Abstract
B-lymphocyte development in the bone marrow is controlled by the coordinated action of transcription factors creating regulatory networks ensuring activation of the B-lymphoid program and silencing of alternative cell fates. This process is tightly connected to malignant transformation because B-lineage acute lymphoblastic leukemia cells display a pronounced block in differentiation resulting in the expansion of immature progenitor cells. Over the last few years, high-resolution analysis of genetic changes in leukemia has revealed that several key regulators of normal B-cell development, including IKZF1, TCF3, EBF1, and PAX5, are genetically altered in a large portion of the human B-lineage acute leukemias. This opens the possibility of directly linking the disrupted development as well as aberrant gene expression patterns in leukemic cells to molecular functions of defined transcription factors in normal cell differentiation. This review article focuses on the roles of transcription factors in early B-cell development and their involvement in the formation of human leukemia.
B-cell development and acute lymphoblastic leukemia
The development of functional B lymphocytes is dependent on the maturation of multipotent progenitors (MPPs) in the bone marrow (BM). The formation of lineage-restricted progenitors is co-coordinated by the action of transcription factors that activate B-lineage genes as well as restrict alternative cell fates. Analysis of genetically modified mouse models has developed our understanding of the regulatory networks at play in specific stages of B-cell differentiation to drive the differentiation process. The interest in this area of investigation has increased over the last few years with the developing insight that the same regulatory networks are modulated through genetic alterations in human hematologic malignancies. Considering the crucial roles for stage- and lineage-specific transcription factors such as PAX5, IKZF1 (IKAROS), TCF3 (E2A, TCFE2A), and EBF1 in the regulation of normal B-lymphocyte differentiation, it can be predicted that disruptions in the balanced action of these proteins represent an underlying cause of phenotypic features such as developmental arrest observed in B-lineage acute lymphoblastic leukemia (B-ALL).
Even if cytogenetic analysis is the preferred tool for classification of B-lineage leukemia in the clinic, immunophenotyping using classical fluorescence-activated cell sorting (FACS) is a useful tool if characteristics of leukemic cells are to be compared with their normal counterparts. Expression of CD34 and DNA nucleotidylexotransferase is more prominently detected in B-cell precursor ALL (BCP-ALL) whereas metalloendopeptidase (CD10) is detected on leukemia cells in 90% of the B-ALL cases, including some of the more differentiated pre-B ALL cells.1,2 Even though the pre-B ALL cells express cytoplasmic immunoglobulin heavy chain (cIgH), detectable surface IgH expression (sIgH) is limited to B-ALL cells.1,2 The detection of cIgH has a limited value in leukemia diagnosis, however, determination of the developmental stage based on IgH expression presents an advantage over the use of surface markers because it is linked to certain functional characteristics of a defined differentiation stage and not just expression of a certain surface marker. Complete lack of immunoglobulin expression, as in B-precursor ALL, suggests that the progenitor cell either has not completed immunoglobulin rearangement or that the recombination event has failed to generate a functional IgH chain gene. The expression of intracellular IgH chain, as detected in pre-B ALL, suggests that even though an IgH chain has been generated, the cells fail to express high levels of immunoglobulin on the cell surface likely as a result of having failed to generate or possibly express a functional Ig-light chain (IgL). Additionally, staging based on immunoglobulin status creates an opportunity to translate data collected from mouse models to increase our understanding of human leukemia because although surface marker expression differs between the human and the mouse, the order of recombination events is thought to progress in a similar manner.3 Using IgH expression for determination of developmental stage, it has been estimated that B-precursor ALL accounts for 65% to 70% of all infant and childhood leukemias and 50% of the B-lineage ALLs in adults, whereas the pre-B ALLs compose about 25% and sIgH-expressing B-ALLs represent in the range of 2% to 5% of the childhood leukemias.1,2 Hence, it is reasonable to suggest that the majority of the B-lineage leukemias display an early block in development at a stage corresponding to the pro-B or early pre-B-cell stage in normal B-cell development.
This review aims to provide an overview of transcription regulatory networks in normal early B-lymphocyte development and their potential involvement in malignant transformation and human leukemia.
Lymphoid priming in multipotent progenitors creates a permissive epigenetic landscape for B-cell development
Even though B-ALL is defined by expansion of B-lymphoid progenitors, some of the most relevant transcription factors in human B-lineage ALL play crucial roles already in noncommitted progenitors by modulation of the epigenetic landscape and stimulation of transcription to initiate lineage priming. The concept of lineage priming in the hematopoietic system was established in the late 1990s, when it was reported that early MPPs express low levels of lineage-restricted genes presumably as a mean to retain certain lineage potentials.4 Functional lymphoid lineage priming is dependent on the transcription factors IKZF1,5 SPI1 (PU.1),6,7 and TCF38,9 acting in a concerted manner to sustain the expression of lymphoid-associated genes. Using reporter transgenic mice to prospectively isolate and functionally validate early progenitors, it was revealed that MPPs expressing high levels of SPI1 were primed toward myelolymphoid development with reduced capacity for development toward eythromyeloid cell fates.7 This could be explained by SPI1 interacting with regulatory elements to create a permissive epigenetic landscape for downstream-acting lineage-restricted transcription factors.10 This idea is supported by the findings that in the absence of functional SPI1, the formation of lymphoid as well as granulocyte/monocyte progenitors is disrupted.11 Within the multipotent progenitor compartment, high expression of the protein tyrosine kinase FLT3 can be used to prospectively isolate progenitors with high lymphomyeloid but limited erythromyeloid lineage potentials.12 These cells express a set of lymphoid lineage genes including Rag1, Tdt, and sterile immunoglobulin transcripts likely reflecting an epigenetic priming to facilitate lymphoid cell fate.13,14 In the absence of fully functional TCF3,8,9 IKZF1,5 MYB,15 or SPI116,17 the expression of Flt3 as well as other lymphoid lineage genes is reduced, revealing that a complex network of transcription factors plays a crucial role in priming for B-lineage development already in the MPP compartment.
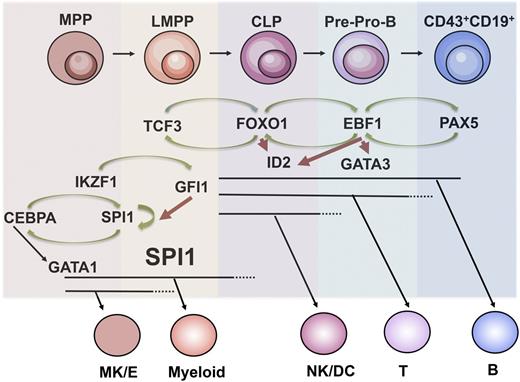
Subsequent differentiation toward B-lymphoid cell fate involves loss of capacity to generate myeloid cells when the progenitors enter the common lymphoid progenitor (CLP) stage.18 The loss of myeloid lineage potential been suggested to involve an interplay between the transcription factors IKZF1, SPI1, and GFI119 (Figure 1). High expression of SPI1 in progenitors results in an adoption of myeloid cell fate while intermediate expression stimulates the formation of B-lineage cells.20 In order to modulate the activity of SPI1, IKZF1 drives transcription from the Gfi1 gene encoding a Zn-finger transcriptional repressor able to directly interact with regulatory elements in the Spi1 gene,19 thereby reducing the transcriptional activity and disrupting the autoregulatory loop driving Spi1 expression. The importance of GFI1 in early B-cell development is supported by the observation that in Gfi1-deficient mice, the generation of the CLP compartment as well as pre-pro-B and pro-B cells is impaired.21 Hence, transcription factors such as IKZF1 and TCF3 stimulate B-cell development through the creation of a proper epigenetic landscape and preservation of lymphoid cell fate options already in MPP compartments.
Regulatory networks in early B-cell development. Schematic drawing over regulatory networks involved in B-cell priming, specification, and commitment. Specific stages and loss of lineage potentials are indicated. Green arrows indicate activation of transcription and red repression of transcription. B, B lymphocyte; CLP, common lymphoid progenitor; G/M, granulocyte/monocyte; LMPP, lymphoid-primed multipotent progenitor; Mk/E, megakaryocyte/erythroid; MPP, multipotent progenitor; NK/DC, natural killer/dendritic; T, T lymphocyte.
Regulatory networks in early B-cell development. Schematic drawing over regulatory networks involved in B-cell priming, specification, and commitment. Specific stages and loss of lineage potentials are indicated. Green arrows indicate activation of transcription and red repression of transcription. B, B lymphocyte; CLP, common lymphoid progenitor; G/M, granulocyte/monocyte; LMPP, lymphoid-primed multipotent progenitor; Mk/E, megakaryocyte/erythroid; MPP, multipotent progenitor; NK/DC, natural killer/dendritic; T, T lymphocyte.
Even though our understanding of early B-lineage development is based mainly on studies of mouse models, many of the basic concepts such as an early separation between myelolymphoid and erythromyeloid cell fate in MPPs22 as well as lineage priming23 appear to be well conserved between mouse and human. A complete disruption of differentiation at the MPP stage would be predicted to cause acute undifferentiated leukemia, a rare condition detected only in about 1% of leukemia patients,1,2 arguing against disruption of lineage priming being a common cause of leukemia formation. However, the relationship between pathological changes in early compartments and features of lineage-specified leukemia cells is complex and alterations in epigenetic programming may well be preserved during the maturation process to impact the behavior of the more differentiated cells.
B-lineage commitment is controlled by regulatory feed-forward loops ensuring activation of lineage-specific genes and repression of alternative cell fates
Once the myeloid lineage potentials are lost, the cells are thought to enter the CLP state with retained potential for development into B, T, natural killer (NK), or dendritic (DC) lineages. The CLP was originally defined as a multipotent cell lacking surface expression of classical lineage markers and with an intermediate expression of LY6A (SCA1) and KIT as well as high expression of IL7 receptor18 and FLT3.24 However, using either the expression of a Rag-125 reporter or the surface expression of LY6D25,26 allowed for the isolation of 2 functionally distinct populations within this cellular compartment. Cells lacking LY6D or Rag1 expression retain all the lymphoid lineage potentials whereas cells positive for any of these markers displayed reduced NK and DC lineage potential.25,26 Development into a LY6D+ stage is critically dependent on TCF326 and the LY6D−CLPs generated in TCF3-deficient mice lack expression of several lymphoid-restricted genes including Rag1 and Dntt27 as well as of the transcription factor FOXO1,27 critically important for normal B-cell development.28,29 Increased expression of Foxo130 in combination with STAT5 activation resulting from IL7R signaling is suggested to activate the transcription of the Ebf1 gene encoding the transcription factor EBF1.31 The importance of the functional interplay between transcription factors and IL7 signaling to promote B-cell specification by induction of Ebf1 expression is supported by the findings that EBF1 can partially rescue B-cell development in mice lacking either IL731 or ZBTB17 (MIZ1), a BTB/POZ domain transcription factor reported crucial for functional IL7 signaling.32
Even though Ebf1 expression can be detected in a variety of tissues, the expression in the hematopoietic system is restricted to the B-lymphoid compartment where it is essential for the activation of the B-lineage–specific program in the earliest B-cell progenitors.33,34 This ability may be linked to that EBF1 has the ability to functionally interact with the SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeling complex35 and thereby activate genes from an epigenetically silenced stage.36 Even though LY6D+ lymphoid progenitors are generated even in the absence of EBF1,37,38 these cells do not express B-lineage genes30 and are not properly lineage-restricted but retain the ability to develop into both NK and DC lineage cells.38 The ability of EBF1 to repress NK cell fate may reside in the ability to suppress the expression of the transcription factor ID2, an important regulator of NK cell development.39 The phenotypic features of EBF1- and FOXO1-deficient LY6D+ lymphoid progenitor cells are highly similar suggesting that they participate in the same regulatory network.30 This was supported by the findings that EBF1 and FOXO1 share regulatory elements in target genes,40 including in the Ebf1 and Foxo1 genes themselves,30 creating an autoregulatory loop driving B-lineage specification through activation of the B-cell–specific gene expression program (Figure 1).
Even if the specification process involves the activation of a large set of B-lineage–restricted genes and initiation of immunoglobulin recombination, these progenitors are not yet stably committed to B-lineage development. In order to achieve stable B-cell commitment EBF1 drives transcription of the Pax5 gene41 to initiate a feed-forward loop between PAX5 and EBF1 that serves to activate a defined set of genes including CD19, allow for completion of IgH-VDJ recombination, and eliminate the potential for alternative cell fates (Figure 1). While the process of specification and commitment is functionally interconnected because Pax5 is an EBF1 target gene,41 it is possible to functionally separate the specification from the commitment process because although PAX5-deficient progenitor cells express a large set of B-lineage–restricted genes,42,43 they are not stably committed.43-46 Even though a process resembling dedifferentiation allows for a limited plasticity toward myeloid lineages,45,47 B-lineage–specified Pax5-deficient cells mainly adopt other lymphoid cell fates converting into T-lineage cells in vitro43 and in vivo.46,47 This plasticity has been suggested to depend on the ability of PAX5 to repress transcription of the Notch1 gene,48 being crucial for early T-cell development. The ability of PAX5 to act as a repressor as well as an activator appears to be context dependent and the protein has been shown capable to functionally interact with both transcriptional activators of the SWI/SNF complex35 and repressors of the Groucho family.49 A similar mechanism is suggested to result in lineage plasticity after the induced loss of EBF1 in B-lineage progenitors50 likely as a result of in increased expression of T-lineage genes including the EBF1 targets Notch-150 and Gata-351 . The interplay between PAX5 and EBF1 is highly complex and involves not only autoregulatory loops but also apparently overlapping and in some regards redundant functions.52 The ability to repress alternative cell fates appears to be dependent on transcription factor dose because increased EBF1 expression is sufficient to cause lineage restriction and prevent formation of myeloid cells from PAX5 deficient hematopoietic progenitors.52 This may be of relevance in the formation of biphenotypic leukemia, a condition detected in about 7% of the ALLs, characterized by combined expression of markers associated with more than 1 hematopoietic lineage.1,2 Reduction of PAX5 dose has been reported to result in expression of myeloid genes in B-cell leukemia models53 and the PAX5 target, BACH2 have been reported to be of importance for myeloid lineage restriction in the CLP compartment.54 Furthermore, oscillations in the levels of PAX5 and EBF1 in MYC-induced lymphoma result in plasticity causing lineage switch between B and myeloid lineages.55 Hence, modulation of regulatory networks may cause disruptions in lineage fidelity possibly contributing to the formation of biphenotypic and mixed lineage malignancies.
Transiting through the pre-B-cell stage demands an interplay between cellular signaling and transcription factor networks
Even though more than half of the B-ALL cases represent BCP-ALL, about 25% of the leukemias are pre-B ALLs, expressing cIgH.1,2 This suggest that while the earliest stages of development has progressed in an apparently normal fashion, subsequent differentiation and initiation of IgL recombination is disturbed. This may be of special relevance for the uncontrolled expansion of pre-B ALL cells because functional IgH recombination is followed by a proliferative burst as a result of combined signaling through the IL7 and the pre-B-cell receptor (pre-BCR).56 The pre-BCR, essential for normal B-cell development, is composed of the newly rearranged IGH chain and the surrogate light chains IgLL1 (λ5, in humans IgLL1 or 14.1) and VPREB as well as signal transducing proteins including CD79A and CD79B.57 The genes encoding the pre-BCR components are direct targets for EBF1 and or PAX540,58,59 and are part of the developmental program initiated already during the specification process.25 Although IL7 signaling is crucial for the expansion of the pro-B- and early pre-B-cell compartment, the progressive development and initiation of IgL recombination demands exit from the cell cycle and reduced IL7 signaling.60,61 The relevance for Il7 signaling in human malignancies is highlighted from the findings that mutations in either the Il7RA per se or other genes involved in the signaling pathway are found in both B- and T-cell leukemia.62 Such activating mutations could potentially impose a developmental block at the pre-B cell stage resulting in both uncontrolled proliferation and impaired IgL recombination. Disruption of IL7R signaling in normal cell development has been suggested to be mediated by a regulatory network where the pre-BCR signal induce increased expression of the transcription factors PAX5 and interferon regulatory factor-4 (IRF-4) as well as stabilization of FOXO1.63 This has several important implications because PAX5 activate transcription of the gene encoding IKZF3 (AIOLOS, IKAROS3)64 suggested to act in collaboration with IKZF1 to displace EBF1 on regulatory elements thereby silencing transcription of the surrogate light chain genes reducing the formation of the pre-BCR complex.65 The crucial role for IKZF1 is highlighted by that deletion of Ikaros in B-cell progenitors result in a complete differentiation block at the pre-B cell stage with impairment in IGL recombination.66 Furthermore, PAX5 is involved in the regulation of the expression of Irf8.64 IRF8 collaborate with IRF4, a direct target for pre-BCR signaling,61 to stimulate the initiation of light chain recombination and progression through the pre-B cell stage.67 The induction of IRF4 has been shown to result in increased expression of the chemokine receptor CXCR4, suggested to cause migration of the pre-B cell away from the IL7-producing stromal cells in the bone marrow thereby reducing IL7 receptor signaling allowing for progression of differentiation.61 Inactivation of the IRF4 and 8 genes is mainly found in myeloid and B-lineage chronic leukemia in humans, however, combined inactivation of these proteins result in the generation of B-lineage ALL in mouse models.68 The pre-BCR signal cause stabilization of FOXO163 thereby promoting reinduction of Rag gene expression28,29 crucial for functional IgL recombination. Hence, progression though the pre-B cell stage involves coordinated activities of transcription factors and specific signaling pathways and disruptions in the balance of these events may well underlie the developmental block observed in pre-B ALL.
Disruption of transcription factor networks link development to disease
While being crucial for normal B-lymphocyte development, several transcription factors with defined roles in B-cell differentiation are directly affected either via translocations, deletions or mutations in B-lineage ALL (Table 1).2,69-108 PAX5107 as well as EBF199 has been found translocated into the IGH loci in B-cell lymphoma resulting in that the transcription of these genes cannot be properly silenced upon terminal B cell differentiation possibly contributing to the developmental arrest observed in the lymphoma cells. While EBF1, with the exception of the EBF1-PDGFRB fusion protein,84 appear to have a rather limited number of fusion partners in B-lineage ALL, PAX5 has been reported to generate fusion proteins with a substantial number of partners including NCOR1, DACH2, GOLGA6, and TAOK1 genes100 (Table 1). TCF3 is involved in the formation of translocations generating fusion proteins with the homeobox transcription factor PBX1109 as well as the transcription factor hepatic leukemia factor (HLF)92 to create powerful oncoproteins.
Genetic changes in human leukemia involving the genes encoding the transcription factors IKZF1, TCF3, EBF1, and PAX5
| Gene . | Malignant phenotype . | Genetic alteration . | Comments . | Reference . |
|---|---|---|---|---|
| IKZF1 | B-lineage ALL | Deletion | Pediatric ALL. In 8%-15% of B-ALL cases. Inactivation of IKZF3 in 3 cases. | Mullighan et al69 |
| Kuiper et al70 | ||||
| Krentz et al71 | ||||
| In adult ALL. In 18%-32% of B-ALL cases. | Safavi et al72 | |||
| Paulsson et al73 | ||||
| Marker for poor prognosis. Detected in 29%-35% of relapsing B-ALL. | Mullighan et al74 | |||
| Yang et al75 | ||||
| Krentz et al71 | ||||
| Van der Veer et al76 | ||||
| Olsson et al77 | ||||
| Asai et al78 | ||||
| Schwab et al79 | ||||
| Dörge et al80 | ||||
| Buitenkamp et al81 | ||||
| Kuiper et al82 | ||||
| Combined with BCR-ABL1. Deleted in 84% of BCR-ABL1 positive ALL in children. | Mullighan et al69 | |||
| Mullighan et al83 | ||||
| Combined in 51% of adult BCR-ABL1 B-ALL cases. | Safavi et al72 | |||
| Combined with activating mutations in FLT3 or Il7R genes. | Roberts et al84 | |||
| Combined with deletion of SH2B3 (encoding LNK) | Roberts et al84 | |||
| Down syndrome. Deletions detected in 35% of ALL patients. | Buitenkamp et al81 | |||
| Mixed phenotype ALL | Deletion | Detected in 13% of patients. | Yan et al85 | |
| B-cell lymphoma | t(3;7)(q27;p12) | Cause deregulated BCL6 expression. | Hosokawa et al86 | |
| TCF3 | B-lineage ALL | Deletion | Pediatric ALL, 1 case. | Mullighan et al69 |
| Inactivation of TCF4 in 2 cases. | Kuiper et al70 | |||
| t(1;19)(q23;p13) | Fusion protein TCF3-PBX1. In 23%-25% of pre-B ALL cases. 5% in BCP-ALL. | Carroll et al87 | ||
| Williams et al88 | ||||
| Hunger et al89 | ||||
| Kamps et al90 | ||||
| Craig and Foon2 | ||||
| Hunger91 | ||||
| t(17;19)(q22;p13) | Fusion protein TCF3-HLF. In 1% of pre-B ALL cases. | Inaba et al92 | ||
| Hunger et al93 | ||||
| Craig and Foon2 | ||||
| Hunger91 | ||||
| inv(19)(p13;q13 | Fusion protein TCF3-FB1 | Brambillasca et al94 | ||
| EBF1 | B-lineage ALL | Deletion | Pediatric ALL. In 4%-6% of B-ALL cases. | Mullighan et al69 |
| Kuiper et al70 | ||||
| In adult ALL. In 4%-7% of B-ALL cases. | Safavi et al72 | |||
| Paulsson et al73 | ||||
| Enriched in relapse, 25% of cases childhood precursor B-ALL | Yang et al75 | |||
| Combined with ETV6-RUNX1. In 11% of ETV6-RUNX1 B-ALL cases. | Mullighan et al69 | |||
| Del (5)(q33;q33) | EBF1-PDGFRB fusion. | Roberts et al84 | ||
| Lengline et al95 | ||||
| Weston et al96 | ||||
| Induced expression of EBF1 inhibitor ZNF423 | Harder et al97 | |||
| Induced expression of EBF1 inhibitor ZFP521 | Yamasaki et al98 | |||
| Mixed phenotype ALL | Deletion | 1 case out of 117 investigated. | Yan et al85 | |
| B-cell lymphoma | t(5;14)(q33;q32) | Translocation to Ig-heavy chain locus. Overexpression. | Bouamar et al99 | |
| PAX5 | B-lineage ALL | Deletion | Pediatric ALL. In 24%-30% of B-ALL cases. | Mullighan et al69 |
| Kuiper et al70 | ||||
| Coyaud et al100 | ||||
| Adult ALL. In 18%-33% of B-ALL cases. | Safavi et al72 | |||
| Paulsson et al73 | ||||
| Coyaud et al100 | ||||
| Combined with TCF3-PBX1. In 41% of B-ALL cases. | Mullighan et al69 | |||
| Combination ETV6-RUNX1. In 28% of B-ALL cases. | Mullighan et al69 | |||
| Down syndrome. Deletions detected in 12% of ALL patients. | Buitenkamp et al81 | |||
| t(9;12)(p13;p13) | Fusion protein PAX5-ETV6, dominant negative. In 1% of pediatric ALL patients. | Strehl et al101 | ||
| Cazzaniga et al102 | ||||
| Fazio et al103 | ||||
| t(7;9)(q11;p13) | Fusion protein PAX5-ENL, dominant negative | Bousquet et al104 | ||
| Coyaud et al100 | ||||
| t(9;15)(p13;q24) | Fusion protein PAX5-PML | Nebral et al105 | ||
| t(3;9)(p14;p13) | Fusion protein PAX5-FOXP1 | Coyaud et al100 | ||
| del(9)(p13;p24) | Fusion protein PAX5-JAK2 | Coyaud et al100 | ||
| t(7;9)(q11;p13) | Fusion protein PAX5-POM121 | Coyaud et al100 | ||
| t(X;9)(q21;p13) | Fusion protein PAX5-DACH2 | Coyaud et al100 | ||
| t(9;17)(p13;p11) | Fusion protein PAX5-NCOR1 | Coyaud et al100 | ||
| t(9;15)(p13;q24) | Fusion protein PAX5-GOLGA6 | Coyaud et al100 | ||
| t(9;7)(p13;p12) | Fusion protein PAX5-LOC392027 | An et al106 | ||
| t(9;12)(p13;p12) | Fusion protein PAX5-5SLCO1B3 | An et al106 | ||
| t(9;20)(p13;q11) | Fusion protein PAX5-ASXL1, -KLF3B, -C20ORF112 | An et al106 | ||
| B-cell lymphoma | t(9;14)(p13;q32) | Translocation to IgH chain locus. Overexpression. | Iida et al107 | |
| Busslinger et al108 |
| Gene . | Malignant phenotype . | Genetic alteration . | Comments . | Reference . |
|---|---|---|---|---|
| IKZF1 | B-lineage ALL | Deletion | Pediatric ALL. In 8%-15% of B-ALL cases. Inactivation of IKZF3 in 3 cases. | Mullighan et al69 |
| Kuiper et al70 | ||||
| Krentz et al71 | ||||
| In adult ALL. In 18%-32% of B-ALL cases. | Safavi et al72 | |||
| Paulsson et al73 | ||||
| Marker for poor prognosis. Detected in 29%-35% of relapsing B-ALL. | Mullighan et al74 | |||
| Yang et al75 | ||||
| Krentz et al71 | ||||
| Van der Veer et al76 | ||||
| Olsson et al77 | ||||
| Asai et al78 | ||||
| Schwab et al79 | ||||
| Dörge et al80 | ||||
| Buitenkamp et al81 | ||||
| Kuiper et al82 | ||||
| Combined with BCR-ABL1. Deleted in 84% of BCR-ABL1 positive ALL in children. | Mullighan et al69 | |||
| Mullighan et al83 | ||||
| Combined in 51% of adult BCR-ABL1 B-ALL cases. | Safavi et al72 | |||
| Combined with activating mutations in FLT3 or Il7R genes. | Roberts et al84 | |||
| Combined with deletion of SH2B3 (encoding LNK) | Roberts et al84 | |||
| Down syndrome. Deletions detected in 35% of ALL patients. | Buitenkamp et al81 | |||
| Mixed phenotype ALL | Deletion | Detected in 13% of patients. | Yan et al85 | |
| B-cell lymphoma | t(3;7)(q27;p12) | Cause deregulated BCL6 expression. | Hosokawa et al86 | |
| TCF3 | B-lineage ALL | Deletion | Pediatric ALL, 1 case. | Mullighan et al69 |
| Inactivation of TCF4 in 2 cases. | Kuiper et al70 | |||
| t(1;19)(q23;p13) | Fusion protein TCF3-PBX1. In 23%-25% of pre-B ALL cases. 5% in BCP-ALL. | Carroll et al87 | ||
| Williams et al88 | ||||
| Hunger et al89 | ||||
| Kamps et al90 | ||||
| Craig and Foon2 | ||||
| Hunger91 | ||||
| t(17;19)(q22;p13) | Fusion protein TCF3-HLF. In 1% of pre-B ALL cases. | Inaba et al92 | ||
| Hunger et al93 | ||||
| Craig and Foon2 | ||||
| Hunger91 | ||||
| inv(19)(p13;q13 | Fusion protein TCF3-FB1 | Brambillasca et al94 | ||
| EBF1 | B-lineage ALL | Deletion | Pediatric ALL. In 4%-6% of B-ALL cases. | Mullighan et al69 |
| Kuiper et al70 | ||||
| In adult ALL. In 4%-7% of B-ALL cases. | Safavi et al72 | |||
| Paulsson et al73 | ||||
| Enriched in relapse, 25% of cases childhood precursor B-ALL | Yang et al75 | |||
| Combined with ETV6-RUNX1. In 11% of ETV6-RUNX1 B-ALL cases. | Mullighan et al69 | |||
| Del (5)(q33;q33) | EBF1-PDGFRB fusion. | Roberts et al84 | ||
| Lengline et al95 | ||||
| Weston et al96 | ||||
| Induced expression of EBF1 inhibitor ZNF423 | Harder et al97 | |||
| Induced expression of EBF1 inhibitor ZFP521 | Yamasaki et al98 | |||
| Mixed phenotype ALL | Deletion | 1 case out of 117 investigated. | Yan et al85 | |
| B-cell lymphoma | t(5;14)(q33;q32) | Translocation to Ig-heavy chain locus. Overexpression. | Bouamar et al99 | |
| PAX5 | B-lineage ALL | Deletion | Pediatric ALL. In 24%-30% of B-ALL cases. | Mullighan et al69 |
| Kuiper et al70 | ||||
| Coyaud et al100 | ||||
| Adult ALL. In 18%-33% of B-ALL cases. | Safavi et al72 | |||
| Paulsson et al73 | ||||
| Coyaud et al100 | ||||
| Combined with TCF3-PBX1. In 41% of B-ALL cases. | Mullighan et al69 | |||
| Combination ETV6-RUNX1. In 28% of B-ALL cases. | Mullighan et al69 | |||
| Down syndrome. Deletions detected in 12% of ALL patients. | Buitenkamp et al81 | |||
| t(9;12)(p13;p13) | Fusion protein PAX5-ETV6, dominant negative. In 1% of pediatric ALL patients. | Strehl et al101 | ||
| Cazzaniga et al102 | ||||
| Fazio et al103 | ||||
| t(7;9)(q11;p13) | Fusion protein PAX5-ENL, dominant negative | Bousquet et al104 | ||
| Coyaud et al100 | ||||
| t(9;15)(p13;q24) | Fusion protein PAX5-PML | Nebral et al105 | ||
| t(3;9)(p14;p13) | Fusion protein PAX5-FOXP1 | Coyaud et al100 | ||
| del(9)(p13;p24) | Fusion protein PAX5-JAK2 | Coyaud et al100 | ||
| t(7;9)(q11;p13) | Fusion protein PAX5-POM121 | Coyaud et al100 | ||
| t(X;9)(q21;p13) | Fusion protein PAX5-DACH2 | Coyaud et al100 | ||
| t(9;17)(p13;p11) | Fusion protein PAX5-NCOR1 | Coyaud et al100 | ||
| t(9;15)(p13;q24) | Fusion protein PAX5-GOLGA6 | Coyaud et al100 | ||
| t(9;7)(p13;p12) | Fusion protein PAX5-LOC392027 | An et al106 | ||
| t(9;12)(p13;p12) | Fusion protein PAX5-5SLCO1B3 | An et al106 | ||
| t(9;20)(p13;q11) | Fusion protein PAX5-ASXL1, -KLF3B, -C20ORF112 | An et al106 | ||
| B-cell lymphoma | t(9;14)(p13;q32) | Translocation to IgH chain locus. Overexpression. | Iida et al107 | |
| Busslinger et al108 |
The formation of oncogenic fusion proteins or complete inactivation of tumor suppressor genes creates rather drastic changes in the regulatory landscape of a developing cell. However, in B-cell development, several lines of investigations suggest that rather subtle changes in transcription factors dose, for instance as imposed by monoallelic inactivation, have critical impact on cell differentiation. The basic-helix-loop-helix containing E-proteins, TCF4, TCF12 and TCF3 are balanced by the expression of the inhibitory ID proteins110 and reduced function of TCF3 as a result of mono allelic inactivation of the Tcf3 gene results in deficiencies in the pro-B cell compartment.111 Reduced Ebf1 dose as a consequence of a heterozygous mutation of the Ebf1 gene result in reduced numbers of pre-B-cells while the earliest stages of development remain largely unaffected.111,112 Several additional transcription factors including SPI1,20 MYB,15 BCL11A113 and IKZF1114 act in a dose dependent manner in early B-lymphocyte development stressing the importance of properly balanced regulatory networks in cell differentiation. Furthermore, heterozygote deletion of Ebf1 in combination with a heterozygote deletion of Tcf3 or Runx1, results in a synergistic reduction in B-cell progenitor formation112,115 and about half of the mice carrying a combined heterozygous loss of Ebf1 and Pax5 develop pre-B cell leukemia before 30 weeks of age while few or no such tumors are observed in the single heterozygous animals.116 This highlights the sensitivity of regulatory networks in B-lymphocyte development to changes in functional transcription factor dose.
A direct link between transcription factor dose and human malignancies became evident when a larger set of childhood leukemias were analyzed by high-resolution single nucleotide polymorphism analysis revealing a high frequency of heterozygote deletions of EBF1, IKZF1 and PAX569,70 (Table 1). The most commonly detected deletion involved PAX5 where almost a third of the childhood B-ALL's carried mutations that could be presumed to result in reduced functional activity of the transcription factor. Reduced PAX5 function could also be detected as a result of a familial mutation in the PAX5 gene predisposing for the development of leukemia.117 The apparent involvement of PAX5 was rather surprising since mouse models had not provided any direct evidence for that B-cell development would be critically dependent on Pax5 dose.111 However, the observation that PAX5 deletions are associated with complex karyotypes and common recurrent translocations69,70 indicates that the impact of reduced PAX5 function is dependent of other oncogenic events. In line with this hypothesis, heterozygote deletion of Pax5 in combination with transgenic expression of a constitutive active STAT5 resulted in increased development of B-lineage leukemia.118 A similar result was obtained after heterozygote inactivation of the Ebf1 gene118 leading to the suggestion that the reduction in EBF1 or PAX5 dose would result in a partial developmental block contributing to the malignant conversion and developmental arrest observed in pro-B cell leukemia. Even though the finding that restoration of PAX5 expression in a leukemia model resulted in differentiation of the transformed cells119 support this idea, a complete block of differentiation as imposed by RAG deficiency, did not result in leukemia formation in collaboration with activated STAT5 as efficiently as heterozygote loss of Pax5 or Ebf1.118 Furthermore, reduced EBF1 dose result in increased DNA damage in nontransformed B-cell progenitors116 suggesting that the role of these proteins in the transformation process may be more complex than initially thought.
Even though mutations in PAX5 are the most frequently detected alteration in the B-lineage transcription factor genes in the primary leukemias, IKZF1 deletions or truncations are the most dramatically enriched abnormality in relapsing cases of childhood B-ALL (Table 1). The relative frequency of IKZF1 mutations increase from about 8% to 15% in primary to 29% to 35% in relapsing leukemia making it a strong independent risk factor in B-ALL.71,74,120 The role of IKZF1 in B-ALL is complex since mouse models lacking fully functional IKZF1 rather develop T-cell leukemia.121 However, when mutations in IKZF1 are combined with a constitutively expressed BCR-ABL1 gene, the development of B-lineage disease is enhanced122,123 revealing the importance of combinatorial events in leukemia development. This is well in line with the observation that a majority of the BCR-ABL1 expressing human ALLs carries mutated functionally impaired or dominant negative IKZF1 alleles.69,70,83 Hence, even though transcription factor dose is involved in the development and progression of B-ALL, many of the effects observed is a result of combined actions of several oncogenic events.
Although the most direct way to reduce functional transcription factor dose is mutation of the coding gene, other mechanisms of action may be implied. Interesting examples are the regulation of MYB function by the regulatory RNA miR-150124 and repression of IRF4 activity by the onco-micro RNA miR-125b in leukemia.125 Modulation can also be achieved through changes in expression of interaction partners such as the EBF repressor protein ZNF423, up regulated in B-ALL cells.97 This protein is normally not expressed in hematopoietic cells, however, deregulated BMP2 signaling appear to be sufficient to cause expression in B-cell progenitors. ZNF521 has a similar function as a repressor of EBF1 activity and this protein is also found in over expressed in human B-ALL often associated with E2A-HLF fusion proteins.98 Fusion proteins may also modulate transcription factor activity as exemplified by that BCR-ABL1 modify the ability of PAX5 to induce expression of the target gene BACH2126 and that the PAX5 co-repressor TLE4 (GRG4)49 is a target for the E2A-HLF protein.127 Even though many functions of key regulatory proteins are unique, there are examples of apparently redundant activities of transcription factors at defined developmental stages. This is exemplified by the finding that combined mutations of IRF4 and IRF867,68 as well as combined mutation of SPI1 and SPIB128 dramatically enhance the developmental block and leukemia formation as compared with the single mutants. Furthermore, even if IKZF1 deletions and truncations appear to be dominant in human leukemia, IKZF3 mutations are detected in B-lineage ALL.69 Thus, the functional dose of a given transcription factor could be modulated by alterations in the activity of other regulatory proteins with overlapping functions. Hence, it is becoming increasingly clear that transcription factor dose may be under the influence of different levels of molecular control in leukemia and an increased understanding of the regulatory networks may well result in that additional genetic alterations in leukemia can be coupled to modulation of transcription factor dose.
Concluding remarks
It is well established that transcription factors play crucial roles in human malignancies by the formation of oncogenic fusion proteins. However, the apparent importance of rather modest disruptions of regulatory networks such as heterozygote deletion of a coding gene highlights the importance of balanced transcription factor activity in blood cell development. Disrupting the interplay between transcription factors may affect the progenitor B-cell directly by inducing cell division or reducing apoptosis. It may also directly contribute to differentiation blockade and possibly lineage infidelity resulting in conversion of the leukemic phenotype or bi-phenotypic malignancies. It can be predicted that the possibility to link development to disease at the molecular level will largely increase our understanding of transformation processes as well as facilitate our possibilities of diagnosing and curing leukemia in the future.
Acknowledgments
This work was supported by grants from the Swedish Cancer Society, the Swedish Research Council, including a Center grant to Hematolinne in Lund, the Knut and Alice Wallenberg Foundation, the Swedish Childhood Cancer Foundation, and Linköping University.
Authorship
Contribution: R.S., M.A.J.P., J.U., and M.S. contributed to the design of the article and the factual content.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mikael Sigvardsson, Faculty of Health Sciences, Linköping University, University Lab 1, Level 13, Linköping, Sweden; e-mail: mikael.sigvardsson@liu.se.