observations supported the notion that PRMT5 overexpression may serve as an important mechanism promoting B-cell immortalization after EBV infection.
Development of a first-in-class small-molecule PRMT5 inhibitor
The initial lack of a crystal structure of hPRMT5 led us to use a comparative modeling and structure-based virtual screening approach to identify compounds that could specifically inhibit PRMT5 activity.
Multiple crystal structures of homologous arginine methyltransferases were used in the hPRMT5 model building process. The crystal structure of rat PRMT1 (rPRMT1) showed highest homology to hPRMT5 and provided the primary template for modeling. Overlay of the in silico model of the hPRMT5 catalytic domain with the rPRMT1 crystal structure25,26 showed a nearly identical alignment (Figure 2A) with a backbone α-carbon root mean squared deviation of 1.14 Å, supporting the conserved nature of the catalytic domains of this enzyme family (Figure 2A-C). Furthermore, we were able to computationally dock S-adenosyl-L-homocysteine (SAH) and substrate arginine residue successfully to the respective binding pockets within the hPRMT5 model, thus validating the catalytic site for the purpose of structure-based drug design (Figure 2C-D).
Establishment of a first-in-class PRMT5-specific inhibitor. (A) View of the crystal structure of rPRMT1 (aa 41-353, Protein Data Bank [PDB] ID 1OR8, gray ribbon representation) superimposed on the C-terminal domain of the hPRMT5 model (aa 310-637, green ribbon representation) showing the conserved nature of the PRMT family catalytic domain. The cofactor SAM and substrate arginine residue binding pockets are shown as red and blue regions, respectively. (B) rPRMT3 crystal structure displaying the cocrystallized SAH and arginine residue (purple carbon stick representation). (C) hPRMT5 model showing docked SAH and arginine residue similarly oriented in comparison with the rPRMT3 crystal structure. (D) Close-up view of SAH and arginine (stick representation; purple carbon) docked to the hPRMT5 model (green ribbon and key residues in green carbon stick representation). (E) CMP5 (stick representation; purple carbon) docked to the hPRMT5 model (green ribbon and key residues in green carbon stick representation) showing initial predicted interaction with residues. For clarity, residues covering the catalytic site face are not shown in the figures. (F) Immunofluorescence staining of JeKo cells (MCL cell line) treated with DMSO, CMP5, or CMP6 using antibodies against symmetrically (S) dimethylated S2Me-H4R8 and S2Me-H3R8. DAPI (4,6 diamidino-2-phenylindole) was used to stain nuclei. (G) Chemical structure of selective PRMT5 inhibitor CMP5 and nonreactive control CMP6. (H) Histone methyltransferase assays were performed as described in “Methods” in the presence of DMSO, CMP5 (10-100 μM), or CMP6 (10-100 μM). (I) View of the crystal structure of the C-terminal domain of hPRMT5 (aa 310-637, PDB ID 4GQB, blue ribbon representation) superimposed on model hPRMT5 (green ribbon representation). (J) Close-up catalytic site view of the superposed hPRMT5 crystal structure (blue ribbon and blue carbon stick representation) and model (green ribbon and green carbon stick representation). Cocrystallized SAM analog and substrate arginine residue are shown as yellow carbon stick and line representation, respectively. (K) View of the docked conformation of CMP5 (yellow carbon stick representation) within the active site of the optimized hPRMT5 crystal structure. Interacting amino acid residues are shown in blue stick format. (L) An equal number (0.5 × 106) of normal B cells (resting or activated) or the indicated DLBCL cell lines (Pfeiffer and SUDHL-2) were treated with increasing concentrations of CMP5, and cell viability was determined by annexin V–propidium iodide (PI) staining and flow cytometry at 24 hours. (M) Normal B cells from 3 separate healthy donors were treated with increasing concentrations of CMP5 and cell death was determined by annexin V–PI staining and flow cytometry at 24, 48, and 72 hours. (N-O) A transformed cell line (60A) and an immortalized cell line (D-9) were treated with increasing concentrations of CMP5 and cell death was determined by annexin V–PI staining and flow cytometry at 24 and 48 hours. Data are shown as the percentage of annexin V− PI− cells (live cells) and are normalized to untreated control. *P < .05; **P < .01. Error bars indicate standard error of the mean (SEM).
Establishment of a first-in-class PRMT5-specific inhibitor. (A) View of the crystal structure of rPRMT1 (aa 41-353, Protein Data Bank [PDB] ID 1OR8, gray ribbon representation) superimposed on the C-terminal domain of the hPRMT5 model (aa 310-637, green ribbon representation) showing the conserved nature of the PRMT family catalytic domain. The cofactor SAM and substrate arginine residue binding pockets are shown as red and blue regions, respectively. (B) rPRMT3 crystal structure displaying the cocrystallized SAH and arginine residue (purple carbon stick representation). (C) hPRMT5 model showing docked SAH and arginine residue similarly oriented in comparison with the rPRMT3 crystal structure. (D) Close-up view of SAH and arginine (stick representation; purple carbon) docked to the hPRMT5 model (green ribbon and key residues in green carbon stick representation). (E) CMP5 (stick representation; purple carbon) docked to the hPRMT5 model (green ribbon and key residues in green carbon stick representation) showing initial predicted interaction with residues. For clarity, residues covering the catalytic site face are not shown in the figures. (F) Immunofluorescence staining of JeKo cells (MCL cell line) treated with DMSO, CMP5, or CMP6 using antibodies against symmetrically (S) dimethylated S2Me-H4R8 and S2Me-H3R8. DAPI (4,6 diamidino-2-phenylindole) was used to stain nuclei. (G) Chemical structure of selective PRMT5 inhibitor CMP5 and nonreactive control CMP6. (H) Histone methyltransferase assays were performed as described in “Methods” in the presence of DMSO, CMP5 (10-100 μM), or CMP6 (10-100 μM). (I) View of the crystal structure of the C-terminal domain of hPRMT5 (aa 310-637, PDB ID 4GQB, blue ribbon representation) superimposed on model hPRMT5 (green ribbon representation). (J) Close-up catalytic site view of the superposed hPRMT5 crystal structure (blue ribbon and blue carbon stick representation) and model (green ribbon and green carbon stick representation). Cocrystallized SAM analog and substrate arginine residue are shown as yellow carbon stick and line representation, respectively. (K) View of the docked conformation of CMP5 (yellow carbon stick representation) within the active site of the optimized hPRMT5 crystal structure. Interacting amino acid residues are shown in blue stick format. (L) An equal number (0.5 × 106) of normal B cells (resting or activated) or the indicated DLBCL cell lines (Pfeiffer and SUDHL-2) were treated with increasing concentrations of CMP5, and cell viability was determined by annexin V–propidium iodide (PI) staining and flow cytometry at 24 hours. (M) Normal B cells from 3 separate healthy donors were treated with increasing concentrations of CMP5 and cell death was determined by annexin V–PI staining and flow cytometry at 24, 48, and 72 hours. (N-O) A transformed cell line (60A) and an immortalized cell line (D-9) were treated with increasing concentrations of CMP5 and cell death was determined by annexin V–PI staining and flow cytometry at 24 and 48 hours. Data are shown as the percentage of annexin V− PI− cells (live cells) and are normalized to untreated control. *P < .05; **P < .01. Error bars indicate standard error of the mean (SEM).
Using a structure-based computational and combinatorial lead optimization method, we used our hPRMT5 catalytic site model to screen the ChemBridge CNS-Set library of 10 000 small molecule compounds (CMPs). Virtual docking of candidate small molecules revealed a select number of structures to associate with low binding energy in S-adenosyl-L-methionine (SAM) cofactor and arginine binding pockets (identified as red and blue regions in Figure 2A, respectively). Initial testing identified 8 CMPs with lowest binding energy (supplemental Figure 5A-B) for screening assays using immunofluorescence for ability to inhibit type I or type II H4R3 methylation in JeKo cells (Figure 2F). Cellular screening assays allowed us to identify CMP5 (Figure 2G; supplemental Figure 5) as the optimal candidate inhibitor for further study. Unlike other CMPs including CMP6 (Figure 2G), with no effect on H4R3 methylation, CMP5 was found to selectively block S2Me-H4R3 by inhibiting PRMT5 methyltransferase activity (Figure 2H) on histone preparations. Moreover, CMP5 was inactive against other type I (PRMT1 and PRMT4) and type II (PRMT7) enzymes, underscoring its specificity toward PRMT5 (Figure 2H).
The initial predicted binding interactions of CMP5 with the hPRMT5 model suggested that the pyridine ring of CMP5 could form π-stacking interaction with Phe327 residue (Figure 2E), which was hypothesized to be critical for directing the ability of PRMT5 to catalyze symmetric dimethylation of arginine,27 also potentially explaining the selectivity of CMP5 for type II PRMT5 not type I PRMTs. When our in silico model was compared with the recently reported crystal structure of hPRMT5 (Figure 2I purple ribbon),28 we found our catalytic domain model to be nearly identical (Figure 2I green ribbon). Additionally, alignment of the amino acid residues within 6 Å of the catalytic site residues of our model produced a lower root mean squared deviation of 1.17 Å, demonstrating a better agreement with the catalytic site residues (Figure 2J). A few differences were also observed from the crystal structure, notably the rotamer placement of Glu444. The orientation of this Glu444 is toward the solvent-accessible surface in the model while the crystal structure orientation is rotated at a dihedral angle of 102°. Due to these differences, CMP5 docked to the hPRMT5 crystal structure showed that the carbazole ring occupied a space toward the cofactor adenosine binding position (Figure 2K) instead of the cofactor methionine subpocket initially predicted with the homology model (Figure 2E).
Cytotoxicity studies showed that treatment with increasing concentrations of CMP5 was selectively toxic to lymphoma cells (Figure 2L,N-O) (P < .05), while demonstrating limited toxicity to normal resting B lymphocytes even after prolonged incubation (48 and 72 hours, Figure 2M). In addition, whereas CMP5 treatment of LCLs led to loss of PRMT5 epigenetic marks, S2Me-H4R3 and S2Me-H3R8, it did not affect asymmetric methylation of H4R3, a type I PRMT histone mark (supplemental Figure 6). Collectively, these data demonstrate that PRMT5 inhibition leads to selective tumor cell death and that PRMT5 is an ideal therapeutic target given its selective overexpression in malignant cells.
Selective PRMT5 inhibition prevents EBV-driven B-cell immortalization and leads to global gene derepression
We performed EBV–B-cell immortalization assays using an shRNA-lentiviral PRMT5 knockdown and CMP5-based approach. Normal B cells were infected with EBV: shRNA (or control scrambled shRNA) was added to cultures on day 8 (Figure 3A-B); CMP5 (or dimethylsulfoxide [DMSO] vehicle control or nonreactive CMP6, Figure 3C) was added on days 4, 7, 14, and 21. Conditions were monitored every 3 to 5 days for proliferation/cell viability and absolute lymphocyte number. Knockdown with shRNA and selective inhibition of PRMT5 activity (Figure 3B-C) prevented EBV infection from driving B-cell survival, suggesting that PRMT5 is essential to support initiation of B-cell immortalization and maintenance of the transformed phenotype.
PRMT5 inhibition prevents EBV-driven B-cell immortalization. (A-B) Purified normal B cells were infected with EBV and cultures were exposed to either lentivirus expressing PRMT5-specific siRNA or scramble RNA control. PRMT5 expression was assessed by confocal microscopy, and cell proliferation was assessed by uptake of [3H]-thymidine and flow cytometry. (C) Purified normal B cells were infected with EBV and, at various time points (days 4, 7, 14, and 21), cultures were exposed to DMSO, highly selective PRMT5 inhibitor (CMP5, 40 µM), or nonreactive control small-molecule CMP6 (40 µM). The effect of PRMT5 inhibition on EBV+ B-cell outgrowth was measured by absolute numbers of CD19+ cells over 35 days. *P < .05. Error bars indicate SEM.
PRMT5 inhibition prevents EBV-driven B-cell immortalization. (A-B) Purified normal B cells were infected with EBV and cultures were exposed to either lentivirus expressing PRMT5-specific siRNA or scramble RNA control. PRMT5 expression was assessed by confocal microscopy, and cell proliferation was assessed by uptake of [3H]-thymidine and flow cytometry. (C) Purified normal B cells were infected with EBV and, at various time points (days 4, 7, 14, and 21), cultures were exposed to DMSO, highly selective PRMT5 inhibitor (CMP5, 40 µM), or nonreactive control small-molecule CMP6 (40 µM). The effect of PRMT5 inhibition on EBV+ B-cell outgrowth was measured by absolute numbers of CD19+ cells over 35 days. *P < .05. Error bars indicate SEM.
RNA sequencing in transformed LCLs (60A) revealed significant derepression of the genome with CMP5 treatment (402 upregulated genes at 12 hours vs 149 downregulated genes) (P < .005) and identified an enrichment for interferon regulatory factor 4 and sterol regulatory element binding protein 1/2 targets (P < .005; supplemental Figure 7). We also performed RNA-seq of PRMT5-shRNA–treated 60A cells and observed significant overlap between genes derepressed by CMP5 and genes derepressed by PRMT5-shRNA (24 genes; P < .005), confirming the specificity of our compound. Although the functional significance of the PRMT5-derepressed program remains to be elucidated, these results confirm the direct role of PRMT5 as a global transcriptional repressor.
Posttranscriptional regulation of PRMT5 expression in EBV-transformed and EBV-immortalized B cells
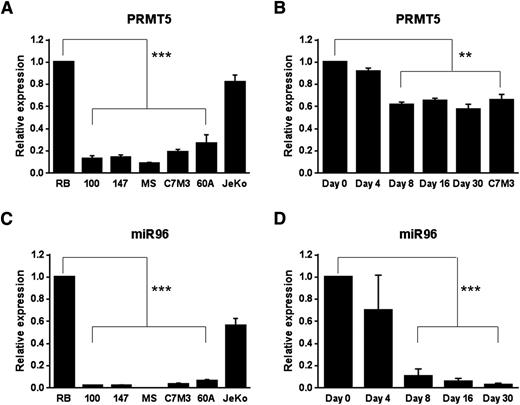
In contrast to what we observed at the protein level, transformed LCLs showed markedly reduced levels of PRMT5 transcript compared with resting B cells (P < .005) (Figure 4A). qRT-PCR on RNA harvested at various times following B-cell infection with EBV showed decrease in PRMT5 transcript as early as day 8 (Figure 4B) (P < .01), a time when protein expression was clearly observed (Figure 1D-E). We have previously reported on the posttranscriptional nature of PRMT5-induced expression via repression of miR96 in MCL.15 Similarly to what we previously observed, we found miR96 levels to be significantly lower in transformed LCLs when compared with resting B lymphocytes (P < .005) (Figure 4C). We also documented a sharp decline in miR96 levels during B-lymphocyte immortalization that appeared as early as day 4, became significant by day 8 postinfection (Figure 4D; P < .005), and remained repressed to full establishment of B-cell immortalization, suggesting a posttranscriptional nature of PRMT5-induced expression via repression of miR96.
PRMT5 transcript and miR96 are silenced in EBV-transformed cells and during B-cell immortalization. (A-B) PRMT5 messenger RNA (mRNA) expression was measured by qRT-PCR in resting B cells (RB), transformed B cells (A) and at various time points after EBV infection of normal B cells (B). C7M3 is a fully transformed LCL (B). The bar graph shows normalized fold expression of PRMT5 mRNA relative to normal B cells using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as internal control. (C-D) miR96 expression was measured by qRT-PCR in RB cells (RB), transformed B cells (C) and at various time points after EBV infection of normal B cells (D). JeKo is a MCL cell line used as control (C). The bar graph shows normalized fold expression of miR96 relative to normal B cells using GAPDH as internal control. **P < .01; ***P < .005. Error bars indicate SEM.
PRMT5 transcript and miR96 are silenced in EBV-transformed cells and during B-cell immortalization. (A-B) PRMT5 messenger RNA (mRNA) expression was measured by qRT-PCR in resting B cells (RB), transformed B cells (A) and at various time points after EBV infection of normal B cells (B). C7M3 is a fully transformed LCL (B). The bar graph shows normalized fold expression of PRMT5 mRNA relative to normal B cells using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as internal control. (C-D) miR96 expression was measured by qRT-PCR in RB cells (RB), transformed B cells (C) and at various time points after EBV infection of normal B cells (D). JeKo is a MCL cell line used as control (C). The bar graph shows normalized fold expression of miR96 relative to normal B cells using GAPDH as internal control. **P < .01; ***P < .005. Error bars indicate SEM.
LMP1-mediated NF-κB activity coordinates a repressive complex to silence miR96 and promote PRMT5 expression
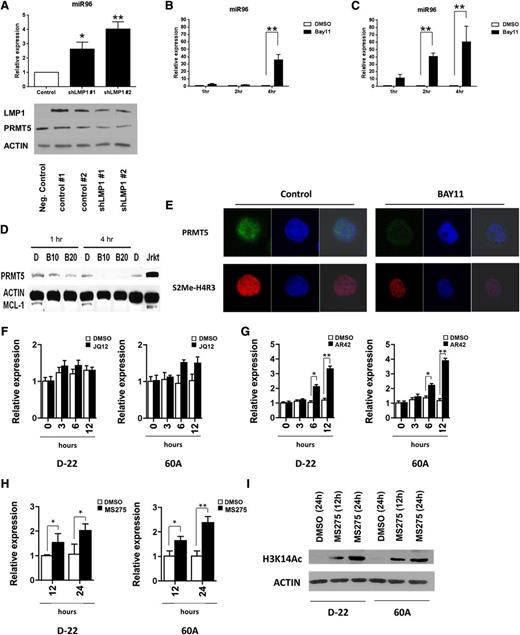
LMP1 promotes constitutive NF-κB activity and nuclear translocation of NF-κB elements to either positively or negatively regulate target gene expression.29-32 Two separate LMP1-specific shRNA preparations led to efficient LMP1 and PRMT5 knockdown (Figure 5A bottom panel) and significant upregulation of miR96 in the LCL 60A (P < .01) (Figure 5A top panel). Additionally, 4-hour incubation of transformed LCLs (SR27 and 60A) with a selective inhibitor of IkB kinase α (BAY11) led to significant (P < .01) upregulation of miR96 (Figure 5B-C) and loss of PRMT5 (Figure 5D) as well as S2Me2-H4R3 (Figure 5E), supporting the hypothesis that LMP1-driven NF-κB activity was contributing to transcriptional repression of miR96. Use of the selective histone deacetylase (HDAC)1,2 inhibitor JQ12 failed to significantly enhance miR96 expression (Figure 5F), whereas use of the broad-spectrum HDAC inhibitor AR42 and the selective HDAC1,3 inhibitor MS275 led to statistically significant (P < .05) enhanced miR96 expression (Figure 5G-H, respectively), supporting the role of HDAC3 as a critical component of a transcriptional repressive complex. Treatment of LCLs with MS275 led to hyperacetylation of H3K14, a known HDAC3 target, confirming the specificity of this inhibitor (Figure 5I).33
LMP1-mediated NF-κB activity coordinates a repressive complex to repress miR96 and promote PRMT5 expression. (A) miR96 expression was measured by qRT-PCR in a transformed LCL (60A) after LMP1 knockdown using 2 separate LMP1-specific shRNA preparations. Western blot showing the effect of LMP1 knockdown on LMP1 and PRMT5 expression is also included (bottom panel). LMP1 knockdown resulted in statistically significant increase of miR96 expression at 48 hours with both preparations. (B-C) Two transformed LCLs (60A and SR27) were incubated with the selective inhibitor of IkB kinase α (BAY11, 10 µM) and miR96 expression was evaluated by qRT-PCR. Exposure to BAY11 (10 µM) resulted in a statistically significant increase of miR96 expression at 4 hours in 60A cells and at 2 and 4 hours in SR27 compared with DMSO control. (D-E) The expression of PRMT5 and epigenetic mark, S2Me-H4R3, was evaluated by western blot and confocal microscopy in 60A cells incubated for 4 hours with BAY11 (10 µM). Actin was used as loading control and MCL-1 as control for BAY11 activity. (F-H) One immortalized (D-22) and 1 transformed LCL (60A) were incubated with a selective HDAC1,2 inhibitor (JQ12, 10 µM) (F), a broad-spectrum HDAC inhibitor (AR42, 1 µM) (G) and with a selective HDAC1,3 inhibitor (MS275, 2 µM) (H), or DMSO control. miR96 expression was evaluated by qRT-PCR at the indicated time points. The bar graph shows normalized fold expression of miR96 relative to untreated cells using GAPDH as internal control. *P < .05; **P < .01. Error bars indicate SEM. (I) Western blot of H3K14Ac in D-22 and 60A cells treated for 12 and 24 hours with MS275 (2 µM). Actin was used as loading control.
LMP1-mediated NF-κB activity coordinates a repressive complex to repress miR96 and promote PRMT5 expression. (A) miR96 expression was measured by qRT-PCR in a transformed LCL (60A) after LMP1 knockdown using 2 separate LMP1-specific shRNA preparations. Western blot showing the effect of LMP1 knockdown on LMP1 and PRMT5 expression is also included (bottom panel). LMP1 knockdown resulted in statistically significant increase of miR96 expression at 48 hours with both preparations. (B-C) Two transformed LCLs (60A and SR27) were incubated with the selective inhibitor of IkB kinase α (BAY11, 10 µM) and miR96 expression was evaluated by qRT-PCR. Exposure to BAY11 (10 µM) resulted in a statistically significant increase of miR96 expression at 4 hours in 60A cells and at 2 and 4 hours in SR27 compared with DMSO control. (D-E) The expression of PRMT5 and epigenetic mark, S2Me-H4R3, was evaluated by western blot and confocal microscopy in 60A cells incubated for 4 hours with BAY11 (10 µM). Actin was used as loading control and MCL-1 as control for BAY11 activity. (F-H) One immortalized (D-22) and 1 transformed LCL (60A) were incubated with a selective HDAC1,2 inhibitor (JQ12, 10 µM) (F), a broad-spectrum HDAC inhibitor (AR42, 1 µM) (G) and with a selective HDAC1,3 inhibitor (MS275, 2 µM) (H), or DMSO control. miR96 expression was evaluated by qRT-PCR at the indicated time points. The bar graph shows normalized fold expression of miR96 relative to untreated cells using GAPDH as internal control. *P < .05; **P < .01. Error bars indicate SEM. (I) Western blot of H3K14Ac in D-22 and 60A cells treated for 12 and 24 hours with MS275 (2 µM). Actin was used as loading control.
ChIP studies with antibodies specific for PRMT5, S2Me-H4R3, and S2Me-H3R8 showed significant enrichment of PRMT5 and its marks at the miR96 promoter in the LCL 60A (Figure 6A) (P < .01). Additionally, knockdown (shRNA) or inhibition of PRMT5 with CMP5 led to loss of recruitment of PRMT5 and its epigenetic marks on the miR96 promoter (Figure 6B-C) (P < .05) and transcriptional derepression of miR96 (Figure 6D-E) (P < .01). ChIP studies were also performed with antibodies against p65, HDAC3, and the epigenetic marks Ac-H4K8, Ac-H3K14, and Ac-H2BK12 in 60A cells treated either with PRMT5-shRNA or CMP5. PRMT5 knockdown and inhibition demonstrated enhanced recruitment of p65, loss of HDAC3, and hyperacetylation of lysine marks on histones H3, H4, and H2B, consistent with restored transcriptional activity of miR96 (Figure 6F-G).34,35 In addition, IP experiments in an immortalized LCL (D-22) and in a transformed LCL (60A) showed physical association between PRMT5 and HDAC3 (Figure 6H top panel) and between HDAC3 and p65 (Figure 6H bottom panel). Treatment with CMP5 led to disruption of this repressive complex (HDAC3, PRMT5) and loss of recruitment at the miR96 promoter with simultaneous enhanced recruitment of p65 with the histone acetyltransferase p300, enhanced acetylation of histone marks, and activation of miR96 transcription (Figure 6I). These studies document, for the first time, how NF-κB–repressive complexes driving lysine deacetylation coordinate with PRMT5 to silence a critical miR that is vital to support its own expression in lymphoma cells.
PRMT5 inhibition leads to miR96 transcriptional derepression. ChIP assays were performed on crosslinked chromatin from transformed B cells (60A and SR27) using either preimmune (PI) or the indicated immune antibodies, and the retained DNA was amplified by qPCR using miR96-specific primers and probe. Fold enrichment with each antibody was calculated relative to the PI sample. (A) ChIP findings showed significant enrichment of PRMT5 and its epigenetic marks (S2Me-H3R8 and S2Me-H4R3) at the miR96 promoter. (B-E) ChIP assays and qPCR showed that PRMT5 knockdown (shRNA) or inhibition (CMP5, 40 μM) in EBV-transformed LCL 60A led to loss of recruitment of the enzyme and its epigenetic marks to the miR96 promoter (B and C, respectively) and transcriptional derepression of miR96 (D and E, respectively). miR197 is a nonbinding miR used as control. (F-G) ChIP assays and qPCR showed that PRMT5 knockdown (shRNA) or inhibition (CMP5, 40 μM) in EBV-transformed LCL 60A led to enhanced recruitment of p65 and p300, loss of HDAC3 recruitment, and hyperacetylation of lysine marks on histones H3, H4, and H2B, changes consistent with restored transcriptional activity of miR96. ChIP experiments were repeated in SR27 cells with similar results. *P < .05; **P < .01. Error bars indicate SEM. (H) Nuclear extract (500 µg) from the immortalized cell line (D-22) and the fully transformed cell line 60A was immunoprecipitated using either preimmune (immunoglobulin G [IgG]) or anti-PRMT5 (top panel) or anti-p65 antibody (bottom panel), and bound proteins were analyzed by western blot analysis using anti-HDAC3, anti-PRMT5, anti-p300, or antip65 antibody. Input represents 50 µg of nuclear extract from indicated cell lines. Results indicate that treatment with PRMT5 inhibitor leads to physical deassociation between PRMT5 and HDAC3 with subsequent physical association between PRMT5 and p300. (I) Proposed schema for PRMT5 regulation of miR96 transcription and mir96 regulation of PRMT5 translation: LMP1-driven NF-κB–repressive complex, which includes PRMT5, p65, and HDAC3, binds to the miR96 promoter and leads to miR96 transcriptional silencing with subsequent enhanced PRMT5 translation. Inhibition of PRMT5 leads to: disruption of the PRMT5/p65/HDAC3-repressive complex, loss of recruitment to the miR96 promoter, enhanced association of p65 with the acetyltransferase p300, and recruitment of this activation complex to miR96 promoter. This leads to hyperacetylation of histone marks and miR96 re-expression which in turn leads to inhibition of PRMT5 translation and subsequent re-expression of critical regulatory/tumor suppressor genes.
PRMT5 inhibition leads to miR96 transcriptional derepression. ChIP assays were performed on crosslinked chromatin from transformed B cells (60A and SR27) using either preimmune (PI) or the indicated immune antibodies, and the retained DNA was amplified by qPCR using miR96-specific primers and probe. Fold enrichment with each antibody was calculated relative to the PI sample. (A) ChIP findings showed significant enrichment of PRMT5 and its epigenetic marks (S2Me-H3R8 and S2Me-H4R3) at the miR96 promoter. (B-E) ChIP assays and qPCR showed that PRMT5 knockdown (shRNA) or inhibition (CMP5, 40 μM) in EBV-transformed LCL 60A led to loss of recruitment of the enzyme and its epigenetic marks to the miR96 promoter (B and C, respectively) and transcriptional derepression of miR96 (D and E, respectively). miR197 is a nonbinding miR used as control. (F-G) ChIP assays and qPCR showed that PRMT5 knockdown (shRNA) or inhibition (CMP5, 40 μM) in EBV-transformed LCL 60A led to enhanced recruitment of p65 and p300, loss of HDAC3 recruitment, and hyperacetylation of lysine marks on histones H3, H4, and H2B, changes consistent with restored transcriptional activity of miR96. ChIP experiments were repeated in SR27 cells with similar results. *P < .05; **P < .01. Error bars indicate SEM. (H) Nuclear extract (500 µg) from the immortalized cell line (D-22) and the fully transformed cell line 60A was immunoprecipitated using either preimmune (immunoglobulin G [IgG]) or anti-PRMT5 (top panel) or anti-p65 antibody (bottom panel), and bound proteins were analyzed by western blot analysis using anti-HDAC3, anti-PRMT5, anti-p300, or antip65 antibody. Input represents 50 µg of nuclear extract from indicated cell lines. Results indicate that treatment with PRMT5 inhibitor leads to physical deassociation between PRMT5 and HDAC3 with subsequent physical association between PRMT5 and p300. (I) Proposed schema for PRMT5 regulation of miR96 transcription and mir96 regulation of PRMT5 translation: LMP1-driven NF-κB–repressive complex, which includes PRMT5, p65, and HDAC3, binds to the miR96 promoter and leads to miR96 transcriptional silencing with subsequent enhanced PRMT5 translation. Inhibition of PRMT5 leads to: disruption of the PRMT5/p65/HDAC3-repressive complex, loss of recruitment to the miR96 promoter, enhanced association of p65 with the acetyltransferase p300, and recruitment of this activation complex to miR96 promoter. This leads to hyperacetylation of histone marks and miR96 re-expression which in turn leads to inhibition of PRMT5 translation and subsequent re-expression of critical regulatory/tumor suppressor genes.
PRMT5 overexpression leads to recruitment and epigenetic repression of tumor suppressors ST7 and PTPROt
Genome-wide mapping performed on chromatin isolated from DLBCL cells (Pfeiffer) immunoprecipitated with S2Me-H3R8 antibody guided our identification of potential direct epigenetic targets of PRMT5 that may be relevant to the pathogenesis of EBV-driven B-cell transformation and identified the promoter of the PTPROt tumor suppressor gene to show 3.5-fold enrichment for S2Me-H3R8 (not shown). PTPROt, a tyrosine phosphatase, is a truncated form of the PTPRO gene product, and regulates the B-cell receptor (BCR) signaling proteins spleen tyrosine kinase (SYK), Lck/Yes novel tyrosine kinase (LYN), sarcoma-family kinases (SRC), and Bruton tyrosine kinase (BTK) in B-cell malignancies.36,37 PTPROt transcript and protein were found to be undetectable (P < .005) in immortalized LCLs (D-9, D-22, and D-27) by qRT-PCR and confocal microscopy, respectively (Figure 7A,C). We also noted a significant decrease in PTPROt transcript as early as day 8 postinfection and continued to decline to near undetectable levels when immortalized LCLs were established (Figure 7B) (P < .005). ChIP experiments with PRMT5 antibody confirmed direct enrichment on the PTPROt promoter (Figure 7D) (P < .005), and PRMT5 inhibition led to PTPROt transcriptional derepression (Figure 7E) (P < .05) and restoration of protein expression (Figure 7E bottom). Restoration of PTPROt led to dephosphorylation of BCR signaling proteins SYK, SRC, and BTK (Figure 7F).
PRMT5 inhibition restores regulation to the BCR pathway. (A-B) PTPROt mRNA expression was measured by qRT-PCR in resting B cells (RB), immortalized B cells generated from 3 separate donors (D-9, D-22, D-27) (A) and at various time points after EBV infection of normal B cells from 2 separate donors (D-33 and D-25) (B). The bar graph shows normalized fold expression of PTPROt mRNA relative to normal B cells using GAPDH as internal control. (C) PTPROt expression assessed by confocal microscopy in resting B cells as well as in the fully immortalized lymphoblastoid cell line (D-9). (D) ChIP assay was performed on crosslinked chromatin from immortalized (D-5, D-22) and transformed B cells (C7M3) using anti-PRMT5 antibody, and the retained DNA was amplified by qPCR using PTPROt-specific primers and probe. Fold enrichment was calculated relative to the input sample. Error bars represent standard deviation of triplicate measurements. (E) PTPROt mRNA expression and protein levels were evaluated by qRT-PCR (top) and confocal microscopy (bottom) in 2 immortalized cell lines (D-5 and D-22) incubated for 24 hours with either DMSO control or a highly selective PRMT5 inhibitor, compound 5 (CMP5, 40 µM). (F) Phosphotyrosine proteins immunoprecipitated from whole-cell extracts of 60A cells incubated for 24 hours with either DMSO or CMP5 (40 µM) were separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and probed with anti-SYK antibody (top left) and anti-pY(416)SRC and SRC antibodies (top right). Whole-cell extracts of 60A cells incubated for 24 hours with wither DMSO, CMP5 (40 µM), or ibrutinib (250 nM) were separated on SDS-PAGE and probed with anti-pBTK antibody, anti-BTK or GAPDH antibodies (bottom). *P < .05; **P < .01; ***P < .005. Error bars indicate SEM.
PRMT5 inhibition restores regulation to the BCR pathway. (A-B) PTPROt mRNA expression was measured by qRT-PCR in resting B cells (RB), immortalized B cells generated from 3 separate donors (D-9, D-22, D-27) (A) and at various time points after EBV infection of normal B cells from 2 separate donors (D-33 and D-25) (B). The bar graph shows normalized fold expression of PTPROt mRNA relative to normal B cells using GAPDH as internal control. (C) PTPROt expression assessed by confocal microscopy in resting B cells as well as in the fully immortalized lymphoblastoid cell line (D-9). (D) ChIP assay was performed on crosslinked chromatin from immortalized (D-5, D-22) and transformed B cells (C7M3) using anti-PRMT5 antibody, and the retained DNA was amplified by qPCR using PTPROt-specific primers and probe. Fold enrichment was calculated relative to the input sample. Error bars represent standard deviation of triplicate measurements. (E) PTPROt mRNA expression and protein levels were evaluated by qRT-PCR (top) and confocal microscopy (bottom) in 2 immortalized cell lines (D-5 and D-22) incubated for 24 hours with either DMSO control or a highly selective PRMT5 inhibitor, compound 5 (CMP5, 40 µM). (F) Phosphotyrosine proteins immunoprecipitated from whole-cell extracts of 60A cells incubated for 24 hours with either DMSO or CMP5 (40 µM) were separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and probed with anti-SYK antibody (top left) and anti-pY(416)SRC and SRC antibodies (top right). Whole-cell extracts of 60A cells incubated for 24 hours with wither DMSO, CMP5 (40 µM), or ibrutinib (250 nM) were separated on SDS-PAGE and probed with anti-pBTK antibody, anti-BTK or GAPDH antibodies (bottom). *P < .05; **P < .01; ***P < .005. Error bars indicate SEM.
ChIP assays using anti-PRMT5 and anti-S2Me-H4R3 antibodies confirmed the suppressor of tumorigenicity 7 (ST7) gene to be a direct target and silenced by PRMT5 in LCLs (supplemental Figure 8A) (P < .01). Western blot analysis and qRT-PCR failed to show measurable ST7 in LCLs, however, abundant ST7 transcript and protein were observed in resting B cells (supplemental Figure 8B-C) (P < .01). PRMT5 knockdown with shRNA lentivirus or inhibition with CMP5 led to transcriptional derepression of ST7 (supplemental Figure 8D). During B-cell immortalization, ST7 levels began to decline at day 16 and eventually became undetectable by day 30 postinfection (supplemental Figure 8E). These observations support the notion that EBV utilizes PRMT5 to target and silence genes with important tumor suppressor properties during B-cell immortalization, and that sustained PRMT5 activity is critical for supporting the malignant phenotype.
Discussion
PRMT5 associates with chromatin remodeling complexes containing HDACs, MBD2, and DNMT3a enzyme to silence the transcription of regulatory genes by catalyzing symmetric dimethylation of arginine residues on histone tails.7,8,15,16,38 PRMT5 is overexpressed in a variety of lymphomas, including MCL and DLBCLs, and PRMT5 knockdown leads to apoptosis by restoration of the RBL2/E2F tumor suppressor pathway, transcriptional silencing of the PRC2 genes (EZH2, SUZ12, EED) and restored expression of proapoptotic EZH2-target genes (CASP10 and DAP1).15,16,22 Furthermore, PRMT5 is necessary for CYCLIN-D1–mediated neoplastic growth and proliferation through transcriptional repression of CUL4.3 Finally, it has been recently shown that PRMT5 can activate NF-κB by physical association and dimethylation of the NF-κB subunit p6518 and that PRMT5 can mediate p53 methylation, promoting G1 arrest in response to DNA damage.4
In this study, we used in vitro and in vivo models of EBV immortalization and transformation to understand how posttranslational changes of histones impact gene expression, the mechanism of PRMT5 dysregulation, and how perturbation of these processes contributes to lymphomagenesis. It has been previously shown that early passages of immortalized LCLs have poor ability to grow in soft agar or to generate tumors in nude mice.39 However, it must be said that both immortalized LCLs and human EBV-LPD tumor cells from hu-PBL-SCID mice have been shown to induce LPD in SCID mice.40 Although in vitro–generated LCLs and fully transformed cell lines might represent different aspects of the same disease, in vitro–generated LCLs express the same viral genes as EBV lymphomas41 ; therefore, we believe that both systems represent a useful and reliable model for studying EBV-mediated B-cell oncogenesis.
We showed that PRMT5, although not expressed in normal or activated B cells, is markedly overexpressed in primary EBV+ lymphomas and LCLs, suggesting that PRMT5 overexpression is a marker of cellular transformation rather than proliferation. In addition to symmetric dimethylation of H4R3, PRMT5 also induces symmetric dimethylation of H3R8 with loss of asymmetric methylation of H4R3, a type I PRMT histone mark, indicating a global chromatin-wide repressive epigenetic change very early after EBV infection of B cells.
Development of a first-in-class small-molecule inhibitor of PRMT5 showed a selective effect on S2Me-H4R3 and S2Me-H3R8 with no effect against type I (PRMT1 and PRMT4) or other type II (PRMT7) enzymes. We also conducted cytotoxicity studies showing selective antitumor activity against LCLs. To further support the specificity of our inhibitors, we showed similar epigenetic changes and cytotoxic effects with shRNA and siRNA designed to knockdown PRMT5. Consistent with these findings, RNA-seq experiments using shRNA or CMP5 supported restored expression of multiple regulatory genes that compared similarly between shRNA and inhibitor-treated LCLs. This first-in-class drug was used here as a tool compound to explore the biological effect of PRMT5 inhibition and as a first step to promote the introduction of an entirely new therapeutic approach for patients with EBV+ lymphomas and other PRMT5-overexpressing B-cell lymphomas. It should also be highlighted that CMP5 and later generation PRMT5 inhibitors require further optimization for in vivo use.
In previous work, we described the posttranscriptional nature of PRMT5 expression via miR regulation and linked dysregulated expression of PRMT5 to aberrant expression of miR-96 in MCL.15 Here, we show that PRMT5 enhances its own expression by participating in a PRMT5/p65/HDAC3 complex that represses miR-96 transcription. Furthermore, although HDAC3 and PRMT5 confer transcriptional repressive activity to p65 at the miR-96 promoter, treatment of lymphoma cells with PRMT5 inhibitors leads to rapid dissociation of this repressive complex and differential association of p65 with p300 with enhanced recruitment to and subsequent transactivation of the miR-96 promoter. These data support the notion of a dual role of p65 as repressor and activator that is dependent on PRMT5 activity.
Our findings link PRMT5 dysregulation and aberrant miR96 expression to LMP1 signaling through NF-κB, identifying cross-talk between viral oncogene activity and host regulatory networks to affect oncogenic pathways driven by PRMT5. It has been previously shown that EBV enhances survival of infected B cells through LMP1 by upregulating the expression of antiapoptotic genes such as BFL142 and BCL2.43 Here, we demonstrate that PRMT5 overexpression is vital to the support of B-cell immortalization, transformation, and maintenance of the malignant phenotype.
Earlier studies indicated that LMP2A functions by altering normal BCR signaling through antigen-independent activation of downstream protein kinases.44-47 Although PRMT5 inhibition does not have any direct effect on the expression of EBV-encoded proteins (supplemental Figure 9), it does affect the expression of multiple cellular regulatory genes, highlighting its relevance as an epigenetic modifier capable of driving host and not viral oncogenic processes. PTPROt is a tyrosine phosphatase that specifically targets BCR-triggered tyrosine kinases.48 Here, we showed, by ChIP, direct PRMT5 recruitment to the PTPROt promoter. Additionally, PRMT5 inhibition led to PTPROt transcriptional derepression and restoration of protein expression which, in turn, led to dephosphorylation of BCR signaling kinases. PTPROt expression decreases lymphoma cell proliferation and induces apoptosis.36 We have also recently established the in vivo function of PTPROt through the generation of a transgenic mouse with B-cell–specific expression of PTPROt, and showed that PTPROt-mediated regulation of p53/Foxm1 suppresses the leukemic phenotype in a chronic lymphocytic leukemia mouse model.49 Our results suggest that PRMT5-mediated downregulation of PTPROt following EBV infection and ensuing dysregulated BCR signaling play a role in supporting the B-cell immortalization process induced by EBV.
It is becoming more evident that dysregulation of histone-modifying enzymes is associated with cancer etiology and pathogenesis. We have taken advantage of the oncogenic potential of EBV and used it to illustrate the global epigenetic repressive role played by PRMT5 during B-cell transformation. We have linked a potent oncogenic virus (EBV) to the overexpression of an epigenetic modifier (PRMT5) and characterized the critical role that PRMT5 plays in promoting its own overexpression through a repressive complex (p65/PRMT5/HDAC3) that transcriptionally silences miR96. Because of selective PRMT5 expression in malignant cells, the central role of this enzyme in regulating its own expression and its activity in transcriptionally silencing multiple regulatory genes, PRMT5 appears to be an ideal therapeutic target for lymphoma. In addition to providing an innovative strategy for EBV+ lymphomas as well as other subtypes of B-cell NHLs, this approach could be used to identify potential novel targets for therapy and will promote exploration of new combination strategies as treatment options for these diseases.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the American Society of Hematology/European Hematology Association (L.Alinari), the Leukemia & Lymphoma Society Translational Research Project (LLS TRP) (R.A.B.), Friends of Jason Gould Foundation (R.A.B., P.L.S., J.T.P.), The Ohio State University Drug Development Institute (R.A.B., C.L.), National Institutes of Health, National Institute of Neurological Disorders and Stroke grant R21NS071346 (R.A.B., C.L.) and National Cancer Institute grant R01CA116093 (S.S., R.A.B.).
Authorship
Contribution: L.Alinari designed and performed research, analyzed data, wrote and reviewed the paper, and approved the final version of the manuscript; K.V.M., F.Y., S.C.-K., and O.E. designed and performed research, analyzed data, reviewed the paper, and approved the final version of the manuscript; V.K., J.-H.C., E.M.S., C.Q., P.L.S., L.K., J.T.P., B.Y., Y.W., and S.R. performed research, analyzed data, and approved the final version of the manuscript; R.L. performed research, analyzed data, reviewed drafts, and approved the final version of the manuscript; A.D.L. and J.E.B. provided reagents, reviewed drafts, and approved the final version of the manuscript; S.P., C.A., and L.Ayers provided samples, performed research, reviewed drafts, and approved the final version of the manuscript; T.M. and S.M. performed research, analyzed data, reviewed the paper, and approved the final version of the manuscript; J.C.B. and S.J. designed research, reviewed drafts, and approved the final version of the manuscript; and S.S., C.L., and R.A.B. designed and supervised research, obtained funding for the work, reviewed drafts, and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Robert A. Baiocchi, Division of Hematology, Department of Internal Medicine, The Ohio State University, B420 Starling Loving Hall, 320 West 10th Ave, Columbus, OH 43210; e-mail: robert.baiocchi@osumc.edu; Chenglong Li, Division of Medicinal Chemistry and Pharmacognosy, The Ohio State University, 612 Riffe Building, 486 W. 12th Ave, Columbus, OH 43210; e-mail: li.728@osu.edu; and Said Sif, Department of Biological and Environmental Sciences, College of Arts and Sciences, Qatar University, PO Box 2713, Doha, Qatar; e-mail: ssif@qu.edu.qa.
References
Author notes
L. Alinari, K.V.M., and F.Y. contributed equally.
S.S., C.L., and R.A.B. contributed equally.


![Figure 2. Establishment of a first-in-class PRMT5-specific inhibitor. (A) View of the crystal structure of rPRMT1 (aa 41-353, Protein Data Bank [PDB] ID 1OR8, gray ribbon representation) superimposed on the C-terminal domain of the hPRMT5 model (aa 310-637, green ribbon representation) showing the conserved nature of the PRMT family catalytic domain. The cofactor SAM and substrate arginine residue binding pockets are shown as red and blue regions, respectively. (B) rPRMT3 crystal structure displaying the cocrystallized SAH and arginine residue (purple carbon stick representation). (C) hPRMT5 model showing docked SAH and arginine residue similarly oriented in comparison with the rPRMT3 crystal structure. (D) Close-up view of SAH and arginine (stick representation; purple carbon) docked to the hPRMT5 model (green ribbon and key residues in green carbon stick representation). (E) CMP5 (stick representation; purple carbon) docked to the hPRMT5 model (green ribbon and key residues in green carbon stick representation) showing initial predicted interaction with residues. For clarity, residues covering the catalytic site face are not shown in the figures. (F) Immunofluorescence staining of JeKo cells (MCL cell line) treated with DMSO, CMP5, or CMP6 using antibodies against symmetrically (S) dimethylated S2Me-H4R8 and S2Me-H3R8. DAPI (4,6 diamidino-2-phenylindole) was used to stain nuclei. (G) Chemical structure of selective PRMT5 inhibitor CMP5 and nonreactive control CMP6. (H) Histone methyltransferase assays were performed as described in “Methods” in the presence of DMSO, CMP5 (10-100 μM), or CMP6 (10-100 μM). (I) View of the crystal structure of the C-terminal domain of hPRMT5 (aa 310-637, PDB ID 4GQB, blue ribbon representation) superimposed on model hPRMT5 (green ribbon representation). (J) Close-up catalytic site view of the superposed hPRMT5 crystal structure (blue ribbon and blue carbon stick representation) and model (green ribbon and green carbon stick representation). Cocrystallized SAM analog and substrate arginine residue are shown as yellow carbon stick and line representation, respectively. (K) View of the docked conformation of CMP5 (yellow carbon stick representation) within the active site of the optimized hPRMT5 crystal structure. Interacting amino acid residues are shown in blue stick format. (L) An equal number (0.5 × 106) of normal B cells (resting or activated) or the indicated DLBCL cell lines (Pfeiffer and SUDHL-2) were treated with increasing concentrations of CMP5, and cell viability was determined by annexin V–propidium iodide (PI) staining and flow cytometry at 24 hours. (M) Normal B cells from 3 separate healthy donors were treated with increasing concentrations of CMP5 and cell death was determined by annexin V–PI staining and flow cytometry at 24, 48, and 72 hours. (N-O) A transformed cell line (60A) and an immortalized cell line (D-9) were treated with increasing concentrations of CMP5 and cell death was determined by annexin V–PI staining and flow cytometry at 24 and 48 hours. Data are shown as the percentage of annexin V− PI− cells (live cells) and are normalized to untreated control. *P < .05; **P < .01. Error bars indicate standard error of the mean (SEM).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/16/10.1182_blood-2014-12-619783/4/m_2530f2.jpeg?Expires=1765882970&Signature=yRTyrjDPJVscLjhMIR5f8tKWgwJIEpsRm9pERbpowDV7c6BA9XdMBIx1Q-vAIGrLjyCBBuabTV6hVXnBRuoyK9xDlAl0dZJyRWUL4g0LyfyrzfaqBM94sNtCEzyIGm1OGFV3wvrYmdppNH1cvGKPl0edBT6txnyz0K3-KVxR~Ngyuzi9UoDbPrNE23mjubibiKuM-ijw45njiq9G4mltViHg8T3xNQx72aK038ICEdbvj7po4OOJjTSY-XiLi6s72E-S85RUF07FjqpJ-UNWYNiN-lVfCV2MRwVpIAwb0WGODHtmQWTT7l28ZZ3~EgCB~HVyJEif8b51Ve0yjl9RAA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. PRMT5 inhibition prevents EBV-driven B-cell immortalization. (A-B) Purified normal B cells were infected with EBV and cultures were exposed to either lentivirus expressing PRMT5-specific siRNA or scramble RNA control. PRMT5 expression was assessed by confocal microscopy, and cell proliferation was assessed by uptake of [3H]-thymidine and flow cytometry. (C) Purified normal B cells were infected with EBV and, at various time points (days 4, 7, 14, and 21), cultures were exposed to DMSO, highly selective PRMT5 inhibitor (CMP5, 40 µM), or nonreactive control small-molecule CMP6 (40 µM). The effect of PRMT5 inhibition on EBV+ B-cell outgrowth was measured by absolute numbers of CD19+ cells over 35 days. *P < .05. Error bars indicate SEM.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/16/10.1182_blood-2014-12-619783/4/m_2530f3.jpeg?Expires=1765882970&Signature=E7Kh2q7Pn4oU10b2QgfNfPGOxkXt3k1EfsVWBUgvdpGANmA3DOIXc5xZJqROoDnxHWaSVU21mRQjNW~J2wewcRCMCqGDjxSdpoQA-GWblK4OZ2cpGhn5cfesHSWcheGv~TLaz7ctOkV7n2k5wYfv3SqZvWIIkpKzYz3E9JQSKTqZUqwqwGc-MV3R7JvV6XfpLx5XyGDuGOrJfR52aqyHCx6sOOIXnbfYo2LfFcj9EPIh4Fn47DmGEYLB-lObi7y31hvjvXC~PzzHhj9yeQTveeTZc2OjcqiZYC-f3k4NT-753p5A27EUYVd4dXGAE06s7xf8lVe3iVIgnlgQKfjYVw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. PRMT5 inhibition leads to miR96 transcriptional derepression. ChIP assays were performed on crosslinked chromatin from transformed B cells (60A and SR27) using either preimmune (PI) or the indicated immune antibodies, and the retained DNA was amplified by qPCR using miR96-specific primers and probe. Fold enrichment with each antibody was calculated relative to the PI sample. (A) ChIP findings showed significant enrichment of PRMT5 and its epigenetic marks (S2Me-H3R8 and S2Me-H4R3) at the miR96 promoter. (B-E) ChIP assays and qPCR showed that PRMT5 knockdown (shRNA) or inhibition (CMP5, 40 μM) in EBV-transformed LCL 60A led to loss of recruitment of the enzyme and its epigenetic marks to the miR96 promoter (B and C, respectively) and transcriptional derepression of miR96 (D and E, respectively). miR197 is a nonbinding miR used as control. (F-G) ChIP assays and qPCR showed that PRMT5 knockdown (shRNA) or inhibition (CMP5, 40 μM) in EBV-transformed LCL 60A led to enhanced recruitment of p65 and p300, loss of HDAC3 recruitment, and hyperacetylation of lysine marks on histones H3, H4, and H2B, changes consistent with restored transcriptional activity of miR96. ChIP experiments were repeated in SR27 cells with similar results. *P < .05; **P < .01. Error bars indicate SEM. (H) Nuclear extract (500 µg) from the immortalized cell line (D-22) and the fully transformed cell line 60A was immunoprecipitated using either preimmune (immunoglobulin G [IgG]) or anti-PRMT5 (top panel) or anti-p65 antibody (bottom panel), and bound proteins were analyzed by western blot analysis using anti-HDAC3, anti-PRMT5, anti-p300, or antip65 antibody. Input represents 50 µg of nuclear extract from indicated cell lines. Results indicate that treatment with PRMT5 inhibitor leads to physical deassociation between PRMT5 and HDAC3 with subsequent physical association between PRMT5 and p300. (I) Proposed schema for PRMT5 regulation of miR96 transcription and mir96 regulation of PRMT5 translation: LMP1-driven NF-κB–repressive complex, which includes PRMT5, p65, and HDAC3, binds to the miR96 promoter and leads to miR96 transcriptional silencing with subsequent enhanced PRMT5 translation. Inhibition of PRMT5 leads to: disruption of the PRMT5/p65/HDAC3-repressive complex, loss of recruitment to the miR96 promoter, enhanced association of p65 with the acetyltransferase p300, and recruitment of this activation complex to miR96 promoter. This leads to hyperacetylation of histone marks and miR96 re-expression which in turn leads to inhibition of PRMT5 translation and subsequent re-expression of critical regulatory/tumor suppressor genes.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/16/10.1182_blood-2014-12-619783/4/m_2530f6.jpeg?Expires=1765882970&Signature=faHvR5kTepHDS4vAwp47XgB9K4V-1o4mPp7flJ6m8SJO-VjdLGzmFKcAMGxXv7etdI6TzLXSYY~uLRnyoT6CWZkPExWhpdpBRpSMnZtexjfBf5-ORDvTcDwuno-JaXS71KHmPlMoFg9~lUSLacnvNTK06j0iHsGpCOFxP5uuOoiO-o15uYBFqdNvx7RkGO0qj9ACxa9qE7lo3RZRWgC0YyDDK7nyXkKi~2o6MNBjZls4dPiqFNxHI7npnaQ144KzAFaWo~M1h9U2P~KIpmn3LI7T1eRRCweodWq~U5G~PL8flQftTqBlU5Lsmgi8zTowYe~kseC7ToDHAQxPqifN3g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
