Key Points
The glucocorticoid receptor coordinately regulates the antiapoptotic BCL2 and proapoptotic BIM genes in pediatric ALL cells in vivo.
GR binding at a novel intronic region is associated with BIM transcription and dexamethasone sensitivity in pediatric ALL cells in vivo.
Abstract
Glucocorticoids are critical components of combination chemotherapy regimens in pediatric acute lymphoblastic leukemia (ALL). The proapoptotic BIM protein is an important mediator of glucocorticoid-induced apoptosis in normal and malignant lymphocytes, whereas the antiapoptotic BCL2 confers resistance. The signaling pathways regulating BIM and BCL2 expression in glucocorticoid-treated lymphoid cells remain unclear. In this study, pediatric ALL patient-derived xenografts (PDXs) inherently sensitive or resistant to glucocorticoids were exposed to dexamethasone in vivo. Microarray analysis showed that KLF13 and MYB gene expression changes were significantly greater in dexamethasone-sensitive than -resistant PDXs. Chromatin immunoprecipitation (ChIP) analysis detected glucocorticoid receptor (GR) binding at the KLF13 promoter to trigger KLF13 expression only in sensitive PDXs. Next, KLF13 bound to the MYB promoter, deactivating MYB expression only in sensitive PDXs. Sustained MYB expression in resistant PDXs resulted in maintenance of BCL2 expression and inhibition of apoptosis. ChIP sequencing analysis revealed a novel GR binding site in a BIM intronic region (IGR) that was engaged only in dexamethasone-sensitive PDXs. The absence of GR binding at the BIM IGR was associated with BIM silencing and dexamethasone resistance. This study has identified novel mechanisms of opposing BCL2 and BIM gene regulation that control glucocorticoid-induced apoptosis in pediatric ALL cells in vivo.
Introduction
Glucocorticoids, ie, dexamethasone and prednisolone, are commonly used to treat lymphoid malignancies including pediatric acute lymphoblastic leukemia (ALL).1,2 The current standard for combination chemotherapy adopted by the Berlin-Frankfurt-Munster (BFM) group involves an initial week of glucocorticoid monotherapy combined with a single intrathecal dose of methotrexate.3,4 Patients who respond poorly to this initial week of treatment (prednisolone poor responders) experience poorer outcome than prednisolone good responders, despite their therapy being intensified due to increased risk of relapse.5 A greater understanding of the mechanisms associated with resistance to glucocorticoids in pediatric ALL would facilitate the design of treatment strategies to overcome resistance and improve outcome.
The glucocorticoid-induced apoptotic response is mediated through the glucocorticoid receptor (GR), a member of the nuclear receptor family of ligand-dependent transcription factors.6,7 On ligand binding, the GR dissociates from the large protein complex that maintains it in an inactive conformation in the cytoplasm. On activation, the GR translocates to the nucleus and binds as a dimer to activate target genes via direct interaction with specific palindromic DNA sequences known as glucocorticoid response elements (GREs).8-10 GR binding can lead to gene transactivation by binding to multiple DNA loci including proximal promoter regions and/or distal sites.9-12 The GR can interact with histone acetyltransferases, such as cAMP-response element binding protein–binding protein (CBP) and P300, as well as other coactivators, to induce histone acetylation.13-17 The GR can also interact with the switch/sucrose nonfermentable chromatin-remodeling complex and the boundary element/insulator-binding protein CTCF to alter chromatin structure.12,18 Histone acetylation and chromatin remodeling can alter nucleosomal packaging to allow increased access of transacting factors and components of the basal transcriptional machinery to the local DNA.19,20 In addition, the GR can also repress gene transcription via DNA-independent interactions with transcription factors such as activator protein-1 and nuclear factor κB.21,22 GR-induced activation or repression of gene transcription controls apoptosis of normal and malignant lymphocytes.
A critical role for BCL2 family proteins in glucocorticoid-induced apoptosis of malignant lymphocytes has been identified by our group and others.23-25 A proapoptotic member of the BCL2 family, BIM, is upregulated on glucocorticoid stimulation, which antagonizes antiapoptotic members such as BCL2, BCL-XL, and MCL-1.26,27 Disequilibrium of pro- and antiapoptotic proteins leads to the activation of BAX, disrupting mitochondrial transmembrane potential and promoting activation of caspases.27-29 However, it is still unclear which specific glucocorticoid-regulated genes are involved in transducing the apoptotic signal, upstream of BIM/BCL2 dynamic interactions.
We have previously shown that the in vivo and in vitro dexamethasone responses of a panel of childhood ALL biopsies established as xenografts in NOD/SCID mice reflected the clinical outcome of the patients from whom they were derived.30-32 In contrast to findings with in vitro-cultured cell lines, dexamethasone resistance in xenografts was not attributed to a dysfunctional GR.25,33,34 We found that there was a clear disparity between dexamethasone-sensitive and -resistant xenografts in their ability to induce transcription of BIM. Although BIM lacks GREs in its promoter, we observed that the epigenetic regulation of the BIM promoter, including histone acetylation and methylation status, were significantly different between sensitive and resistant xenografts.35,36 In this study, gene modulation on in vivo dexamethasone treatment was assessed by microarray analysis, and whole-genome GR binding sites and histone acetylation status were detected using chromatin immunoprecipitation (ChIP) and ChIP sequencing (ChIP-seq) analyses. The combination of these techniques provided an understanding of dexamethasone-induced signaling cascades in ALL cells in vivo. We revealed a novel signaling pathway of KLF13-mediated BCL2 repression in sensitive xenografts and identified a novel GR binding site in a distal region of the BIM locus that is likely to regulate BIM expression. These findings provide novel links between the GR and pro/antiapoptotic proteins, which are likely to be important in the understanding of mechanisms of glucocorticoid resistance in lymphoid malignancies.
Materials and methods
ALL xenograft model and primary patient samples
The process by which continuous xenografts from childhood ALL biopsies have been established in immunodeficient NOD/SCID mice has been previously described in detail.35 Full details are provided in the supplemental Methods, available on the Blood Web site. All investigations using human tissue were performed in accordance with the principles embodied in the declaration of Helsinki. All animal studies had previous approval from the Animal Care and Ethics Committee of the University of New South Wales whereas experiments that used patient biopsy material were approved by the Human Research Ethics Committees of the South East Sydney and Illawara Area Health Service and the University of New South Wales.
In vitro and in vivo dexamethasone treatment, sample preparation, and analysis
ALL xenograft cells were inoculated by tail-vein injection into NOD/SCID mice, and engraftment was monitored weekly as previously described.25 For in vivo efficacy studies mice were randomized and treated with either dexamethasone (15 mg/kg) or vehicle control by intraperitoneal injection when the %huCD45+ cells in the peripheral blood reached 1% (see below and supplemental Methods). For cell and molecular biology experiments, mice were treated when there were >70% %huCD45+ cells in the peripheral blood and culled 8 hours thereafter. Cell suspensions of spleens were prepared and mononuclear cells were enriched to >97% human by density gradient centrifugation.
For gene expression and microarray studies, RNA was extracted immediately after harvesting. The microarray was performed using Illumina HumanWG-6 v3 Expression BeadChips and analyzed as previously described.37-40 The data were deposited in National Center for Biotechnology Information's Gene Expression Omnibus41 (accession no. GSE57795). Full details are provided in the supplemental Methods.
For ChIP and ChIP-seq experiments, spleen-harvested cells were fixed with 1% formaldehyde for 10 minutes at room temperature immediately after harvesting. Nuclei were extracted from fixed cells by 10-minute incubation in lysis buffer (0.2% NP40 in 10 mM Tris buffer, pH 8.0) followed by centrifugation at 1250 g for 5 minutes at 4°C. The nuclei samples were snap frozen and stored at −80°C for further use.
ChIP and ChIP-seq
ChIP and ChIP-seq were carried out as previously described.35,42-44 Details of transcriptional factor binding sites, primer design, and ChIP/ChIP-seq protocols are provided in the supplemental Methods. The ChIP-seq data were deposited in National Center for Biotechnology Information's Gene Expression Omnibus41 (accession no. GSE58266).
Assessment of dexamethasone sensitivity
In vitro dexamethasone sensitivity was assessed using the assessment of mitochondrial activity by Alamar blue assay. The in vivo dexamethasone sensitivity was determined by the leukemia growth delay (LGD) using established methods.31 Full details are provided in the supplemental Methods.
Reverse transcription-polymerase chain reaction and western blotting
Real-time quantitative reverse-transcription PCR (RT2-PCR) and western blot analysis were carried out as previously described.35 Full details are provided in the supplemental Methods.
Lentiviral gene knockdown
KLF13 gene knockdown was performed using pLKO.1 lentiviral constructs (Sigma-Aldrich, St. Louis, MO) in Nalm6 cells. Full details are provided in the supplemental Methods.
GR-DNA binding enzyme-linked immunosorbent assay
GR-DNA binding affinity in nuclear extracts was quantified by TransAM Transcription Factor enzyme-linked immunosorbent assay (Active Motif, Carlsbad, CA), as previously described.25 Full details are provided in the supplemental Methods.
Luciferase reporter assay
Luciferase reporter assays were performed using pGL2B and pGL2P vectors from Promega (Madison, WI), and luminescence was detected using the Dual-Glo system from Promega. Full details are provided in the supplemental Methods.
Statistics
Microarray and in vivo ChIP studies were performed using spleen-harvested cells from 3 randomized ALL-engrafted mice at each condition of treatment.45 In vitro gene expression and time course ChIP studies were performed with 3 independent experiments. Quantitative variables of normally distributed data were compared by the Student t test, and non-normally distributed data were compared by the Mann-Whitney U test. All statistical tests were 2-sided, and P < .05 was considered statistically significant.
Results
Xenograft selection and dexamethasone response
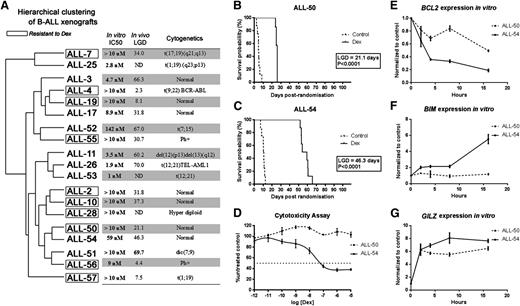
We previously established a panel of dexamethasone-sensitive and -resistant ALL xenografts derived from primary biopsy specimens of B-cell precursor ALL patients,25,30,31 as well as a panel of xenografts derived from prednisolone poor responders and prednisolone good responders (Table 1).46 To select a molecularly matched pair of xenografts to study mechanisms associated with dexamethasone resistance, we first performed microarray analysis of basal gene expression profiles (GEPs) of all xenografts. Unsupervised hierarchical clustering of the xenografts and 3 key characteristics (in vitro and in vivo dexamethasone sensitivity and cytogenetics) are shown in Figure 1A. As expected, some xenografts (eg, ALL-11, -26, and -53) clustered according to dexamethasone sensitivity. However, in more than half of the cases, basal GEPs did not predict dexamethasone responses. Four pairs of xenografts closely matched in basal GEPs but differing in their responses to dexamethasone treatment were identified (ie, ALL-7/-25; ALL-52/-55; ALL-50/-54; and ALL-51/-56). To avoid bias toward specific cytogenetic mutations, ALL-50 and ALL-54 were chosen for further detailed study because they exhibited normal cytogenetics.
Patient demographics and in vivo/in vitro responses to dexamethasone
| Xenograft . | Age at diagnosis, (month)/sex . | Cytogenetics . | CR1 duration (months) . | Current clinical status . | In vitro IC50 . | In vivo LGD (days) . | In vitro stratification . | In vivo stratification . |
|---|---|---|---|---|---|---|---|---|
| ALL-28 | 20/M | Hyperdiploid | +94 | CR1 | >10 µM | ND | Resistant | ND |
| ALL-50 | 131/M | normal | 30 | DOD | >10 µM | 21.1 | Resistant | Resistant |
| ALL-55 | 176/M | t(9;22) | +36 | CR1 | >10 µM | 30.7 | Resistant | Resistant |
| ALL-56 | 120/M | t(9;22) | 55 | CR2 | 9.0 nM | 4.4 | Sensitive | Resistant |
| ALL-57 | 72/F | t(1;19) | +27 | CR1 | >10 µM | 7.5 | Resistant | Resistant |
| ALL-26 | 43/F | t(12;21) | +96 | CR1 | 1.9 nM | 70.0 | Sensitive | Sensitive |
| ALL-51 | 19/M | dic(7;9) | +92 | CR1 | >10 µM | 69.7 | Resistant | Sensitive |
| ALL-52 | 138/M | t(7;15) | +80 | CR1 | 142 nM | 67.0 | Sensitive | Sensitive |
| ALL-53 | 87/M | t(12;21) | +102 | CR1 | 1.0 nM | ND | Sensitive | ND |
| ALL-54 | 89/M | normal | +93 | CR1 | 59 nM | 46.3 | Sensitive | Sensitive |
| Xenograft . | Age at diagnosis, (month)/sex . | Cytogenetics . | CR1 duration (months) . | Current clinical status . | In vitro IC50 . | In vivo LGD (days) . | In vitro stratification . | In vivo stratification . |
|---|---|---|---|---|---|---|---|---|
| ALL-28 | 20/M | Hyperdiploid | +94 | CR1 | >10 µM | ND | Resistant | ND |
| ALL-50 | 131/M | normal | 30 | DOD | >10 µM | 21.1 | Resistant | Resistant |
| ALL-55 | 176/M | t(9;22) | +36 | CR1 | >10 µM | 30.7 | Resistant | Resistant |
| ALL-56 | 120/M | t(9;22) | 55 | CR2 | 9.0 nM | 4.4 | Sensitive | Resistant |
| ALL-57 | 72/F | t(1;19) | +27 | CR1 | >10 µM | 7.5 | Resistant | Resistant |
| ALL-26 | 43/F | t(12;21) | +96 | CR1 | 1.9 nM | 70.0 | Sensitive | Sensitive |
| ALL-51 | 19/M | dic(7;9) | +92 | CR1 | >10 µM | 69.7 | Resistant | Sensitive |
| ALL-52 | 138/M | t(7;15) | +80 | CR1 | 142 nM | 67.0 | Sensitive | Sensitive |
| ALL-53 | 87/M | t(12;21) | +102 | CR1 | 1.0 nM | ND | Sensitive | ND |
| ALL-54 | 89/M | normal | +93 | CR1 | 59 nM | 46.3 | Sensitive | Sensitive |
CR1, first complete remission; CR2, second complete remission; DOD, dead of disease; LGD, leukemia growth delay (days); ND, not determined; resistant, IC50 > 1 μM, LGD < 40 days; sensitive, IC50 < 1 µM, LGD > 40 days; +, no event.
Hierarchical clustering of xenografts and their dexamethasone sensitivities in vivo and in vitro. (A) Unsupervised cluster dendrogram of the 37 879 probes across 19 BCP-ALL xenografts. The 10 framed xenografts are resistant to dexamethasone, according to their LGD in vivo and IC50 in vitro. The 9 unframed xenografts are sensitive to dexamethasone. (B-C) Kaplan-Meier curves of EFS following treatment with dexamethasone 15 mg/kg intraperitoneally Monday throughout Friday for 4 weeks (solid line) or vehicle control (dashed line) for (B) ALL-50 and (C) ALL-54. LGD calculated as the difference between median EFS of treated and control groups. (D) ALL-50 and ALL-54 were treated with increasing concentrations of dexamethasone, and viability was assessed by Alamar Blue assay after 48-hour incubation. Values are expressed as a percentage of the untreated control. Data represent mean ± standard error of the mean for n = 3 experiments. (E-G) ALL-50 and ALL-54 cultured cells in vitro were exposed to dexamethasone (1 µmol/L) and were harvested at times up to 16 hours and processed for real-time RT-PCR quantitation. Data are expressed relative (fold) to solvent-treated controls at each time point. Each data point represents the mean ± standard error of the mean from 3 independent experiments. Dex, dexamethasone; IC50, the dexamethasone half maximal inhibitory concentration; LGD, leukemia growth delay; ND, not determined.
Hierarchical clustering of xenografts and their dexamethasone sensitivities in vivo and in vitro. (A) Unsupervised cluster dendrogram of the 37 879 probes across 19 BCP-ALL xenografts. The 10 framed xenografts are resistant to dexamethasone, according to their LGD in vivo and IC50 in vitro. The 9 unframed xenografts are sensitive to dexamethasone. (B-C) Kaplan-Meier curves of EFS following treatment with dexamethasone 15 mg/kg intraperitoneally Monday throughout Friday for 4 weeks (solid line) or vehicle control (dashed line) for (B) ALL-50 and (C) ALL-54. LGD calculated as the difference between median EFS of treated and control groups. (D) ALL-50 and ALL-54 were treated with increasing concentrations of dexamethasone, and viability was assessed by Alamar Blue assay after 48-hour incubation. Values are expressed as a percentage of the untreated control. Data represent mean ± standard error of the mean for n = 3 experiments. (E-G) ALL-50 and ALL-54 cultured cells in vitro were exposed to dexamethasone (1 µmol/L) and were harvested at times up to 16 hours and processed for real-time RT-PCR quantitation. Data are expressed relative (fold) to solvent-treated controls at each time point. Each data point represents the mean ± standard error of the mean from 3 independent experiments. Dex, dexamethasone; IC50, the dexamethasone half maximal inhibitory concentration; LGD, leukemia growth delay; ND, not determined.
Figure 1B-D illustrates the in vivo and in vitro dexamethasone sensitivities of ALL-50 and ALL-54. Kaplan-Meier survival plots of mouse event-free survival (EFS) revealed a longer LGD of 46.3 days in ALL-54 (Figure 1C) compared with 21.1 days for ALL-50 (Figure 1B). The results of the cytotoxicity assays in vitro were consistent with the in vivo data and showed profound resistance in ALL-50 (IC50 >10 μM) compared with ALL-54 (IC50 = 59 nM), a difference of >200-fold (Figure 1D). Next, we detected that the GR in both xenografts translocated from the cytoplasm to nucleus (supplemental Figure 1) and bound consensus GRE oligos by GR-DNA binding enzyme-linked immunosorbent assay (supplemental Figure 2) following dexamethasone stimulation. The entire GR cDNA from ALL-50, ALL-54, Nalm6, CEM-WT, and CEM-MTXR3 cells was sequenced following RT-PCR amplification. Consistent with our previous results,47 CEM-WT cells were heterozygous for a wild-type codon TTA and a mutated codon TTT at 2259 bp from the translation start site, whereas CEM-MTXR3 cells exhibited loss of heterozygosity and only expressed the mutated codon (supplemental Figure 3). In contrast, ALL-50, ALL-54, and Nalm6 cells only expressed the entire wild-type GR sequence. These results indicate that the distinct responses of ALL-50 and -54 to dexamethasone were not due to a dysfunctional GR, in contrast to the observation in most glucocorticoid-resistant cell lines.25,33
We then assessed the expression of candidate dexamethasone-regulated genes following treatment of ex vivo-cultured ALL-50 and -54 cells and observed downregulation of BCL2 and upregulation of BIM in ALL-54 (sensitive) but markedly less so in ALL-50 (resistant) (Figure 1E-F). In contrast, GILZ, a primary target of the GR and not directly involved in apoptosis, was upregulated in both xenografts on dexamethasone treatment (Figure 1G). These results suggested that coordinated regulation of BIM and BCL2 controls dexamethasone-induced apoptosis of ALL xenograft cells.
Microarray analysis of in vivo dexamethasone-induced gene expression
To further understand the basis for differential in vivo glucocorticoid sensitivity of pediatric ALL xenografts, microarray analysis of gene expression was carried out on ALL-50 and ALL-54 cells harvested 8 hours after treatment. Principal component analysis demonstrated that the transcriptomes of the control and dexamethasone-treated ALL-50 and ALL-54 samples clustered into distinct groups, indicating reproducible gene expression differences between dexamethasone-sensitive and -resistant xenografts (Figure 2A). Limma analysis identified 457 genes that were significantly differentially regulated by dexamethasone between ALL-50 and ALL-54 (FDR <0.05; Figure 2B). Among these genes, 9 were previously reported to have functional relevance to either GR signaling or epigenetic modulation (eg, histone acetylation by P300/CBP; a library of GR-, CBP-, and P300-related genes is shown in supplemental Table 1). It was not anticipated that BIM (BCL2L11) would be selected by Limma analysis because its peak of induction was around 16 hours (Figure 1F).
Microarray analysis of in vivo dexamethasone-induced gene expression. ALL-50 and ALL-54 engrafted mice were treated with dexamethasone 15 mg/kg or vehicle control, and cells harvested after 8 hours. GEPs of spleen-harvested cells were evaluated using microarray. The data for each condition are presented with samples from 3 randomized ALL-engrafted mice. (A) Principal component analysis of global GEPs of the ALL-50 and ALL-54 samples before (circles) and after (squares) treatment with dexamethasone. (B) Heatmap of differentially expressed genes between ALL-50 and ALL-54 after dexamethasone treatment. A list of 457 genes was generated and ranked using Limma with a formula of (x54.Dex-x54.Con)/2-(x50.Dex-x50.Con)/2 (false discovery rate [FDR] <0.05). (C) Nine genes related to either GR signaling or epigenetic modulation were selected from the Limma gene list. Relative gene expressions of the 9 selected genes and BIM (BCL2L11) in ALL-50 and ALL-54 during dexamethasone treatment were presented in a heatmap. (D) Heatmap of the dexamethasone-induced fold changes of 10 genes in a panel of 10 xenografts including 5 dexamethasone-resistant xenografts (ALL-28/-50/-55/-56/-57) and 5 dexamethasone-sensitive xenografts (ALL-26/-51/-52/-53/-54). Hierarchical clustering of the 10 genes was performed based on their fold changes on dexamethasone treatment. (E-F) Scatter plots of dexamethasone-induced fold changes of KLF13 and MYB expression in the 2 groups of dexamethasone-resistant and -sensitive xenografts. *P < .05 (Mann-Whitney U test). (G) Correlation curve of dexamethasone-induced fold changes of MYB and BCL2 expression levels in the 10 xenografts. (H) Correlation curve of dexamethasone-induced fold changes of BCL2 and BIM expression levels in the 10 xenografts. Con, vehicle control; Dex, dexamethasone.
Microarray analysis of in vivo dexamethasone-induced gene expression. ALL-50 and ALL-54 engrafted mice were treated with dexamethasone 15 mg/kg or vehicle control, and cells harvested after 8 hours. GEPs of spleen-harvested cells were evaluated using microarray. The data for each condition are presented with samples from 3 randomized ALL-engrafted mice. (A) Principal component analysis of global GEPs of the ALL-50 and ALL-54 samples before (circles) and after (squares) treatment with dexamethasone. (B) Heatmap of differentially expressed genes between ALL-50 and ALL-54 after dexamethasone treatment. A list of 457 genes was generated and ranked using Limma with a formula of (x54.Dex-x54.Con)/2-(x50.Dex-x50.Con)/2 (false discovery rate [FDR] <0.05). (C) Nine genes related to either GR signaling or epigenetic modulation were selected from the Limma gene list. Relative gene expressions of the 9 selected genes and BIM (BCL2L11) in ALL-50 and ALL-54 during dexamethasone treatment were presented in a heatmap. (D) Heatmap of the dexamethasone-induced fold changes of 10 genes in a panel of 10 xenografts including 5 dexamethasone-resistant xenografts (ALL-28/-50/-55/-56/-57) and 5 dexamethasone-sensitive xenografts (ALL-26/-51/-52/-53/-54). Hierarchical clustering of the 10 genes was performed based on their fold changes on dexamethasone treatment. (E-F) Scatter plots of dexamethasone-induced fold changes of KLF13 and MYB expression in the 2 groups of dexamethasone-resistant and -sensitive xenografts. *P < .05 (Mann-Whitney U test). (G) Correlation curve of dexamethasone-induced fold changes of MYB and BCL2 expression levels in the 10 xenografts. (H) Correlation curve of dexamethasone-induced fold changes of BCL2 and BIM expression levels in the 10 xenografts. Con, vehicle control; Dex, dexamethasone.
Expression of the 9 Limma-selected genes plus BIM are presented in heatmap format in Figure 2C. We next extended the study of these 10 genes to the panel of 10 xenografts including 5 each of dexamethasone-sensitive and -resistant xenografts (Table 1). The genes down- or upregulated in ALL-54 still clustered together in this extended xenograft panel (Figure 2D). These results were further analyzed using Limma and the Mann-Whitney U test, where we found the changes in expression of KLF13, NCOA7, and MYB were significantly greater in the group of 5 sensitive xenografts in comparison with the 5 resistant xenografts (FDR <0.05; Table 2; Figure 2E-F). Furthermore, the differential regulation of MYB, a known activator of BCL2,48-50 was significantly correlated with BCL2 regulation on dexamethasone treatment in the 10 xenografts (P < .05; Figure 2G). In addition, the downregulation of BCL2 was significantly correlated with BIM upregulation (P < .05; Figure 2H). These results again supported coordinated regulation of these genes in controlling the dexamethasone responses of ALL cells.
Comparison of gene regulation in 5 dexamethasone-sensitive and 5 dexamethasone-resistant xenografts by Limma
| Gene . | LogFC (S vs R) . | Direction (S vs R) . | P value . | FDR . |
|---|---|---|---|---|
| KLF13 | 0.7958 | Up | .01 | 0.02* |
| NCOA7 | 1.0064 | Up | .02 | 0.02* |
| MYB | −0.8249 | Down | .02 | 0.02* |
| BCL2 | −0.6976 | Down | .09 | 0.08 |
| TCF12 | −0.5206 | Down | .16 | 0.11 |
| NRIP1 | −0.7081 | Down | .18 | 0.11 |
| BCL2L11 | 0.1959 | Up | .28 | 0.14 |
| CEBPB | 0.4671 | Up | .44 | 0.19 |
| KHDRBS3 | −0.1222 | Down | .80 | 0.29 |
| KLF8 | 0.0742 | Up | .81 | 0.29 |
| Gene . | LogFC (S vs R) . | Direction (S vs R) . | P value . | FDR . |
|---|---|---|---|---|
| KLF13 | 0.7958 | Up | .01 | 0.02* |
| NCOA7 | 1.0064 | Up | .02 | 0.02* |
| MYB | −0.8249 | Down | .02 | 0.02* |
| BCL2 | −0.6976 | Down | .09 | 0.08 |
| TCF12 | −0.5206 | Down | .16 | 0.11 |
| NRIP1 | −0.7081 | Down | .18 | 0.11 |
| BCL2L11 | 0.1959 | Up | .28 | 0.14 |
| CEBPB | 0.4671 | Up | .44 | 0.19 |
| KHDRBS3 | −0.1222 | Down | .80 | 0.29 |
| KLF8 | 0.0742 | Up | .81 | 0.29 |
R, dexamethasone-resistant xenografts; S, dexamethasone-sensitive xenografts.
Significantly different between sensitive and resistant xenografts.
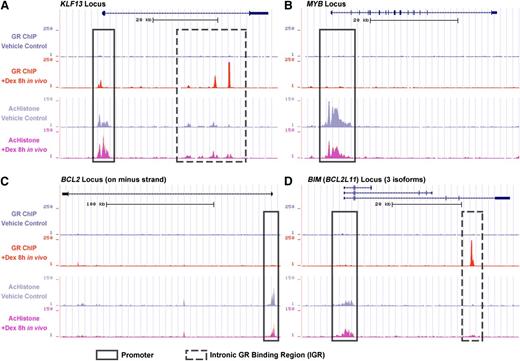
GR binding and histone acetylation in xenograft in vivo by ChIP-seq study
To identify genes directly targeted by the GR, we performed ChIP-seq with ALL-54 cells harvested from engrafted mice 8 hours after treatment with dexamethasone or vehicle control. We found GR binding sites at the KLF13 promoter (Figure 3A), which indicated that the GR directly triggered KLF13 transcription. We also detected intronic GR binding regions (IGRs) in the KLF13 locus, which could be distal regulation sites of the GR. Similar to KLF13, GR binding sites were also found on the promoters and IGRs of the NCOA7 and GILZ loci (supplemental Figure 4). Furthermore, GR binding was always accompanied by an increase in histone acetylation on both the promoter and IGRs. No GR binding sites were found on the MYB, BCL2, and NRIP1 loci (Figure 3B-C; supplemental Figure 4), but a decrease in histone acetylation was observed on their promoters, consistent with downregulation of these genes. No GR binding sites were identified at the BIM promoter (Figure 3D), but interestingly, a novel IGR was recognized, which was accompanied by increased histone acetylation at the BIM promoter and IGR following dexamethasone treatment. The BIM IGR may be responsible for long-distance regulation of BIM transcription.
ChIP-seq data of GR binding and histone acetylation. ALL-54 engrafted mice were treated with dexamethasone 15 mg/kg or vehicle control and cells were harvested after 8 hours. ChIP sequencing identified GR binding and histone acetylation on the (A) KLF13 locus, (B) MYB locus, (C) BCL2 locus, and (D) BIM locus as determined using the University of California Santa Cruz Genome Browser. The frames with solid lines refer to promoter regions and downstream regions near the transcription start site. The frames with dashed lines refer to the intronic/distal GR binding regions. Dex, dexamethasone; AcHistone, histone acetylation.
ChIP-seq data of GR binding and histone acetylation. ALL-54 engrafted mice were treated with dexamethasone 15 mg/kg or vehicle control and cells were harvested after 8 hours. ChIP sequencing identified GR binding and histone acetylation on the (A) KLF13 locus, (B) MYB locus, (C) BCL2 locus, and (D) BIM locus as determined using the University of California Santa Cruz Genome Browser. The frames with solid lines refer to promoter regions and downstream regions near the transcription start site. The frames with dashed lines refer to the intronic/distal GR binding regions. Dex, dexamethasone; AcHistone, histone acetylation.
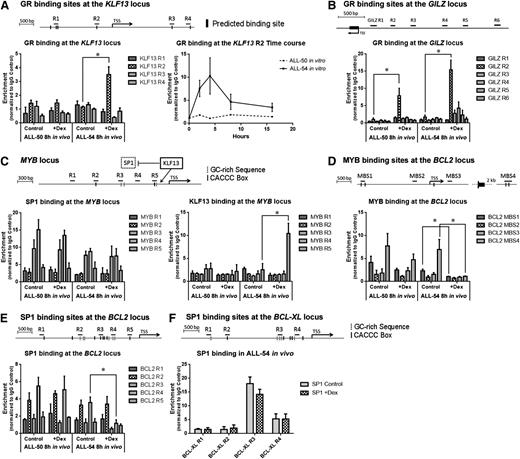
Mechanism of transcriptional regulation of BCL2
The above ChIP-seq data were confirmed and expanded to compare mechanisms of gene regulation in a sensitive and a resistant ALL xenograft. Because the ChIP-seq data indicated that BCL2 is not a direct target of the GR, we studied the possible roles of KLF13 and MYB in BCL2 gene regulation. We designed primers to target the in silico predicted GREs of the KLF13 locus as shown in Figure 4A. Consistent with the ChIP-seq data, GR binding at region 2 of the KLF13 promoter was prominent in the dexamethasone-sensitive ALL-54 cells 8 hours after in vivo dexamethasone treatment, but not in the dexamethasone-resistant ALL-50 cells. An in vitro time course ChIP study revealed a peak of GR binding at the KLF13 promoter in ALL-54 within 4 hours of dexamethasone treatment (Figure 4A). However, no GR binding was observed on the KLF13 promoter of ALL-50. In contrast, the GILZ promoter served as a positive control, where GR binding was detected in both ALL-50 and ALL-54 following dexamethasone treatment in vivo (Figure 4B).
ChIP assay on KLF13 to BCL2 signaling pathway. ChIP was performed using spleen-harvested cells from ALL-engrafted mice treated with dexamethasone 15 mg/kg or vehicle control for 8 hours. The data for each point were generated with samples from 3 randomized ALL-engrafted mice. Time course ChIP study was performed with 3 independent experiments in vitro. (A) GR binding at the KLF13 locus in ALL-50 and ALL-54 following dexamethasone treatment in vivo and time course studies of GR binding at region 2 of the KLF13 locus in ALL-54 in vitro. Black bars refer to GR binding sites predicted in silico. (B) GR binding at the GILZ promoter in ALL-50 and ALL-54 following dexamethasone treatment in vivo. (C) KLF13 and SP1 binding at the MYB promoter in ALL-50 and ALL-54 following dexamethasone treatment in vivo. Dashed bars refer to GC-rich sequences; solid bars refer to CACCC box. (D) c-MYB/v-MYB binding at the BCL2 locus in ALL-50 and ALL-54 following dexamethasone treatment in vivo. Black bars refer to MYB binding sites predicted in silico. (E-F) SP1 binding at the (E) BCL2 and (F) BCL-XL promoter in ALL-50 and ALL-54 following dexamethasone treatment in vivo. TSS, transcription start site; Dex, dexamethasone. *P < .05.
ChIP assay on KLF13 to BCL2 signaling pathway. ChIP was performed using spleen-harvested cells from ALL-engrafted mice treated with dexamethasone 15 mg/kg or vehicle control for 8 hours. The data for each point were generated with samples from 3 randomized ALL-engrafted mice. Time course ChIP study was performed with 3 independent experiments in vitro. (A) GR binding at the KLF13 locus in ALL-50 and ALL-54 following dexamethasone treatment in vivo and time course studies of GR binding at region 2 of the KLF13 locus in ALL-54 in vitro. Black bars refer to GR binding sites predicted in silico. (B) GR binding at the GILZ promoter in ALL-50 and ALL-54 following dexamethasone treatment in vivo. (C) KLF13 and SP1 binding at the MYB promoter in ALL-50 and ALL-54 following dexamethasone treatment in vivo. Dashed bars refer to GC-rich sequences; solid bars refer to CACCC box. (D) c-MYB/v-MYB binding at the BCL2 locus in ALL-50 and ALL-54 following dexamethasone treatment in vivo. Black bars refer to MYB binding sites predicted in silico. (E-F) SP1 binding at the (E) BCL2 and (F) BCL-XL promoter in ALL-50 and ALL-54 following dexamethasone treatment in vivo. TSS, transcription start site; Dex, dexamethasone. *P < .05.
KLF13 has been shown to deactivate gene transcription by competing for DNA binding sites with SP1.51,52 Therefore, we studied KLF13 and SP1 binding at the MYB and BCL2 promoters following dexamethasone treatment. First, we designed primers to target CACCC boxes and GC-rich sequences on the MYB and BCL2 promoters (Figure 4C,E). The structures of the SP1 and KLF13 proteins are similar, containing 3 highly homologous C-terminal zinc finger motifs, but SP1 preferentially binds GC-rich sites, whereas KLF13 prefers the sequence CACCC.51 SP1 binding in cells exposed to vehicle control was observed in regions 3 and 4 of the MYB promoter (Figure 4C). Importantly, on dexamethasone treatment, KLF13 bound to region 5 of the MYB promoter in ALL-54 but not ALL-50 cells (Figure 4C), which indicates that KLF13 deactivated the SP1-triggered MYB transcription in ALL-54. An in vitro time course study of ALL-54 revealed a clear temporal relationship between the levels of KLF13 and MYB proteins, with MYB protein downregulation occurring after KLF13 upregulation (supplemental Figure 5).
Because MYB has been shown to suppress apoptosis by inducing BCL2 expression in multiple cell types,48-50 we assessed MYB binding sites on the BCL2 locus following dexamethasone treatment and included MYB binding sites that were predicted in silico (MBS1) as well as other published binding sites (MBS2-4).50 MYB was associated with MBS1 and MBS4 at the BCL2 locus before dexamethasone treatment, and this binding was virtually eradicated following dexamethasone treatment in ALL-54 but not ALL-50 (Figure 4D). Besides MYB, SP1 binding at region 4 of the BCL2 promoter was also significantly decreased following dexamethasone treatment of ALL-54 but not ALL-50 (Figure 4E). The decrease in MYB and SP1 binding at the BCL2 promoter specifically in ALL-54 is consistent with inhibition of BCL2 transcription (Figure 1E). For these experiments, the BCL-XL promoter served as a control, where SP1 binding was also detected but not modulated by dexamethasone treatment in ALL-54 (Figure 4F).
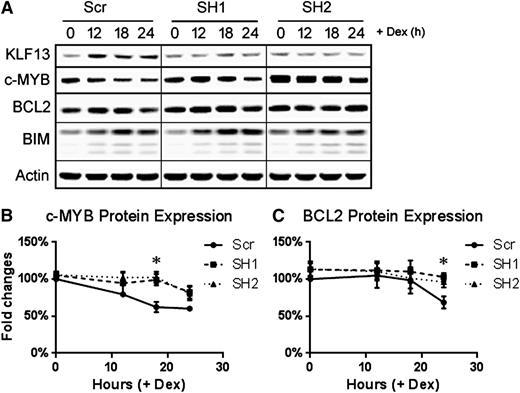
To further verify the role of KLF13 in regulating BCL2, we knocked down KLF13 expression in Nalm6 cells using a lentiviral shRNA vector. As shown in Figure 5A, knocking down KLF13 with 2 different shRNAs blocked KLF13 expression in Nalm6 cells after dexamethasone treatment compared with the scrambled shRNA control. A time course of protein expression revealed that c-MYB (Figure 5A-B) and BCL2 (Figure 5A,C) levels were maintained at 18 and 24 hours, respectively, following dexamethasone treatment in KLF13 knockdown samples compared with the significant decrease of c-MYB and BCL2 in the control samples. In contrast, BIM expression was upregulated in all transduced lines (Figure 5A).
KLF13 knockdown in Nalm6 cells. Nalm6 cells were transduced with either scrambled shRNA or 2 distinct KLF13 shRNAs using lentivirus. (A) Time course of KLF13, c-MYB, BCL2, BIM, and Actin protein expression following 1 µM dexamethasone treatment in vitro. (B) Quantified expression of c-MYB normalized to actin and then to Scr 0-hour samples. N = 3; *P < .05. (C) Quantified expression of BCL2 normalized to actin and then to Scr 0-hour samples. N = 3; *P < .05. Dex, dexamethasone; Scr, scrambled shRNA; SH1, KLF13 shRNA 1; SH2, KLF13 shRNA 2.
KLF13 knockdown in Nalm6 cells. Nalm6 cells were transduced with either scrambled shRNA or 2 distinct KLF13 shRNAs using lentivirus. (A) Time course of KLF13, c-MYB, BCL2, BIM, and Actin protein expression following 1 µM dexamethasone treatment in vitro. (B) Quantified expression of c-MYB normalized to actin and then to Scr 0-hour samples. N = 3; *P < .05. (C) Quantified expression of BCL2 normalized to actin and then to Scr 0-hour samples. N = 3; *P < .05. Dex, dexamethasone; Scr, scrambled shRNA; SH1, KLF13 shRNA 1; SH2, KLF13 shRNA 2.
Mechanism of BIM transcriptional regulation by the GR
Because it was previously unclear how BIM transcription was upregulated by glucocorticoids in lymphoid cells, we next studied GR and RNA Polymerase II (RNA Pol II) binding and histone acetylation at the BIM locus using conventional ChIP assays. As shown in Figure 6A, 9 pairs of primers were designed to cover up to 3 kb of BIM promoter and intron 1. First, we found a strong increase in histone acetylation and RNA Pol II binding at the BIM promoter and first intron in ALL-54 but not ALL-50 8 hours after dexamethasone treatment in vivo (Figure 6B-E), which was consistent with the AcHistone ChIP-seq data shown in Figure 3D. This was further confirmed by a time course ChIP study in vitro with up to 16 hours of dexamethasone treatment, showing the dexamethasone-inducing modulation of GR and RNA Pol II binding and histone acetylation on the BIM and GILZ promoters in the 2 xenografts (supplemental Figure 6). Overall, these data indicate that the BIM promoter was only activated in the dexamethasone-sensitive xenograft ALL-54 both in vitro and in vivo.
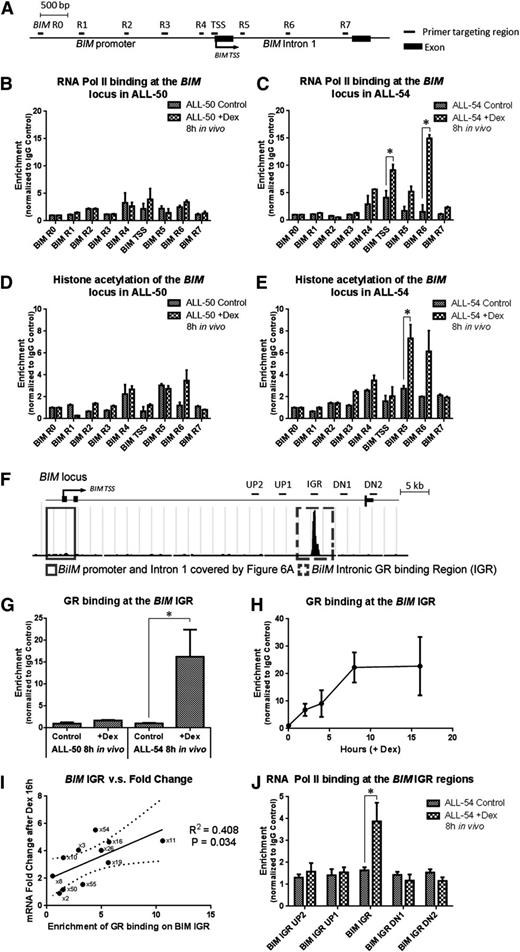
Dexamethasone-induced modulation of the BIM locus. (A) Schematic demonstrating the primers used in the ChIP study covering the BIM promoter and first intron. (B-C) RNA Pol II binding and (D-E) histone acetylation at the BIM locus were determined in (B,D) ALL-50 and (C,E) ALL-54 following 8-hour treatment with dexamethasone or vehicle control in vivo. The data for each point were generated with samples from 3 randomized ALL-engrafted mice. (F) Schematic showing GR binding at the BIM IGR (intronic GR binding region) from the ChIP-seq data. Four pairs of primers were designed to cover 5 to 10 kb up- and downstream of the BIM IGR region. (G) GR binding at the BIM IGR in ALL-50 and ALL-54 following treatment with dexamethasone or vehicle control in vivo (n = 3). (H) Time course study of GR binding at the BIM IGR in ALL-54 following dexamethasone treatment in vitro (n = 3). (I) Regression curve of dexamethasone-induced fold changes of BIM expression vs enrichment of GR binding at the BIM IGR in 11 xenografts, P = .03. BIM expression was detected by RT-PCR using 3 independent experiments in vitro. (J) RNA Pol II binding at the BIM IGR as well as up- and downstream regions in ALL-54 following 8-hour treatment with dexamethasone or vehicle control in vivo (n = 3). TSS, transcription start site; Dex, dexamethasone. *P < .05.
Dexamethasone-induced modulation of the BIM locus. (A) Schematic demonstrating the primers used in the ChIP study covering the BIM promoter and first intron. (B-C) RNA Pol II binding and (D-E) histone acetylation at the BIM locus were determined in (B,D) ALL-50 and (C,E) ALL-54 following 8-hour treatment with dexamethasone or vehicle control in vivo. The data for each point were generated with samples from 3 randomized ALL-engrafted mice. (F) Schematic showing GR binding at the BIM IGR (intronic GR binding region) from the ChIP-seq data. Four pairs of primers were designed to cover 5 to 10 kb up- and downstream of the BIM IGR region. (G) GR binding at the BIM IGR in ALL-50 and ALL-54 following treatment with dexamethasone or vehicle control in vivo (n = 3). (H) Time course study of GR binding at the BIM IGR in ALL-54 following dexamethasone treatment in vitro (n = 3). (I) Regression curve of dexamethasone-induced fold changes of BIM expression vs enrichment of GR binding at the BIM IGR in 11 xenografts, P = .03. BIM expression was detected by RT-PCR using 3 independent experiments in vitro. (J) RNA Pol II binding at the BIM IGR as well as up- and downstream regions in ALL-54 following 8-hour treatment with dexamethasone or vehicle control in vivo (n = 3). TSS, transcription start site; Dex, dexamethasone. *P < .05.
In the ChIP-seq study (Figure 3D), we identified a GR binding site at the BIM IGR that we proposed to be a long-range regulatory domain. We next performed conventional ChIP and detected GR binding at the IGR in ALL-54 but not ALL-50 following dexamethasone treatment in vivo (Figure 6F-G). Time course ChIP studies revealed a gradual increase in GR binding at the IGR in ALL-54 up to 8 hours after dexamethasone treatment in vitro (Figure 6H). Extending this study to 11 xenografts, we found a significant correlation between GR-IGR binding and BIM induction in vitro following dexamethasone treatment (R2 = 0.41, P = .034; Figure 6I). Next, we identified RNA Pol II binding at the IGR but not at 5 to 10 kb up- or downstream of the IGR (Figure 6J), indicating a role for the GR in recruiting RNA Pol II to trigger gene transcription.
BIM IGR enhances GRE-dependent gene transcription
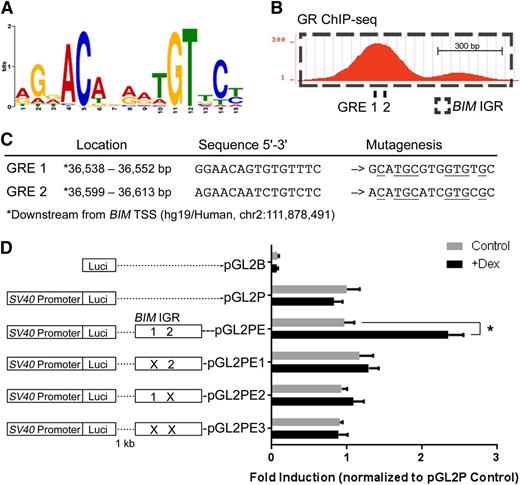
The GR binding motif (Figure 7A) was identified from the in vivo GR ChIP-seq data, similar to motifs previously identified on the basis of in vitro models.53-55 Next, we found 2 potential GREs containing the conserved GR binding motif at the first GR-binding peak within the BIM IGR (Figure 7B). Conventional ChIP detected no significant GR binding at the region of the 3′ minor peak in Figure 7B (data not shown). To study the function of the potential GREs, we cloned the BIM IGR with GREs 1 and 2 in wild-type and mutated sequences 1 kb downstream of the luciferase gene in the pGL2P reporter vector (Figure 7C) and carried out reporter assays in the Nalm6 ALL cell line. Dexamethasone treatment significantly augmented luciferase expression exclusively in the construct containing the wild-type BIM IGR (pGL2PE) (Figure 7D). Mutating the individual GREs (pGL2PE1 and 2) or both (pGL2PE3) abolished dexamethasone-induced luciferase expression. These results indicate that the BIM IGR functions as an enhancer in response to GR binding at its GREs.
The BIM IGR functions as a dexamethasone-inducible enhancer of transcription. (A) The GR binding motif identified by GR ChIP-seq in in vivo dexamethasone-treated ALL-54 cells. (B) Two potential GREs at the BIM IGR were revealed by sequence analysis, both located within the first peak of the GR ChIP-seq signal. (C) Locations and sequences of the potential GREs, which were mutated as shown (mutated residues are underlined). (D) The effects of single or dual GRE mutations on dexamethasone-induced reporter gene activity in Nalm6 cells. Reporter plasmid with firefly luciferase and control plasmid with renilla luciferase were cotransfected into Nalm6 cells. Firefly luminescence was determined after 16-hour treatment of dexamethasone and normalized to the renilla luminescence signal. The fold inductions were then calculated by normalizing to pGL2P control. N = 5; *P < .05.
The BIM IGR functions as a dexamethasone-inducible enhancer of transcription. (A) The GR binding motif identified by GR ChIP-seq in in vivo dexamethasone-treated ALL-54 cells. (B) Two potential GREs at the BIM IGR were revealed by sequence analysis, both located within the first peak of the GR ChIP-seq signal. (C) Locations and sequences of the potential GREs, which were mutated as shown (mutated residues are underlined). (D) The effects of single or dual GRE mutations on dexamethasone-induced reporter gene activity in Nalm6 cells. Reporter plasmid with firefly luciferase and control plasmid with renilla luciferase were cotransfected into Nalm6 cells. Firefly luminescence was determined after 16-hour treatment of dexamethasone and normalized to the renilla luminescence signal. The fold inductions were then calculated by normalizing to pGL2P control. N = 5; *P < .05.
Discussion
The mechanism of dexamethasone-induced apoptosis in ALL cells has been studied using a variety of cell types in vitro and in vivo.7,25,32-36 In this study, we performed microarray, ChIP, and ChIP-seq assays using in vivo dexamethasone-treated human pediatric ALL xenografts. Many genes have been previously published for their roles in GR-induced apoptosis. However, in our microarray study, most of these genes were modulated similarly in ALL-50 and ALL-54 by dexamethasone in vivo, including some well-recognized genes such as CEBPB, FOXO3, FOS, HSP90AA1, JUN, NCOA1/2, NFKB1, POU2F1/2, SMAD3, and STAT5A.7-10,17,56,57 This disparity could be due to differences between experimental model systems compared with our use of in vivo treated patient-derived xenografts, in which we observed a similar trend of activation of GR downstream signaling pathways in sensitive and resistant xenografts.
To identify the genes that were differentially regulated between dexamethasone-sensitive and -resistant xenografts, we next performed a 1-step Limma analysis on the data from 4 conditions, ie, ALL-50 and ALL-54 with and without dexamethasone treatment. Among the identified genes, KLF13, MYB, BCL2, and BIM were especially interesting. By combining ChIP and ChIP-seq data, we demonstrated that GR binding at the KLF13 locus and BIM IGR in dexamethasone-sensitive ALL-54 represented 2 distinct signaling pathways. The former induced KLF13 expression, which subsequently inhibited SP1-triggered MYB transcription and transcription of BCL2, a downstream target of MYB. GR binding at the BIM IGR appears to act as an enhancer of BIM transcription, which may play a key role in glucocorticoid-induced apoptosis of ALL cells.23-27,35 In contrast to ALL-54, the lack of GR binding at the KLF13 and BIM loci in ALL-50 correlated with its resistance to dexamethasone.
In an effort to further understand the underlying basis for defective GR binding at the BIM IGR in dexamethasone-resistant xenografts in the context of wild-type GR expression, we analyzed the abundance of DNaseI hypersensitive sites (DHSs) at the BIM IGR in 78 normal and malignant human cell types from Encyclopedia of DNA Elements (ENCODE)/OpenChrom (Duke University)58 using the University of California Santa Cruz Genome Browser.59 All 18 datasets of normal and malignant B and T lymphocytes showed high or medium DHS intensity at the BIM IGR, whereas 56 of 60 other cell types had low/nil DHS intensity (supplemental Table 2 and Figure 7). These findings indicate that the BIM IGR exists in an open chromatin conformation exclusively in lymphoid cells, which may allow the GR and other transcription factors to bind and enhance BIM transcription, thereby contributing to the acute sensitivity of normal and malignant lymphocytes to glucocorticoid-induced apoptosis. Next, we analyzed potential transcription factor binding data generated from a large collection of ChIP-seq experiments by ENCODE.58 As shown in supplemental Figure 8, there is potential binding of 2 transcription factors at the first GR-binding peak in the BIM IGR in which the 2 GREs are located, whereas 9 transcription factors could bind at the second GR-binding peak. In particular, transcription factors such as CTCF could contribute to modifying the DNA structure to bridge the BIM promoter and the regulatory domains.60
Although GR binding at the BIM IGR may regulate gene transcription via long-range chromatin remodeling, the transcription factor(s) involved in activating the BIM promoter remain to be defined. We previously identified reduced Foxo3a binding at the BIM promoter in dexamethasone-resistant vs -sensitive ALL xenografts.35 In this study, we observed a decrease in phosphorylated-Foxo3a protein (inactive form) in the cytoplasm of ALL-54 on dexamethasone treatment in vitro and a concomitant increase in Foxo3a (active form) in the nucleus, neither of which were observed in ALL-50 (supplemental Figure 9). These data support the published mechanism of Foxo3a in recruiting histone acetyltransferases and triggering BIM transcription.16,61,62
In conclusion, in this study, we compared the signal pathways involved in BIM/BCL2 gene regulation in dexamethasone-sensitive and -resistant ALL xenografts in vivo and identified novel mechanisms of opposing BIM and BCL2 gene regulation that control glucocorticoid-induced apoptosis in pediatric ALL cells in vivo.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Chintanu Sarma and Santi Suryani for assistance in analyzing the microarray gene expression data. The authors acknowledge the ENCODE Consortium and the Duke, University of North Carolina-Chapel Hill, University of Texas-Austin, and European Bioinformatics Institute ENCODE groups for generating DNaseI HS datasets and Transcription Factor ChIP-seq (161 factors) datasets. The Children’s Cancer Institute Australia is affiliated with University of New South Wales Australia and The Sydney Children’s Hospitals Network.
This research was funded by grants from the National Health and Medical Research Council of Australia and the Cancer Council New South Wales. V.A.B. was supported by fellowships from the Leukaemia Foundation of Australia and the Steven Walter Foundation. R.B.L. is supported by a fellowship from the National Health and Medical Research Council of Australia.
Authorship
Contribution: D.J., V.A.B., D.B., J.A.I.T., J.E.P., and R.B.L. designed the study; D.J., V.A.B., D.B., J.A.I.T., N.A.Y., J.W.H.W., and K.K. generated and analyzed the data; and D.J. and R.B.L. interpreted the data and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard B. Lock, Children’s Cancer Institute Australia for Medical Research, Lowy Cancer Research Centre, UNSW, PO Box 81, Randwick, NSW 2031, Australia; e-mail: rlock@ccia.unsw.edu.au.

![Figure 2. Microarray analysis of in vivo dexamethasone-induced gene expression. ALL-50 and ALL-54 engrafted mice were treated with dexamethasone 15 mg/kg or vehicle control, and cells harvested after 8 hours. GEPs of spleen-harvested cells were evaluated using microarray. The data for each condition are presented with samples from 3 randomized ALL-engrafted mice. (A) Principal component analysis of global GEPs of the ALL-50 and ALL-54 samples before (circles) and after (squares) treatment with dexamethasone. (B) Heatmap of differentially expressed genes between ALL-50 and ALL-54 after dexamethasone treatment. A list of 457 genes was generated and ranked using Limma with a formula of (x54.Dex-x54.Con)/2-(x50.Dex-x50.Con)/2 (false discovery rate [FDR] <0.05). (C) Nine genes related to either GR signaling or epigenetic modulation were selected from the Limma gene list. Relative gene expressions of the 9 selected genes and BIM (BCL2L11) in ALL-50 and ALL-54 during dexamethasone treatment were presented in a heatmap. (D) Heatmap of the dexamethasone-induced fold changes of 10 genes in a panel of 10 xenografts including 5 dexamethasone-resistant xenografts (ALL-28/-50/-55/-56/-57) and 5 dexamethasone-sensitive xenografts (ALL-26/-51/-52/-53/-54). Hierarchical clustering of the 10 genes was performed based on their fold changes on dexamethasone treatment. (E-F) Scatter plots of dexamethasone-induced fold changes of KLF13 and MYB expression in the 2 groups of dexamethasone-resistant and -sensitive xenografts. *P < .05 (Mann-Whitney U test). (G) Correlation curve of dexamethasone-induced fold changes of MYB and BCL2 expression levels in the 10 xenografts. (H) Correlation curve of dexamethasone-induced fold changes of BCL2 and BIM expression levels in the 10 xenografts. Con, vehicle control; Dex, dexamethasone.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/2/10.1182_blood-2014-05-576470/4/m_273f2.jpeg?Expires=1769880127&Signature=RL58aDYkGxlDxTrNYUSmxey7qgcsbMlpAb2bsfQ1uOOWM9uQ5rBnXPVqpyp1z-PucLPVaXY4KcySkQz7OSzFKRgDKLQIBll0NrU4DX644KgN~IcpyWzMpC2MSLEuhwbt7-YhHJTul-sxLqQhhSNX8giEVb3qxW7EHHfBJhxkFDfDVNnXoTeuvoi1oQgljvKra~zO-if3ZserjqSYK2-tgLrLolRSIPgXqwUcKd5fhPPrWbUaW3nLKgGJ3r5mqNgoNzklxUsHy6vGWWkMi2u9rMyAxDq98SZaTGu9p64QUesIG-6etg5Oxx89T1qWW3TBGz8TGfSrDYa1BFwW2~9Tiw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal