Key Points
CML patients with advanced-phase myeloid disease frequently show decreased IKAROS protein in primitive cells.
Expression of a dominant-negative IKAROS isoform expands primitive human CML cells and enhances their differentiation into basophils.
Abstract
Without effective therapy, chronic-phase chronic myeloid leukemia (CP-CML) evolves into an acute leukemia (blast crisis [BC]) that displays either myeloid or B-lymphoid characteristics. This transition is often preceded by a clinically recognized, but biologically poorly characterized, accelerated phase (AP). Here, we report that IKAROS protein is absent or reduced in bone marrow blasts from most CML patients with advanced myeloid disease (AP or BC). This contrasts with primitive CP-CML cells and BCR-ABL1–negative acute myeloid leukemia blasts, which express readily detectable IKAROS. To investigate whether loss of IKAROS contributes to myeloid disease progression in CP-CML, we examined the effects of forced expression of a dominant-negative isoform of IKAROS (IK6) in CP-CML patients’ CD34+ cells. We confirmed that IK6 disrupts IKAROS activity in transduced CP-CML cells and showed that it confers on them features of AP-CML, including a prolonged increased output in vitro and in xenografted mice of primitive cells with an enhanced ability to differentiate into basophils. Expression of IK6 in CD34+ CP-CML cells also led to activation of signal transducer and activator of transcription 5 and transcriptional repression of its negative regulators. These findings implicate loss of IKAROS as a frequent step and potential diagnostic harbinger of progressive myeloid disease in CML patients.
Introduction
Chronic myeloid leukemia (CML) is an attractive model for analyzing the clonal evolution of human malignancies, because the disease is typically diagnosed when the initial Ph+/BCR-ABL1+ chronic phase (CP) clone is still capable of normal multilineage myeloid and B-lymphoid differentiation.1,2 However, genomic instability causes the continuous generation of new subclones,3-5 and, in the absence of targeted therapy,6 progression to a differentiation-arrested acute leukemia (blast crisis [BC]) is inevitable.2,7 BC-CML is often preceded by a clinically recognized period referred to as accelerated phase (AP), in which the clone expands more rapidly and clonal basophils become more prominent. However, molecular events that underpin myeloid disease progression from CP-CML to AP-CML have not been determined.
IKAROS is essential for lymphoid cell development in mice, and suppression of its activity in this species causes the development of lethal T-cell tumors.8-11 In human hematopoietic cells, somatic mutations that target the IKZF1 locus and result in either loss of IKAROS protein or expression of dominant-negative isoforms are a common feature of BCR-ABL1+ B-cell acute lymphoblastic leukemia (B-ALL) and lymphoid BC-CML.12-14 Mutations in IKZF1 resulting in loss of IKAROS expression or the production dominant-negative isoforms have also been documented in occasional cases of human myeloid leukemias.12,15-18
In a preliminary screen, we tested a set of complementary DNAs (cDNAs) for their ability to recapitulate features of disease progression in CP-CML CD34+ cells. This set included IK6, one of the known dominant-negative isoforms of IKAROS, which produced a marked growth promoting effect on transduced CP-CML cells. This led us to examine the expression of IKAROS in bone marrow (BM) biopsy specimens from a series of CML patients. This showed loss of IKAROS protein to be a frequent feature of myeloid disease progression. To investigate whether loss of IKAROS activity may change the biology of CP-CML cells, we analyzed the cellular and molecular consequences of IK6-mediated disruption of normal IKAROS in CD34+ CML cells. The results show that this manipulation gives these cells an intrinsically enhanced ability to expand their numbers without losing their ability to differentiate, particularly into mature basophils.
Materials and methods
Human cells
CD34+ cells (>85% pure) were isolated immunomagnetically (EasySep, STEMCELL Technologies) from CP-CML patients and normal adult donors of cells collected for allogeneic transplantation. Archival BM trephine biopsy specimens from contributing centers were stained for IKAROS protein and detected using an alkaline phosphate–labeled secondary, fast red chromogen (Bond Polymer Refine Red Detection, Leica) and hematoxylin counterstain. These procedures were approved by the research ethics board of the University of British Columbia and other participating institutions. Research was conducted in accordance with the Declaration of Helsinki.
Lentiviral constructs and gene transfer
cDNAs (supplemental Table 1 available on the Blood Web site) were cloned into a lentiviral vector containing a green or yellow fluorescent protein (GFP or YFP) cDNA.19,20 Virus was produced21 and cells for in vitro studies transduced as described previously.22 After another 2 days in vitro, transduced (GFP+ or YFP+) cells were isolated by fluorescence-activated cell sorting (FACS). Xenografted cells were similarly transduced but in an overall time of 6 hours and cells then transplanted immediately (ie, without further selection).
Confocal microscopy
Stained cells (see “Flow cytometry,” below) were cytospun onto coated slides and mounted with Vectashield containing 4,6 diamidino-2-phenylindole (VECTOR Laboratories). Images were acquired with a FluoView confocal laser scanning microscope (Fv10i, Olympus) and processed with ImageJ.
Cultures
Equal numbers of FACS-selected test- and control-transduced cells (3000-10 000) were cultured and harvested as described previously23 with the extra inclusion of feeders engineered to produce human Flt3-ligand. Viable (propidium iodide [PI] negative) GFP+ and YFP+ nonadherent cells were counted separately by flow cytometry using counting beads (AccuCheck, Life Technologies). Morphological analyses were performed on Wright-Giemsa–stained cytospin preparations. Colony-forming cell (CFC) assays were performed as described previously.23 All CD34+ cells from CML #1 have previously been shown to be BCR-ABL1 positive.24 CFCs from CML #2 to #5 were assessed by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) for BCR-ABL1,24 and only BCR-ABL1+ (leukemic) CFCs were included in the analyses. CFCs were genotyped for IK6 by qRT-PCR.
Flow cytometry
For surface phenotype analyses, cells were blocked and then stained with fluorochrome-labeled antibodies (supplemental Table 2) and PI. For intracellular IKAROS detection, cells were first permeabilized in detergent-buffered paraformaldehyde (eBioscience) and then stained as above using appropriate secondary antibodies. For analysis of intracellular signaling proteins, cells were washed twice in Iscove’s medium, incubated at 37°C for 3 hours, fixed in 1.6% paraformaldehyde (room temperature for 10 minutes), stained with cell surface antibodies, washed twice in phosphate-buffered saline, resuspended in 80% ethanol at −80°C, washed twice in phosphate-buffered saline, stained on ice with antibodies to intracellular proteins (supplemental Table 2), and then washed and stained with appropriate secondary antibodies. Analyses were performed using a Fortessa flow cytometer (BD). FlowJo software (Tree Star) was used to obtain median fluorescent intensity values.
qRT-PCR
cDNA was prepared using SuperScript VILO (Life Technologies) from RNA isolated on spin columns (Qiagen). Primers were designed to amplify across at least 1 intron and generate a product of 80 to 100 bp using Primer3 software (http://frodo.wi.mit.edu/). qRT-PCR was performed using Fast SYBR green (Life Technologies) on a 7500 Fast Real-Time PCR analyzer (Applied Biosystems). Transcript levels were normalized to GUSB and ABL1 and mean ∆Ct values determined.
Xenotransplantation
Eight- to 12-week-old NOD-Rag1nullIL-2Rγcnull mice producing human interleukin-3, granulocyte-macrophage colony-stimulating factor, and Steel factor (NRG-3GS mice derived from NSG-3GS mice25 ) were irradiated with 900 cGy of 137Cs γ-rays at 3.75 cGy/min prior to being injected IV each with 105 control- and 105 IK6-transduced CD34+CD38− CP-CML cells. A total of 106 live (PI−) BM aspirate cells were stained and analyzed as described previously.26 These procedures approved by the University of British Columbia.
Tyrosine kinase inhibitor sensitivity assays
Transduced and resorted CD34+ CP-CML cells were cultured in 96-well plates (200 cells/100 µL per well) in Iscove’s medium containing 30% fetal bovine serum plus the same growth factors as the CFC assays with added Flt3-ligand at 50 ng/mL and varying concentrations of imatinib (Novartis) or dasatinib (Bristol-Meyers Squibb). Viable (PI−) GFP+ and YFP+ cells were enumerated on day 6 by flow cytometry. All 50% inhibition/inhibitory concentration values were calculated using Prism software.
Statistics
Values shown are the mean ± standard error of the mean (SEM). Significant differences were determined using Student t test.
Results
Forced expression of IK6, c-MYC, NUP98-HOXA9, or KRAS T58I in primary CD34+ CP-CML cells expands their numbers in vitro
The present studies were prompted by a preliminary series of experiments exploring the ability of candidate cDNAs to produce a disease progression phenotype in transduced CD34+ cells isolated from a CP-CML patient. For this screen, we started with 15 vectors designed to target a spectrum of molecular pathways previously implicated in aggressive human leukemias through effects on intracellular signaling, transcriptional regulation, and DNA damage repair (supplemental Table 1). To look for an effect of each cDNA, we transduced CML cells in parallel with a test and a control vector (each encoding a different fluorochrome, GFP or YFP) and set up a series of cultures, in each case containing an equal number of test and control cells. The cells were seeded onto human growth factor–producing mouse fibroblast feeders (previously found to support the production of differentiated granulocytes and monocytes from normal CD34+ cells for several weeks), and outputs from test and control cells were compared over time (Figure 1A). To avoid the need to genotype these outputs, we used cells from a selected CP-CML patient in which all of the primitive compartment had been previously found to be BCR-ABL1 positive.24 As a potential indicator of a progressed disease phenotype, we looked for enhanced outputs of primitive and differentiating cells from the test- vs control-transduced cells.
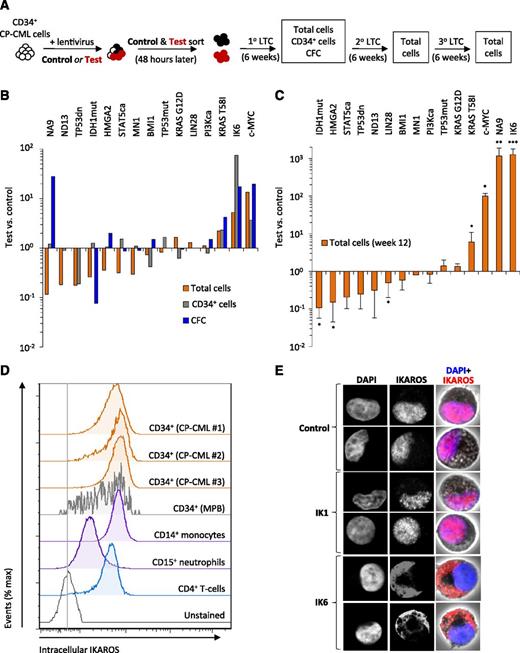
NUP98-HOXA9, MYC, KRAS T58I, and IK6 promote the growth of CP-CML cells. (A) Experimental design used to assess the effects of 15 candidate cDNAs on the growth of CD34+ CP-CML cells in cultures initiated with equal numbers of test- and control-transduced cells (3-10 × 103, depending on the experiment). (B) Total cells, CD34+ cells and CFCs in 6-week cultures. Values shown are the mean ratios of the numbers of test to control cells (identified by expression of GFP or YFP) or the matching CFC numbers measured in 4 replicate primary cultures. (C) Total cells present in 6-week secondary cultures (1 primary culture equivalent per secondary culture; total of 12 weeks of culture). Values shown are the mean ± SEM of ratios of the numbers of test to control cells. (D) Representative flow cytometric profiles of intracellular IKAROS protein levels in different subsets of CP-CML cells from 4 patients and CD34+ cells isolated from mobilized peripheral blood (MPB) samples from 2 normal donors. (E) Confocal microscopy images of representative single CP-CML cells, either untransduced (control) or following transduction with wild-type IKAROS (IK1) or IK6, and stained with 4,6 diamidino-2-phenylindole (DAPI, blue) and an antibody reactive with both wild-type and IK6 isoforms (red). *P < .05; **P < .01; ***P < .001. 1°, primary; 2°, secondary; 3°, tertiary; IDH1mut, IDH1 R132H; IK6, dominant-negative IKAROS, IK6 isoform; LTC, long-term culture; NA9, NUP98-HOXA9; ND13, NUP98-HOXD13; PI3Kca, PIK3CA H1047R; STAT5ca, STAT5A H298R/S710F27 ; TP53dn, TP53 dominant-negative GSE5628 ; TP53mut, TP53 R273C.
NUP98-HOXA9, MYC, KRAS T58I, and IK6 promote the growth of CP-CML cells. (A) Experimental design used to assess the effects of 15 candidate cDNAs on the growth of CD34+ CP-CML cells in cultures initiated with equal numbers of test- and control-transduced cells (3-10 × 103, depending on the experiment). (B) Total cells, CD34+ cells and CFCs in 6-week cultures. Values shown are the mean ratios of the numbers of test to control cells (identified by expression of GFP or YFP) or the matching CFC numbers measured in 4 replicate primary cultures. (C) Total cells present in 6-week secondary cultures (1 primary culture equivalent per secondary culture; total of 12 weeks of culture). Values shown are the mean ± SEM of ratios of the numbers of test to control cells. (D) Representative flow cytometric profiles of intracellular IKAROS protein levels in different subsets of CP-CML cells from 4 patients and CD34+ cells isolated from mobilized peripheral blood (MPB) samples from 2 normal donors. (E) Confocal microscopy images of representative single CP-CML cells, either untransduced (control) or following transduction with wild-type IKAROS (IK1) or IK6, and stained with 4,6 diamidino-2-phenylindole (DAPI, blue) and an antibody reactive with both wild-type and IK6 isoforms (red). *P < .05; **P < .01; ***P < .001. 1°, primary; 2°, secondary; 3°, tertiary; IDH1mut, IDH1 R132H; IK6, dominant-negative IKAROS, IK6 isoform; LTC, long-term culture; NA9, NUP98-HOXA9; ND13, NUP98-HOXD13; PI3Kca, PIK3CA H1047R; STAT5ca, STAT5A H298R/S710F27 ; TP53dn, TP53 dominant-negative GSE5628 ; TP53mut, TP53 R273C.
In this preliminary screen, 4 of the 15 cDNAs tested had significantly marked and sustained growth-promoting effects on the transduced CP-CML cells (Figure 1B-C and supplemental Figure 1).27,28 Three of these 4 cDNAs (encoding c-MYC, NUP98-HOXA9, and KRAS T58I) have been previously implicated in advanced-phase myeloid disease in humans.7,29-31 The fourth cDNA identified was IK6, a dominant-negative isoform of IKAROS that is frequently associated with Ph+/BCR-ABL1+ ALL and lymphoid BC-CML but rarely with myeloid disease. The effects of IK6 in this experiment thus appeared of particular interest, not only because they were unexpected but also because IK6 produced the most potent of the effects seen (Figure 1C).
To further understand the basis of the effect obtained with IK6, we first analyzed different subsets of CP-CML cells by intracellular flow cytometry to determine their levels of expression of IKAROS protein. These studies identified abundant levels of IKAROS protein in CD34+ CP-CML cells, indistinguishable from those seen in their counterparts isolated from samples of mobilized peripheral blood cells from normal adults (Figure 1D). Confocal examination showed that forced expression of IK6 in CP-CML cells caused all IKAROS protein to be retained in the cytoplasm, in contrast to the normal localization of IKAROS in the nucleus observed in both untransduced cells and cells transduced with wild-type IKAROS (IK1) (Figure 1E). These findings demonstrate an ability of IK6 to disrupt the nuclear transcription factor activity of IKAROS, as previously shown for Ph+ B-ALL and other human IK6-expressing leukemias.32,33
Together, these findings demonstrate that IKAROS protein is present in primitive CP-CML cells and that blockade of its normal ability to translocate to the nucleus by forced expression of IK6 has a marked and sustained growth-promoting effect on these cells.
Loss of IKAROS protein is a common feature of AP- and myeloid BC-CML
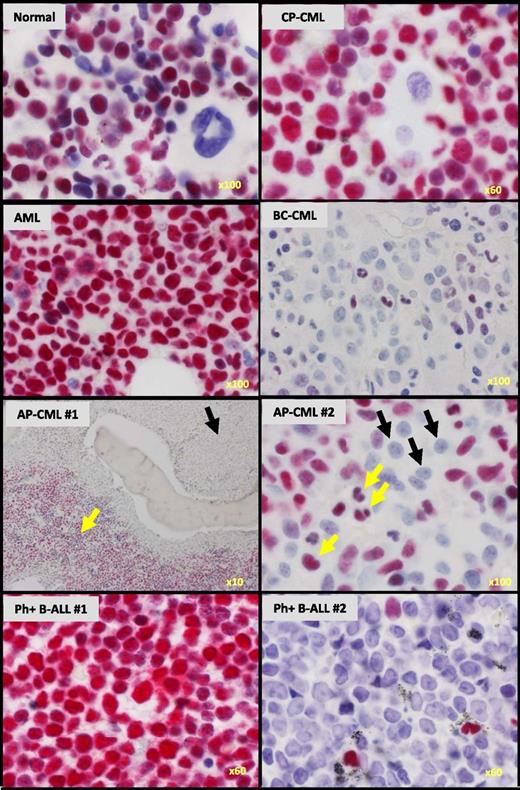
We next asked whether loss of IKAROS activity might be a clinically relevant feature of cells from CML patients with progressive myeloid disease. As controls, we first stained BM trephine biopsy sections from normal and CP-CML patients for IKAROS protein. These experiments confirmed that IKAROS protein is present in all normal and CP-CML cell types with the exception of megakaryocytes (Figure 2). In contrast, we found IKAROS protein to be greatly reduced or undetectable in the nuclei of blast cells present in sections of BM from 30 of 40 AP-CML or myeloid BC-CML patients (Figure 2, supplemental Figure 2, and supplemental Table 3). In 13 of these (33% overall), cells with a blast morphology were either completely negative (9 cases, 23% overall) or showed uniformly weak nuclear staining (4 cases, 10% overall). In the other 17 affected cases (43% overall), both of these abnormal patterns of IKAROS staining were seen in the same section, with some blast cells showing no staining and other blast cells showing weakly positive nuclear staining (as illustrated in supplemental Figure 2). Particularly striking were cases of AP-CML where IKAROS was undetectable in the expanded population of blast cells seen in the paratrabecular region but still obvious in the residual maturing CP-CML cells localized in the intertrabecular regions (Figure 2). In contrast to these results, uniformly strong nuclear IKAROS staining was seen in the BM of 58 out of 59 cases of BCR-ABL1–negative acute myeloid leukemia (AML) (AP/BC-CML vs AML, P < .0001). Similarly stained BM sections from 3 patients with Ph+ B-ALL showed either uniformly strong or uniformly absent IKAROS (Figure 2), consistent with the common acquisition of IKZF1 mutations in this disease that result in either expression of dominant-negative, but readily detected, IKAROS isoforms or a complete failure of IKAROS expression.13
IKAROS protein is reduced in BM blasts from CML patients with advanced-phase myeloid disease, but not from patients with BCR-ABL1–negative AML. Immunohistochemical staining for IKAROS protein (magenta) in representative BM trephine biopsy sections counterstained with hematoxylin from normal BM and patients with CML in CP, AP, and myeloid BC as indicated, as well as patients with BCR-ABL1–negative AML and BCR-ABL1+ (Ph+) B-ALL. AP-CML #1 shows loss of IKAROS staining in the primitive cells in the paratrabecular regions (black arrow) in contrast to strong IKAROS staining in the residual maturing CP-CML cells (yellow arrow). AP-CML #2 similarly shows loss of IKAROS in blasts (black arrows), but not in residual maturing cells (yellow arrows). The original magnification is shown on the bottom right of each panel.
IKAROS protein is reduced in BM blasts from CML patients with advanced-phase myeloid disease, but not from patients with BCR-ABL1–negative AML. Immunohistochemical staining for IKAROS protein (magenta) in representative BM trephine biopsy sections counterstained with hematoxylin from normal BM and patients with CML in CP, AP, and myeloid BC as indicated, as well as patients with BCR-ABL1–negative AML and BCR-ABL1+ (Ph+) B-ALL. AP-CML #1 shows loss of IKAROS staining in the primitive cells in the paratrabecular regions (black arrow) in contrast to strong IKAROS staining in the residual maturing CP-CML cells (yellow arrow). AP-CML #2 similarly shows loss of IKAROS in blasts (black arrows), but not in residual maturing cells (yellow arrows). The original magnification is shown on the bottom right of each panel.
These findings show that loss or reduced expression of IKAROS protein is a common feature of AP and myeloid BC-CML.
Forced expression of IK6 consistently causes a reproducibly sustained expansion of primitive BCR-ABL1+ cells from patients with CP-CML
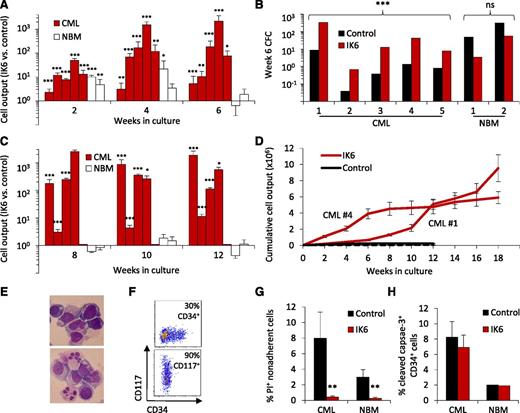
To determine whether IK6 has a consistent and disease-specific growth-promoting effect, we then applied the same design as shown in Figure 1A to the CD34+ cells obtained from an additional 4 CP-CML patients and 2 normal BM samples. In each CP-CML case, the results confirmed the initial finding of an enhanced output of mature (nonadherent) cells and leukemic CFCs from the IK6-transduced cells (10- to 30-fold increase after 6 weeks; Figure 3A-B). For 4 of the 5 CP-CML samples tested, secondary cultures initiated with cells harvested from the corresponding primary cultures showed that this IK6-mediated growth-promoting effect was sustained (Figure 3C). In the fifth case, there was no growth of either IK6- or control-transduced cells in the secondary cultures. The growth-promoting effects of IK6 continued to be evident in tertiary cultures of 2 of the 4 samples, whereas the control-transduced cells were no longer detectable in either of these (Figure 3D). In the other cases, there was no further growth of control or IK6-transduced cells beyond the secondary cultures. Of note, the tertiary cultures in which IK6+ CML cells continued to proliferate showed a progressive increase in the proportion of cells displaying a blast morphology and expressing surface markers characteristic of primitive cells (CD34 or CD117/KIT; Figure 3E-F).
IK6 consistently expands primitive CP-CML cell numbers more than normal cells in vitro. (A) Total mature (nonadherent) cells present in 2-, 4-, and 6-week-old cultures initiated with CD34+ cells (5-10 × 103 each, depending on the experiment) from 5 CP-CML patients and 2 normal BM samples according to the design shown in Figure 1A. Values are the mean ± SEM of the ratios of the number of test (GFP+) vs control (YFP+) cells measured in 4 replicates for each sample. P values: IK6 vs control. (B) Numbers of CFCs present after 6 weeks in the individual cultures shown in panel A, as enumerated in a 14-day methylcellulose assay and expressed as CFCs per 103 starting cells. (C) Total nonadherent cells in secondary 6-week cultures (each initiated with one primary culture equivalent). Cells from CP-CML patient #5 showed no growth in secondary cultures. Values are as in panel A. (D) Cumulative cell outputs in serial primary, secondary, and tertiary cultures initiated with cells from CP-CML patients #1 and #4. Cells from CML patients #2 and #3 showed no further growth in tertiary cultures. Values shown are the mean ± SEM. (E) Representative cytospin preparations of nonadherent cells present in tertiary cultures generated from CP-CML #1 (top) and #4 (bottom). (F) Representative flow cytometry plots corresponding to panel E showing analysis of nonadherent cells present in tertiary cultures generated from CP-CML #1 (top) and #4 (bottom). (G) Proportion of apoptotic (PI+) IK6- and control-derived CP-CML and normal BM nonadherent cells from 2- to 3-week-old cultures. Values shown are the mean ± SEM from 3 replicate cultures for each of the experiments shown in panel A. P values: IK6 vs control. (H) Proportion of apoptotic (cleaved caspase 3+) IK6- and control-derived CP-CML and normal BM CD34+ cells in 2- to 3-week-old cultures. Values shown are the mean ± SEM (as indicated) of results for 3 replicate cultures from 4 CP-CML samples and 1 normal BM sample. *P < .05; **P < .01; ***P < .001. NBM, normal bone marrow.
IK6 consistently expands primitive CP-CML cell numbers more than normal cells in vitro. (A) Total mature (nonadherent) cells present in 2-, 4-, and 6-week-old cultures initiated with CD34+ cells (5-10 × 103 each, depending on the experiment) from 5 CP-CML patients and 2 normal BM samples according to the design shown in Figure 1A. Values are the mean ± SEM of the ratios of the number of test (GFP+) vs control (YFP+) cells measured in 4 replicates for each sample. P values: IK6 vs control. (B) Numbers of CFCs present after 6 weeks in the individual cultures shown in panel A, as enumerated in a 14-day methylcellulose assay and expressed as CFCs per 103 starting cells. (C) Total nonadherent cells in secondary 6-week cultures (each initiated with one primary culture equivalent). Cells from CP-CML patient #5 showed no growth in secondary cultures. Values are as in panel A. (D) Cumulative cell outputs in serial primary, secondary, and tertiary cultures initiated with cells from CP-CML patients #1 and #4. Cells from CML patients #2 and #3 showed no further growth in tertiary cultures. Values shown are the mean ± SEM. (E) Representative cytospin preparations of nonadherent cells present in tertiary cultures generated from CP-CML #1 (top) and #4 (bottom). (F) Representative flow cytometry plots corresponding to panel E showing analysis of nonadherent cells present in tertiary cultures generated from CP-CML #1 (top) and #4 (bottom). (G) Proportion of apoptotic (PI+) IK6- and control-derived CP-CML and normal BM nonadherent cells from 2- to 3-week-old cultures. Values shown are the mean ± SEM from 3 replicate cultures for each of the experiments shown in panel A. P values: IK6 vs control. (H) Proportion of apoptotic (cleaved caspase 3+) IK6- and control-derived CP-CML and normal BM CD34+ cells in 2- to 3-week-old cultures. Values shown are the mean ± SEM (as indicated) of results for 3 replicate cultures from 4 CP-CML samples and 1 normal BM sample. *P < .05; **P < .01; ***P < .001. NBM, normal bone marrow.
Normal CD34+ BM cells transduced with IK6 also showed an initial minor enhanced output of cells by comparison with controls in the same cultures, but this effect had largely disappeared by 6 weeks (Figure 3A). IK6 expression in normal BM cells was also not accompanied by an enhanced output of either CFCs or cells able to grow in secondary cultures (Figure 3B-C).
To determine whether an effect on apoptosis was contributing to the enhanced output of primitive cells obtained from the IK6-transduced cells, we examined the frequency of PI+ total cells and cleaved caspase-3+ CD34+ cells in the cultures of test and control-transduced CP-CML and normal BM cells. Paired analysis showed that IK6 expression slightly, but significantly, reduced the proportion of apoptotic (PI+) mature cells derived from both CML and normal BM progenitors (P < .01 for both; Figure 3G). However, in neither case was there any change in the frequency of primitive (CD34+) cells that showed evidence of apoptosis based on cleaved caspase-3 expression (Figure 3H). Forced expression of IK6 in CP-CML CD34+ cells also did not alter their sensitivity to imatinib or dasatinib (supplemental Figure 3).
Together, these findings show that IK6 expression consistently enhances the expansion of primitive CML cells and their output of differentiating granulopoietic progeny in vitro. They also demonstrate that this effect on CP-CML cells is both more marked and more prolonged than the transient response obtained from similarly transduced normal BM CD34+ cells.
IK6 enhances the output of maturing basophils and eosinophils
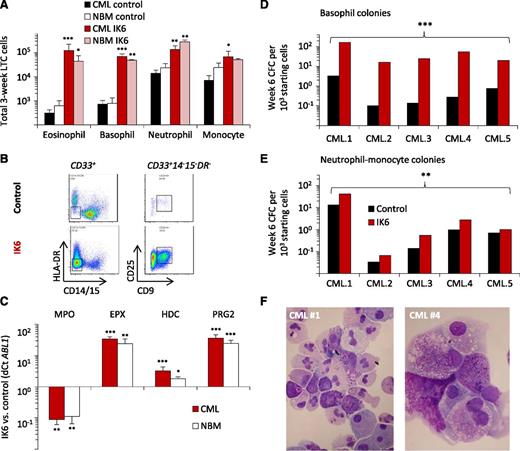
Cytospin preparations of the cells obtained from the cultures initiated with IK6-transduced cells of either CP-CML or normal BM origin showed a striking increase in morphologically defined basophils and eosinophils when compared with their control-transduced counterparts (Figure 4A). This finding was confirmed by both flow cytometric and transcript analyses of these cells (Figure 4B-C).
IK6 enhances basophil production from both normal and CP-CML progenitors. (A) Numbers of different mature cell types in the nonadherent fraction of 3-week cultures initiated with IK6- or control-transduced CD34+ cells from CP-CML or normal BM samples (as assessed in Wright-Giemsa–stained cytospin preparations). Values shown are the mean ± SEM from 3 replicate cultures from 5 CML and 2 normal BM samples; P values: IK6 vs control. (B) Representative flow cytometric analyses of nonadherent cells from 3-week cultures initiated with IK6- or control-transduced CD34+ cells from 5 CP-CML and 2 normal BM samples. Basophils and eosinophils were defined by a CD33+14−15−HLA−DR−25+9+/− phenotype. (C) Lineage-specific transcripts in the nonadherent cells shown in panel A; mean ± SEM of results from 3 replicate cultures from 5 CP-CML and 2 normal BM samples, P values: IK6 vs control. (D-E) Types of colonies obtained in assays of IK6- and control-transduced CP-CML cells harvested from 6-week cultures. The differentiated cell content of the colonies was confirmed in Wright-Giemsa–stained cytospin preparations of harvested colonies. P values: IK6 vs control. (F) Representative cytospin preparation of the cells in colonies generated in CFC assays from plating of nonadherent cells present in the tertiary cultures generated from CP-CML #1 and CP-CML #4. *P < .05; **P < .01; ***P < .001. EPX, eosinophil peroxidase (eosinophil specific); HDC, histidine decarboxylase (basophil specific); MPO, myeloperoxidase (neutrophil specific); NBM: normal bone marrow; PRG2, proteoglycan 2 (basophil/eosinophil).
IK6 enhances basophil production from both normal and CP-CML progenitors. (A) Numbers of different mature cell types in the nonadherent fraction of 3-week cultures initiated with IK6- or control-transduced CD34+ cells from CP-CML or normal BM samples (as assessed in Wright-Giemsa–stained cytospin preparations). Values shown are the mean ± SEM from 3 replicate cultures from 5 CML and 2 normal BM samples; P values: IK6 vs control. (B) Representative flow cytometric analyses of nonadherent cells from 3-week cultures initiated with IK6- or control-transduced CD34+ cells from 5 CP-CML and 2 normal BM samples. Basophils and eosinophils were defined by a CD33+14−15−HLA−DR−25+9+/− phenotype. (C) Lineage-specific transcripts in the nonadherent cells shown in panel A; mean ± SEM of results from 3 replicate cultures from 5 CP-CML and 2 normal BM samples, P values: IK6 vs control. (D-E) Types of colonies obtained in assays of IK6- and control-transduced CP-CML cells harvested from 6-week cultures. The differentiated cell content of the colonies was confirmed in Wright-Giemsa–stained cytospin preparations of harvested colonies. P values: IK6 vs control. (F) Representative cytospin preparation of the cells in colonies generated in CFC assays from plating of nonadherent cells present in the tertiary cultures generated from CP-CML #1 and CP-CML #4. *P < .05; **P < .01; ***P < .001. EPX, eosinophil peroxidase (eosinophil specific); HDC, histidine decarboxylase (basophil specific); MPO, myeloperoxidase (neutrophil specific); NBM: normal bone marrow; PRG2, proteoglycan 2 (basophil/eosinophil).
Similar results were obtained when the progeny of the IK6- and control-transduced CP-CML cells present in the 6-week cultures were compared for their content of basophil progenitors. Flow cytometric analysis showed the IK6-transduced cells had produced more CD34+133low cells, which are enriched for basophil progenitors34 (supplemental Figure 4A). CFC assays further showed a markedly increased output of cells that generated basophil colonies (Figure 4D). IK6 expression also enhanced the 6-week outputs in these cultures of neutrophil/macrophage CFCs (Figure 4E), although this latter effect was less pronounced than the enhanced output of basophil CFCs. In the secondary cultures generated from 4 of the 5 CP-CML samples studied, basophils were again the predominant mature cell type produced, although neutrophils and monocytes were also evident. In addition, the cells obtained from the 2 productive tertiary cultures of IK6-transduced CP-CML cells generated colonies of mature basophils as well as colonies of granulocytes and macrophages in methylcellulose cultures (Figure 4F), thus demonstrating that these cells had not irreversibly lost an ability to differentiate.
These results point to an ability of IK6 to induce additional key features of AP-CML; ie, an enhanced production of basophils as well as an expanded output of primitive cells. Notably, these effects occur in the absence of a noticeable impairment in their ability to terminally differentiate, a feature that characterizes progression to myeloid BC.
IK6 enhances the output of basophils at multiple stages of CP-CML progenitor differentiation
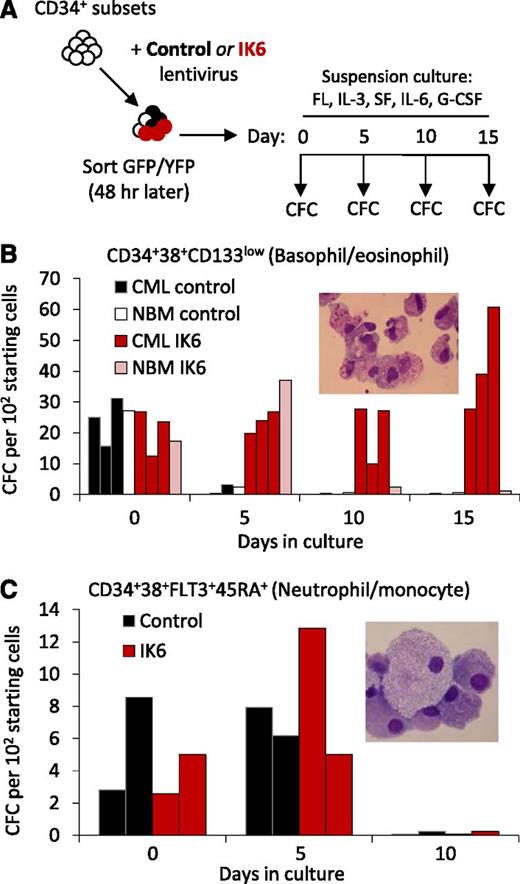
To investigate the differentiation stage(s) targeted by IK6 to cause an increase in basophil (and eosinophil) production, we introduced IK6 into defined progenitor subsets enriched in neutrophil/monocyte or basophil and eosinophil CFCs34,35 (Figure 5A). Of interest, basophil and eosinophil progenitors were already present at a higher frequency in unmanipulated CP-CML samples compared with normal BM (supplemental Figure 4B). IK6 expression did not alter the number of colonies obtained from either phenotype when the cells were plated immediately after transduction. However, after 5 days in growth factor–supplemented suspension cultures, both CP-CML and normal BM basophil and eosinophil progenitors transduced with IK6 produced many more progeny daughter CFCs than similarly cultured control-transduced cells. However, this effect of IK6 was more prolonged on CP-CML cells than on their normal BM counterparts (Figure 5B). In sharp contrast, IK6 had no effect on the number of CFCs obtained in similarly cultured neutrophil/monocyte progenitors of either normal BM or CP-CML origin (Figure 5C). These findings reveal an ability of IK6 to directly and specifically expand committed basophil progenitors, which is markedly enhanced in CP-CML target cells.
IK6 enhances basophil production from primitive CP-CML cells at multiple stages of differentiation. (A) Experimental design used to test the effect of IK6 on the CFCs produced from 2 different subsets of CD34+CD38+ cells after 0 to 15 days in the same suspension culture conditions used for lentiviral gene transfer. (B) Numbers of colonies produced from IK6- and control-transduced CD34+CD38+CD133low cells (enriched for basophil and eosinophil progenitors) obtained by FACS from 3 CP-CML patients and a pool of 3 normal BM samples and cultured as described in panel A. The inset shows a representative cytospin of cells in the colonies. (C) Numbers of colonies produced from IK6- and control-transduced CD34+CD38+FLT3+CD45RA+ cells (enriched for neutrophil/monocyte progenitors) obtained by FACS from 2 CP-CML patients, as described in panel A. The inset shows a representative cytospin of cells in the colonies.
IK6 enhances basophil production from primitive CP-CML cells at multiple stages of differentiation. (A) Experimental design used to test the effect of IK6 on the CFCs produced from 2 different subsets of CD34+CD38+ cells after 0 to 15 days in the same suspension culture conditions used for lentiviral gene transfer. (B) Numbers of colonies produced from IK6- and control-transduced CD34+CD38+CD133low cells (enriched for basophil and eosinophil progenitors) obtained by FACS from 3 CP-CML patients and a pool of 3 normal BM samples and cultured as described in panel A. The inset shows a representative cytospin of cells in the colonies. (C) Numbers of colonies produced from IK6- and control-transduced CD34+CD38+FLT3+CD45RA+ cells (enriched for neutrophil/monocyte progenitors) obtained by FACS from 2 CP-CML patients, as described in panel A. The inset shows a representative cytospin of cells in the colonies.
Overexpression of wild-type IKAROS suppresses the activity of primitive CP-CML cells and partially reverses the IK6-induced increase in basophil outputs
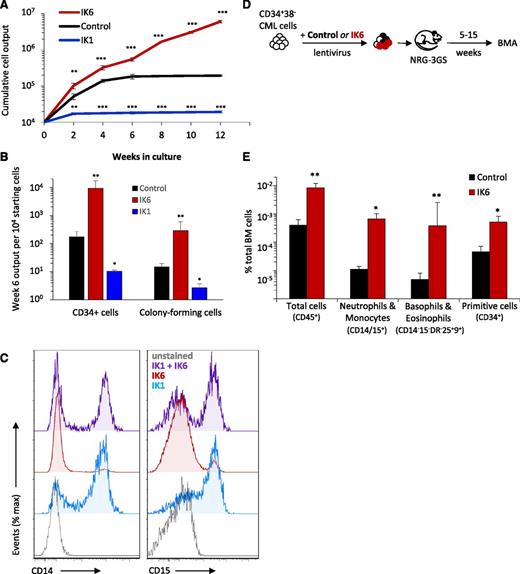
To further clarify the effects of IK6 expression, we compared its effects on CD34+ CP-CML cells with those induced by forced expression of wild-type IKAROS (IK1) using a similar IK1 transduction approach and experimental design as shown in Figure 1A. In contrast to the effects of IK6, forced expression of IK1 resulted in a rapid and sustained decrease in cell outputs in both primary and secondary cultures as compared with those obtained from the cultured control-transduced cells (Figure 6A). The number of CD34+ cells and CFCs produced from the IK1-transduced cells in the same cultures was also 5- to 10-fold lower that the values obtained for the control-transduced cells (Figure 6B). Importantly, cotransduction of CD34+ CP-CML cells with IK1 as well as IK6 partially reversed the effects of IK6 on basophil and eosinophil production (Figure 6C). These results demonstrate that increased levels of IK1 and IK6 in primitive CP-CML cells have opposing as well as opposite effects on their growth and granulopoietic differentiation control.
Forced expression of wild-type IKAROS (IK1) in CP-CML cells inhibits total cell outputs and progenitor expansion and compromises the effects of IK6, which include promoting the growth of CP-CML cells in vivo. (A) Cumulative cell outputs in serial primary and secondary cultures (6 weeks + 6 weeks) initiated with cells from CP-CML patient #1 transduced with IK6, IK1, or a control vector. Values shown are the mean ± SEM of results from 3 replicate cultures. P values: IK1 or IK6 vs control. (B) Numbers of CD34+ cells and CFCs (as enumerated in a 14-day methylcellulose assay) present after 6 weeks in the individual cultures shown in panel A. Values shown are the mean ± SEM of results from 3 replicate cultures, expressed as CD34+ cells or CFCs per 104 starting cells. P values: IK1 or IK6 vs control. (C) Representative flow cytometric profiles of cell-surface expression of CD14 (monocyte) and CD15 (neutrophil) on nonadherent cells in 2- to 3-week cultures initiated with CD34+ CP-CML cells transduced with IK1, IK6, or a combination of IK1 and IK6 (total of 4 CP-CML samples tested showing similar results). (D) Experimental design used to assess the effects of IK6 on the growth of CD34+ CP-CML cells in immunodeficient mice cotransplanted with equal numbers of IK6 (GFP+)- and control (YFP+)-transduced CD34+CD38− cells from CP-CML #1 (105 cells of each per mouse). (E) Levels of BM chimerism (total human and subsets thereof) obtained 5 weeks posttransplant. Values shown are the mean ± SEM of results from 5 individually assessed mice. P values: IK6 vs control. *P < .05; **P < .01; ***P < .001. BMA, bone marrow aspirate; IK1, wild-type (full-length) IKAROS.
Forced expression of wild-type IKAROS (IK1) in CP-CML cells inhibits total cell outputs and progenitor expansion and compromises the effects of IK6, which include promoting the growth of CP-CML cells in vivo. (A) Cumulative cell outputs in serial primary and secondary cultures (6 weeks + 6 weeks) initiated with cells from CP-CML patient #1 transduced with IK6, IK1, or a control vector. Values shown are the mean ± SEM of results from 3 replicate cultures. P values: IK1 or IK6 vs control. (B) Numbers of CD34+ cells and CFCs (as enumerated in a 14-day methylcellulose assay) present after 6 weeks in the individual cultures shown in panel A. Values shown are the mean ± SEM of results from 3 replicate cultures, expressed as CD34+ cells or CFCs per 104 starting cells. P values: IK1 or IK6 vs control. (C) Representative flow cytometric profiles of cell-surface expression of CD14 (monocyte) and CD15 (neutrophil) on nonadherent cells in 2- to 3-week cultures initiated with CD34+ CP-CML cells transduced with IK1, IK6, or a combination of IK1 and IK6 (total of 4 CP-CML samples tested showing similar results). (D) Experimental design used to assess the effects of IK6 on the growth of CD34+ CP-CML cells in immunodeficient mice cotransplanted with equal numbers of IK6 (GFP+)- and control (YFP+)-transduced CD34+CD38− cells from CP-CML #1 (105 cells of each per mouse). (E) Levels of BM chimerism (total human and subsets thereof) obtained 5 weeks posttransplant. Values shown are the mean ± SEM of results from 5 individually assessed mice. P values: IK6 vs control. *P < .05; **P < .01; ***P < .001. BMA, bone marrow aspirate; IK1, wild-type (full-length) IKAROS.
IK6 enhances CP-CML cell outputs in transplanted immunodeficient mice
To determine whether the effects of IK6 would also be seen in an in vivo setting, we coinjected NRG-3GS mice IV with equal numbers of IK6 (GFP+)- and control- (YFP+) transduced CD34+CD38− CP-CML cells and then monitored the number of human cells in the BM of the transplanted mice over the following 15 weeks (Figure 6D). The results again showed a significant enhancing effect of IK6 expression on the number of cells regenerated in the mice. Notably, these included the number of regenerated CD34+ cells as well as the size of the regenerated neutrophil, monocyte, basophil, and eosinophil populations (Figure 6E). These findings provide further evidence for a marked, cell-autonomous stimulating effect of IK6 expression on the cell-output activity of primitive CML cells.
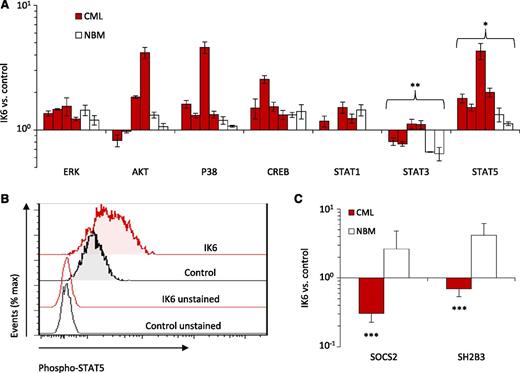
IK6 enhances STAT5 signaling in CD34+ CP-CML cells
We then used flow cytometry and qRT-PCR to look for molecular pathways that are differentially activated in primitive (CD34+) IK6-transduced CP-CML cells. We found that in vitro–generated CD34+ progeny contained consistently higher levels of activated signal transducer and activator of transcription 5 (STAT5) by comparison with those derived from either matched control-transduced cells or IK6-transduced normal BM cells. Effects of IK6 on other signaling intermediaries in CD34+ CP-CML cells were more heterogeneous (Figure 7A-B). In light of these findings, we hypothesized that the expression of IK6 in CP-CML CD34+ cells may disrupt mechanisms regulating JAK2-STAT5 activity in them. Consistent with this postulate was the finding that IK6-expressing CD34+ CP-CML cells contained decreased levels of SOCS2 and SH2B3 (LNK) transcripts, both of which encode important known negative regulators of JAK-STAT signaling. Notably, none of these molecular effects were apparent in the CD34+ IK6-transduced normal BM cells (Figure 7C). These findings raise the possibility that wild-type IKAROS may play a role in downmodulating the activation of STAT5 in CD34+ CP-CML cells.
IK6 enhances the activation of STAT5 in CD34+ CML cells. (A) Phosphoprotein analysis of CD34+ cells from 2- to 4-week cultures initiated with IK6- or control-transduced CD34+ cells shown individually for 4 CP-CML and 2 normal BM samples except for the STAT1 analysis, which was performed on 3 CP-CML and 1 normal BM sample. Values shown for each protein are the mean ± SEM of the ratios of the median fluorescent intensity values obtained for the IK6- compared with control-transduced cells in 3 replicate experiments. P values were generated by comparing the effects of IK6 on CP-CML vs normal BM cells. (B) Representative flow cytometry histograms taken from one of the CML experiments shown in panel A. (C) Comparison of transcript differences in IK6- and control-transduced CD34+ cells harvested from 2- to 4-week cultures for 5 CP-CML and 2 normal BM samples. Values shown are the mean ± SEM of the ratios of normalized transcript levels in the progeny of IK6- as compared with control-transduced cells obtained from 3 to 5 replicate cultures (with the exception of one of the normal BM samples where only 1 replicate could be set up). P values were generated by comparing the effects of IK6 on CP-CML vs normal BM cells. ***P < .001, **P < .01, *P < .05. NBM, normal bone marrow.
IK6 enhances the activation of STAT5 in CD34+ CML cells. (A) Phosphoprotein analysis of CD34+ cells from 2- to 4-week cultures initiated with IK6- or control-transduced CD34+ cells shown individually for 4 CP-CML and 2 normal BM samples except for the STAT1 analysis, which was performed on 3 CP-CML and 1 normal BM sample. Values shown for each protein are the mean ± SEM of the ratios of the median fluorescent intensity values obtained for the IK6- compared with control-transduced cells in 3 replicate experiments. P values were generated by comparing the effects of IK6 on CP-CML vs normal BM cells. (B) Representative flow cytometry histograms taken from one of the CML experiments shown in panel A. (C) Comparison of transcript differences in IK6- and control-transduced CD34+ cells harvested from 2- to 4-week cultures for 5 CP-CML and 2 normal BM samples. Values shown are the mean ± SEM of the ratios of normalized transcript levels in the progeny of IK6- as compared with control-transduced cells obtained from 3 to 5 replicate cultures (with the exception of one of the normal BM samples where only 1 replicate could be set up). P values were generated by comparing the effects of IK6 on CP-CML vs normal BM cells. ***P < .001, **P < .01, *P < .05. NBM, normal bone marrow.
Discussion
Recent studies have uncovered a high degree of clonal complexity in AML, and have highlighted divergent cellular behaviors of evolving subclones, including early clones in which maturation is not grossly perturbed.36-39 However, experimental systems suitable for elucidating early changes that contribute to the genesis of de novo acute leukemia in normal human hematopoietic cells have proven difficult to develop. Genetic manipulation of CP-CML patients’ cells represents an attractive model, as clinical phenotypes associated with clonal evolution are well established in this disease, including the stepwise transitioning from CP to BC via an intermediate AP characterized by an expansion of primitive cells but without evidence of an arrest in their ability to differentiate.40
Here, we describe the power of a stromal-based culture system to reveal the effects of specific genetic alterations on the proliferative and differentiation responses of primitive (CD34+) CP-CML cells. From a preliminary survey of multiple genetic perturbations implicated in human leukemogenesis, we identified 4 with reproducibly sustained growth-promoting effects on primitive CP-CML cells. One of these was IK6, a dominant-negative isoform of IKAROS that suppresses IKAROS activity by preventing its entry into the nucleus,32,33,41 as confirmed here. In fact, IK6 proved the most potent stimulator of primitive CP-CML cell expansion in this system but did not block the ability of their progeny to terminally differentiate. IK6 expression also promoted the production of basophils and eosinophils from CP-CML cells. This is of interest, as in patients with CML, an increased basophil count is both a bad prognostic indicator and a feature of AP-CML.42,43 A role for IKAROS in limiting basophil differentiation has recently been identified in the mouse.44 In addition, we found that IK6 expression in CD34+ CP-CML cells enhances activation of STAT5, which has also been implicated as a biomarker of disease progression in CML patients.45,46
Our finding that IK6-transduced CP-CML CD34+ cells from patients could produce terminally differentiated myeloid cells for up to 18 weeks in vitro and for at least 15 weeks in vivo (in transplanted immunodeficient mice) stands in contrast to a recent study in which the effects of forced expression of BCR-ABL1 and IK6 in human cord blood cells were found to produce a myeloid leukemia in transplanted mice.47 The different outcome in that study likely reflects major differences in the system used; ie, naive human cord blood cells transduced with a retroviral BCR-ABL1 vector in contrast to our focus on BCR-ABL1+ cells obtained directly from adult patients with clinically determined CP-CML disease. The disparity in leukemia initiation between the 2 models is reminiscent of a previous study where forced expression of both BCR-ABL1 and BMI1 in naive human cord blood cells produced lymphoblastic leukemias, whereas the outcome of forced expression of BMI1 in CML patients’ cells was limited to a mild growth-enhancing effect without a block in differentiation.48 Potential explanations for these differences include the supraphysiological STAT5 activation resulting from γ-retroviral–mediated BCR-ABL1 expression, which drives erythroid differentiation in the transduced cells27,49,50 and fails to mimic the differentiation stage-associated changes (decreases) in BCR-ABL1 expression characteristic of CP-CML cells.51,52 Together, however, these different models both point to a cooperativity between IK6 and BCR-ABL1 in promoting disease progression in CML.
AP-CML has long been recognized as a transition phase heralding BC-CML in some patients. However, failure to identify biological or molecular changes specific to AP-CML has kept its acceptance as a distinct entity controversial. Reported similarities between the gene expression profiles of AP and myeloid BC cells have also given added weight to the alternative concept of a simpler biphasic model of disease progression in CML.53 Our finding here that IK6 produces a stable phenotype in primitive CP-CML cells that mimics features of AP now argues strongly in favor of a disease progression model in which AP can constitute a distinct entity intermediate between CP and BC. This concept is further supported by our discovery that IKAROS protein is frequently already absent or decreased in the blasts of AP-CML patients as well as those with myeloid BC. Together, these results point to a loss of IKAROS activity playing a frequent role in the progression of CML to a more advanced phase.
Genomic deletions resulting in loss of functional IKAROS expression are a hallmark of BCR-ABL1–positive B-ALL but have only rarely been reported in patients with myeloid BC-CML.12-14,16-18 Given the high frequency of reduced or absent IKAROS protein expression that we observed in AP-CML and myeloid BC-CML, it seems unlikely that an underlying genomic deletion is responsible. Rather, the paucity of genomic abnormalities in published cases suggests either epigenetic silencing of IKZF1 expression or accelerated loss of IKAROS protein by posttranslational mechanisms is responsible. In support of the former, serial analysis of IKZF1 transcripts53 has shown a stepwise decrease in patient samples tracked from CP through AP to BC-CML (supplemental Figure 5). However, more definitive elucidation of the mechanisms involved, possibly from future analyses of the transcriptional regulation of the IKZF1 locus in normal and leukemic human cells, will clearly be of interest in this regard.
In summary, this work reveals a previously unknown role of IKAROS in regulating human basophil and eosinophil differentiation and points to a loss of IKAROS activity as a potential pathologic harbinger of advancing-phase myeloid disease in CML patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank G. Edin, M. Hale, the Flow Cytometry Core Facility of the Terry Fox Laboratory, and the Animal Resource Center of the BC Cancer Agency for technical assistance. They are grateful to A. Weng, K. Humphries (Terry Fox Laboratory, Vancouver), G. Sauvageau (Institute for Research in Immunology and Cancer, Montreal), and R. Levine (Memorial Sloan Kettering Cancer Center, New York) for sharing DNA plasmids and T. Holyoake (University of Glasgow, UK) and the Hematology Cell Banks of British Columbia, the Fred Hutchison Cancer Research Centre, the University of Western Australia, McMaster University Medical Centre, and the Oregon Health & Science University for providing patients’ cells.
This work was supported by grants from the Terry Fox Foundation, the Canadian Institutes of Health (CIHR), and the Natural Sciences and Engineering Research Council of Canada. P.A.B. held a Kay Kendall Leukaemia Fund Intermediate Fellowship from the United Kingdom. D.J.H.F.K. held CIHR and Vanier Canada Graduate Scholarships, P.H.M. held a CIHR Banting Studentship and formerly a CIHR Transplantation Studentship, S.B. held a University of British Columbia Graduate Studentship and a Vanier Canada Graduate Scholarship, E.B. held a CIHR Regenerative Medicine Graduate Fellowship, and N.K. held a MITACS Elevate Postdoctoral Fellowship.
Authorship
Contribution: P.A.B. designed and performed experiments, analyzed data, and wrote the manuscript; D.J.H.F.K., P.H.M., I.S., and G.R. performed and optimized xenograft analyses and animal husbandry protocols; S.B. performed cloning; E.B. performed culture experiments; N.K. performed confocal analyses; J.T., B.J.D., M.M.L., K.R.L., and J.P.R. provided patient trephine biopsy samples; K.H. and W.N.E. undertook IKAROS immunohistochemistry studies; and C.J.E. directed the project and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Connie J. Eaves, Terry Fox Laboratory, 675 West 10th Avenue, Vancouver, BC V5Z 1L3, Canada; e-mail: ceaves@bccrc.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal