Abstract
Megakaryopoiesis is a complex, stepwise process that takes place largely in the bone marrow. At the apex of the hierarchy, hematopoietic stem cells undergo a number of lineage commitment decisions that ultimately lead to the production of polyploid megakaryocytes. On average, megakaryocytes release 1011 platelets per day into the blood that repair vascular injuries and prevent excessive bleeding. This differentiation process is tightly controlled by exogenous and endogenous factors, which have been the topics of intense research in the hematopoietic field. Indeed, a skewing of megakaryocyte commitment and differentiation may entail the onset of myeloproliferative neoplasms and other preleukemic disorders together with acute megakaryoblastic leukemia, whereas quantitative or qualitative defects in platelet production can lead to inherited platelet disorders. The recent advent of next-generation sequencing has prompted mapping of the genomic landscape of these conditions to provide an accurate view of the underlying lesions. The aims of this review are to introduce the physiological pathways of megakaryopoiesis and to present landmark studies on acquired and inherited disorders that target them. These studies have not only introduced a new era in the fields of molecular medicine and targeted therapies but may also provide us with a better understanding of the mechanisms underlying normal megakaryopoiesis and thrombopoiesis that can inform efforts to create alternative sources of megakaryocytes and platelets.
Introduction
This review emphasizes how unraveling genomic lesions that underpin malignant and nonmalignant disorders of megakaryopoiesis and thrombopoiesis provides clues on the physiological mechanisms that control normal megakaryocyte differentiation, maturation, and platelet function. This knowledge, moreover, informs and influences the field of therapeutic megakaryopoiesis and platelet production, as discussed in the reviews by Sim et al, Eto and Kunishima, and Woolthuis and Park in this review series.
Key genes in normal megakaryopoiesis and platelet production
Megakaryocyte progenitors arise from the lineage restriction of hematopoietic stem cell–derived bipotent megakaryocyte-erythroid progenitors. Proliferating megakaryocyte progenitors terminally differentiate to megakaryocytes, which in turn undergo endomitosis and cytoplasmic maturation. Overall, the process of megakaryocyte development and maturation is called megakaryopoiesis. Mature megakaryocytes in turn produce platelets. Platelet biogenesis (called thrombopoiesis) mainly, but not exclusively,1 occurs through the organization of cytoplasmic extensions (proplatelets) that fragment and are released as platelets into the bloodstream (Figure 1A).
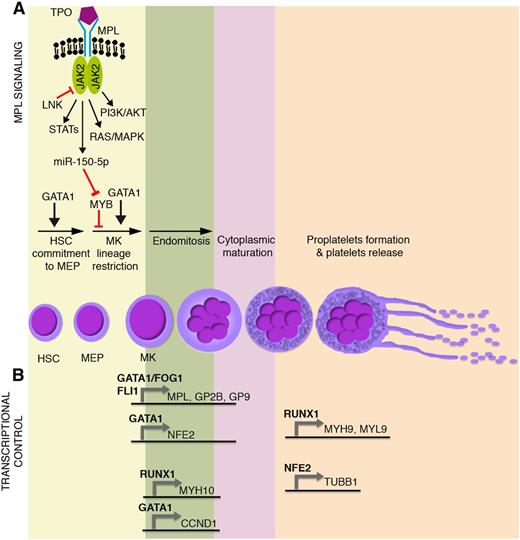
Main molecular mechanisms affected in malignant megakaryopoiesis and platelet function defects. Schematic representation of (A) TPO/MPL signaling pathway together with (B) the most relevant transcriptional regulators of megakaryocyte commitment, differentiation, and maturation. GP2B (CD41) is a component of the GPIIb-IIIa glycoprotein complex. GP9 (CD42a), GP1BA (CD42b), and GP1BB (CD42c) are components of the GPIb-IX-V glycoprotein complex. HSC, hematopoietic stem cell; MEP, megakaryocyte-erythroid progenitor; MK, megakaryocyte.
Main molecular mechanisms affected in malignant megakaryopoiesis and platelet function defects. Schematic representation of (A) TPO/MPL signaling pathway together with (B) the most relevant transcriptional regulators of megakaryocyte commitment, differentiation, and maturation. GP2B (CD41) is a component of the GPIIb-IIIa glycoprotein complex. GP9 (CD42a), GP1BA (CD42b), and GP1BB (CD42c) are components of the GPIb-IX-V glycoprotein complex. HSC, hematopoietic stem cell; MEP, megakaryocyte-erythroid progenitor; MK, megakaryocyte.
Megakaryocyte-erythroid progenitor commitment toward the megakaryocyte lineage and maturation are tightly regulated through transcriptional and epigenetic mechanisms, which act in concert with extrinsically induced signal transduction events. Thrombopoietin (TPO) and its receptor, MPL, play a primary role in the extrinsic control of megakaryopoiesis. MPL is a homodimeric type I receptor that requires the tyrosine kinase Janus kinase 2 (JAK2) for signaling. Importantly, the TPO-MPL axis governs hematopoietic stem cell proliferation and megakaryocyte commitment, but it is dispensable for megakaryocyte maturation and platelet production1,2 (Figure 1A).
Several transcription factors drive megakaryopoiesis and platelet production. Among them, GATA1 is essential for proper erythroid and megakaryocyte differentiation, as demonstrated by the impaired maturation of GATA1– embryonic stem cell–derived erythro-megakaryocytic progenitors3 (Figure 1A). Indeed, germ line missense mutations in GATA1 underlie X-linked thrombocytopenia and anemia.4 Other transcription factors act in a coordinated, balanced manner to drive megakaryocyte-erythroid progenitor fate decision. The proto-oncogene MYB is a major regulator of this process. MYB favors erythropoiesis by inducing the expression of the erythroid transcription factor KLF1.5 In this way, MYB affects the functional antagonism between KLF1 that supports erythropoiesis and the transcription factor FLI1 that supports megakaryopoiesis.6,7 In addition, MYB induces the microRNA miR-486-3p, which in turn lowers the expression of the transcription factor MAF,8 which promotes megakaryopoiesis. The overall result is that, although it is required for the erythropoiesis, MYB constrains the megakaryocyte-erythroid progenitor commitment toward the megakaryocyte lineage. This role is experimentally supported by MYB knockdown or loss-of-function experiments, which show an enhanced megakaryopoiesis and platelet production.5,9 Interestingly, TPO-MPL signaling mimics experimental MYB knockdown, because MPL activation induces the expression of miR-150-5p, which in turn lowers MYB expression,10 thus enhancing megakaryopoiesis11 (Figure 1A). Strikingly, the inactivation of MYB transcriptional activity induces platelet production at supraphysiological levels, even in MPL−/− mice (ie, in the absence of MPL signaling).9
During megakaryocyte lineage commitment, MYB downregulation enables FLI1 expression. FLI1 acts in concert with GATA1-FOG1 and ETS112,13 to transactivate the expression of several megakaryocyte-specific and platelet-specific receptors such as MPL, ITGA2B (ie, GP2B or CD41), and GP9 (ie, CD42a) (Figure 1B).
A number of transcription factors are similarly responsible for the transcriptional control of megakaryocyte maturation and platelet production (Figure 1B). Among them, RUNX1 affects ploidization and proplatelet formation during megakaryocyte maturation by regulating MYH9, MYL9, and MYH10 expression14,15 (Figure 1B). Similarly, GATA1 regulates polyploidization by driving expression of the gene coding for cyclin D1 (CCND1).16 GATA1 also transactivates the expression of the transcription factor p45 NFE2.17 NFE2 in turn induces the expression of several genes critical for proplatelet formation and platelet shedding such as TUBB1 (Figure 1B), which encodes for the tubulin β-1 chain, a major component of microtubules.18 Accordingly, p45 NFE2−/− mice lack circulating platelets.19
Increasing knowledge of both extrinsic cues and intrinsic mechanisms that underpin megakaryopoiesis and thrombopoiesis broadens the panel of targets that could be used to overcome some issues such as yield in therapeutic, large-scale platelet production. For example, interleukin (IL)-1α was recently discovered to drive platelet production by megakaryocyte rupture. This mechanism is an alternative to TPO-induced thrombopoiesis via proplatelet formation, is activated in response to acute platelet needs, and can provide a 20-fold higher yield of platelets.1 This recent insight is emblematic of how unraveling the mechanisms underlying physiological megakaryopoiesis and platelet production can point to new ways for developing and optimizing large-scale platelet production protocols.
The genomic landscape of malignant megakaryopoiesis
This section is focused on the genomic lesions causing malignant megakaryopoiesis that most clearly inform us about normal megakaryopoiesis. Preleukemic disorders characterized by a skewed megakaryopoiesis—namely, Philadelphia chromosome (Ph)-negative myeloproliferative neoplasms (MPNs), 5q- syndrome, and refractory anemia with ring sideroblasts associated with thrombocytosis (RARS-T)—will be reviewed first. Next, acute megakaryoblastic leukemia (AMKL) will be discussed as representing a possible scenario for the progression of the aforementioned preleukemic disorders to acute myeloid leukemia (AML). The diseases presented here share a common feature: the abnormal expansion of megakaryopoiesis. The identification of the underlying molecular defects capitalizes on a few mechanisms that entail an enhancement of megakaryopoiesis and therefore represent a target for overcoming some limitations (eg, the yield) in therapeutic megakaryocyte and platelet production.
Ph-negative MPNs
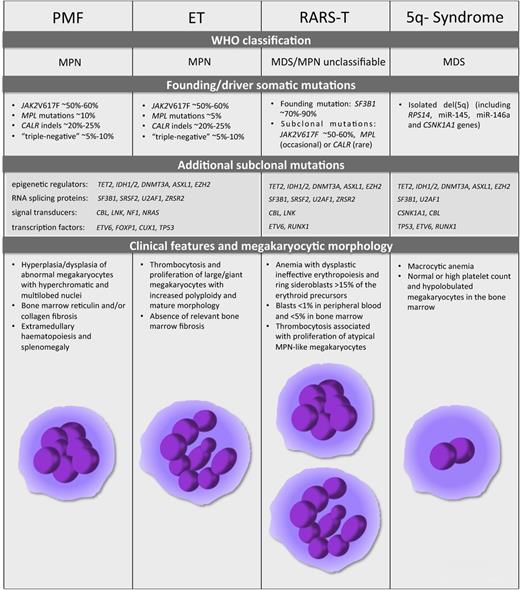
Classical Ph-negative MPNs are a heterogeneous group of clonal hematopoietic disorders characterized by an excessive production of mature myeloid cells and a tendency to progress to AML. The 2008 World Health Organization classification identifies three main diseases—polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF)—based on an expanded lineage in MPN patients.20 PV is characterized by erythrocytosis, whereas ET and PMF have abnormal megakaryopoiesis with alterations in platelet count. In particular, ET patients exhibit thrombocytosis (platelet count >450 × 109/L) and megakaryocyte proliferation in the absence of relevant bone marrow fibrosis, whereas PMF patients display bone marrow with megakaryocyte hyperplasia and dysplasia along with reticulin and/or collagen fiber deposition (Figure 2).
PMF, ET, RARS-T, and 5q- syndrome. The World Health Organization classification, the mutational landscape, and the distinctive clinical and morphologic features for the 4 malignant diseases are described. The schematic representation of megakaryocyte morphology, as a hallmark distinctive of PMF, ET, RARS-T, and 5q- syndrome, is shown at the bottom of the figure.
PMF, ET, RARS-T, and 5q- syndrome. The World Health Organization classification, the mutational landscape, and the distinctive clinical and morphologic features for the 4 malignant diseases are described. The schematic representation of megakaryocyte morphology, as a hallmark distinctive of PMF, ET, RARS-T, and 5q- syndrome, is shown at the bottom of the figure.
Given the role of TPO as the main driver of megakaryocyte commitment and differentiation, most efforts to depict the genomic landscape of MPNs have focused on identifying genetic lesions that alter TPO signaling. The identification of a somatic gain-of-function point mutation in the nonreceptor tyrosine kinase JAK221-24 is considered a major milestone for the MPN field, because the JAK2V617F mutation is present in ∼95% of PV patients and in ∼50% to 60% of ET and PMF patients (Figure 2).21-24 JAK2 associates with the receptors for TPO, erythropoietin, and granulocyte colony-stimulating factor. Importantly, this amino acid substitution abrogates the negative regulatory function of the JH2 domain of JAK2, leading to constitutive JAK2 activation and subsequent cytokine-independent signaling. In vivo studies have provided a deeper insight into the mechanism by which the same lesion can cause 3 different phenotypes and suggest that gene dosage affects the disease phenotype.25 Indeed, Li et al26 demonstrated that acquisition of the homozygous JAK2V617F mutation drives a switch from an ET-like to a PV-like phenotype in knockin mice.
Somatic MPL mutations exist in ∼5% to 10% of ET and PMF patients27-30 (Figure 2), whereas no THPO mutations have been detected in MPNs thus far.31 MPL genetic lesions are most often gain-of-function mutations (eg, MPLW515L and MPLW515K) that lead to cytokine-independent MPL activation and TPO hypersensitivity.27,28 Other somatic mutations in MPL have been reported at lower frequencies in MPN patients (MPLW515A, MPLW515R, and MPLS505N).28-30
In 2013, whole-exome sequencing revealed that somatic mutations in the calreticulin (CALR) gene occurs in 14% to 35% of PMF patients and in 25% to 27% of ET patients, all of which are JAK2 and MPL unmutated32,33 (Figure 2). CALR controls the folding of newly synthesized glycoproteins and acts as a major Ca2+ binding and storage protein in the endoplasmic reticulum. CALR somatic lesions frequently consist of frameshift mutations in exon 9, resulting in the loss of the C-terminal KDEL endoplasmic reticulum retention signal and a partial dislocation of the CALR protein.32 Although more than 50 different CALR insertions/deletions (indels) have been identified, only 2 of them—del52 (type 1) and ins5 (type 2)—collectively account for ∼85% of CALR mutations. Both CALRdel52 and CALRins5 mutants drive the expansion of megakaryopoiesis and thrombopoiesis leading to a human ET-like disease in a retroviral mouse model.34 Indeed, CALR mutants drive the constitutive activation of JAK2-STAT signaling through MPL35 with consequent TPO hypersensitivity. These data are consistent with the observation that JAK-STAT signaling is also aberrantly activated in patients with CALR mutations.32,36 Important differences between CALRdel52- and CALRins5-driven phenotypes in mice are detected, with CALRdel52 additionally expanding the stem cell compartment and causing myelofibrosis and more severe thrombocytosis than CALRins5.34,35 Of note, these features suggest that other molecular mechanisms besides the MPL pathway, such as Ca2+-dependent signaling, could be differently perturbed by CALR mutants.
However, approximately 10% of PMF and ET patients do not carry mutations in any of the 3 mutations (JAK2, MPL, CALR) described above (Figure 2). Somatic lesions in other genes involved in JAK-STAT signaling have been found in MPN patients. For instance, loss-of-function mutations in LNK and CBL, which encode a JAK2 inhibitor and a JAK2/MPL/EPOR–directed E3 ubiquitin ligase, respectively, have been identified in 3% to 6% of MPN patients and may co-occur alongside the JAK2V617F mutation.25,37 Additional lesions have also been associated with MPN pathogenesis and progression25,37,38 (Figure 2).
Taken together, these findings identify constitutive activation of MPL signaling as a mechanism shared by JAK2, MPL, and CALR driver mutations that affects megakaryopoiesis and thrombopoiesis. In addition, these finding further elucidate the role of key modulators of MPL-driven megakaryopoiesis. These data therefore pinpoint novel targets for the manipulation and tuning of the MPL signaling pathway to improve the yield of large-scale platelet generation systems as discussed in other reviews in this series. It is also noteworthy that, even though PMF and ET patients carry the same driver mutations (ie, JAK2, MPL, and CALR), their megakaryocytes show considerable morphologic differences (Figure 2). Therefore, additional mechanisms could be engaged to explain the differences in the skewing of megakaryocyte maturation between PMF and ET. Interestingly, the investigation of these mechanisms could also provide novel insights into the intrinsic and extrinsic signals that control physiological megakaryocyte maturation.
Genome-wide association studies have identified germ line susceptibility genes predisposing to MPN development, such as the JAK2 haplotype called 46/1 (or GGCC) and its associated single nucleotide polymorphism (rs10974944), which represents the main predisposition factor for both sporadic and familial MPNs with acquired JAK2V617F mutations.25 Additional MPN-predisposing variants have recently been identified. Among them, it is worth mentioning a single nucleotide polymorphism (rs9376092) close to the MYB locus associated with reduced MYB expression and strongly related to an ET rather than a PV phenotype in JAK2V617F-positive MPNs.39 These findings strongly suggest that MYB expression levels can affect the ET vs PV phenotype determination in JAK2V617F-positive MPNs, consistent with the role of MYB in megakaryocyte vs erythroid lineage choice illustrated above.5,8
Recent genome-wide transcriptome studies have further characterized the molecular mechanisms underlying MPNs by profiling CD34+ hematopoietic stem and progenitor cells40-44 and have highlighted the abnormal expression of coding and noncoding transcripts involved in megakaryopoiesis and platelet production in PMF and ET patients (eg, NFE2, CD9, CXCR4, WT1, and miR-34a-5p).36,40 For instance, miR-155-5p is overexpressed in PMF CD34+ hematopoietic stem and progenitor cells and targets the polycomb repressive complex 2 component JARID2 to support the abnormal megakaryocyte lineage expansion seen in these patients.45 Similarly, miR-28, which targets MPL messenger RNA and inhibits megakaryocyte terminal differentiation, is overexpressed in MPN megakaryocytes and platelets that frequently display MPL downregulation.46,47
These results provide important insights into how miRNA-mediated posttranscriptional control of gene expression affects megakaryopoiesis and thrombopoiesis in physiological and pathological conditions. They also suggest that modulation of miRNA expression might represent an important step to improve platelet production for therapeutic purposes.
As in MPNs, the MPL signaling pathway is also deregulated in hereditary thrombocytosis, a familial polyclonal disorder that results from germ line mutations in the THPO and MPL genes48 (Table 1). Hereditary thrombocytosis-associated THPO mutations generally lead to increased messenger RNA translation, which eventually results in cytokine overproduction. Of note, germ line and sporadic mutations in hereditary thrombocytosis and MPNs, respectively, occur in the same key megakaryocytic genes (MPL and JAK2), even though different residues are hit (Table 1).
Genes involved in hereditary thrombocytosis
| Gene . | Protein . | Chromosome location . | Mutation . | Inheritance . | Molecular mechanism . |
|---|---|---|---|---|---|
| THPO | Thrombopoietin (TPO) | 3q27 | G>C in the splice donor site of intron 3 | Autosomal dominant | The mutation disrupts the inhibitory uORF leading to increase in mRNA translation. |
| G>T substitution in 5′UTR | Autosomal dominant | The mutation disrupts the inhibitory uORF leading to increase in mRNA translation. | |||
| Deletion of single G in 5′UTR | Autosomal dominant | The mutation disrupts the inhibitory uORF leading to increase in mRNA translation. | |||
| A>G mutation in intron 3 | Autosomal dominant | The A>G substitution in the +1 position of the splice donor of intron 3 leads to increase in mRNA translation. | |||
| T>C at the splice donor site of intron 2 | Autosomal dominant | The splice donor mutation leads to exon 2 skipping and loss of inhibitory 5′UTR sequence, leading to increased TPO expression. | |||
| MPL | Thrombopoietin receptor, MPL | 1p34 | S505N | Autosomal dominant | TPO-independent constitutive activation of MPL. |
| K39N (also called “MPL-Baltimore”) | Autosomal dominant with incomplete penetrance | This mutation is associated with incomplete processing and a reduction in MPL protein. | |||
| P106L | Autosomal recessive | The MPL P106L shows constitutive activity that can be further stimulated by TPO. | |||
| W515R | Autosomal dominant | The mutation increases MPL activity but to a lesser extent than the somatic MPLW515K/L/A mutations observed in MPNs. | |||
| JAK2 | Janus kinase 2, JAK2 | 9p24 | V617I | Autosomal dominant | JAK2V617I has weak constitutive signaling compared with JAK2V617F because of reduction in the threshold for cytokine-induced activation. |
| R867Q | Autosomal dominant | The mutation in JH1 domain abolishes a salt bridge, which is present in the inactive JH1 and is lost upon activation. | |||
| S755R/R938Q | Autosomal dominant | The S755R mutation interferes with an important salt bridge and inhibits the function of JH2 domain. The R938Q is located in a sensitive region of the JH1 near the ATP loop and substrate access site. | |||
| R564Q | Autosomal dominant | JAK2 R564Q exhibits similar levels of increased kinase activity compared with JAK2V617F but fewer growth-promoting effects. | |||
| H608N | Autosomal dominant | The mutation in this region may abrogate the function of the inhibitory JH2 domain, resulting in increased kinase activity. |
| Gene . | Protein . | Chromosome location . | Mutation . | Inheritance . | Molecular mechanism . |
|---|---|---|---|---|---|
| THPO | Thrombopoietin (TPO) | 3q27 | G>C in the splice donor site of intron 3 | Autosomal dominant | The mutation disrupts the inhibitory uORF leading to increase in mRNA translation. |
| G>T substitution in 5′UTR | Autosomal dominant | The mutation disrupts the inhibitory uORF leading to increase in mRNA translation. | |||
| Deletion of single G in 5′UTR | Autosomal dominant | The mutation disrupts the inhibitory uORF leading to increase in mRNA translation. | |||
| A>G mutation in intron 3 | Autosomal dominant | The A>G substitution in the +1 position of the splice donor of intron 3 leads to increase in mRNA translation. | |||
| T>C at the splice donor site of intron 2 | Autosomal dominant | The splice donor mutation leads to exon 2 skipping and loss of inhibitory 5′UTR sequence, leading to increased TPO expression. | |||
| MPL | Thrombopoietin receptor, MPL | 1p34 | S505N | Autosomal dominant | TPO-independent constitutive activation of MPL. |
| K39N (also called “MPL-Baltimore”) | Autosomal dominant with incomplete penetrance | This mutation is associated with incomplete processing and a reduction in MPL protein. | |||
| P106L | Autosomal recessive | The MPL P106L shows constitutive activity that can be further stimulated by TPO. | |||
| W515R | Autosomal dominant | The mutation increases MPL activity but to a lesser extent than the somatic MPLW515K/L/A mutations observed in MPNs. | |||
| JAK2 | Janus kinase 2, JAK2 | 9p24 | V617I | Autosomal dominant | JAK2V617I has weak constitutive signaling compared with JAK2V617F because of reduction in the threshold for cytokine-induced activation. |
| R867Q | Autosomal dominant | The mutation in JH1 domain abolishes a salt bridge, which is present in the inactive JH1 and is lost upon activation. | |||
| S755R/R938Q | Autosomal dominant | The S755R mutation interferes with an important salt bridge and inhibits the function of JH2 domain. The R938Q is located in a sensitive region of the JH1 near the ATP loop and substrate access site. | |||
| R564Q | Autosomal dominant | JAK2 R564Q exhibits similar levels of increased kinase activity compared with JAK2V617F but fewer growth-promoting effects. | |||
| H608N | Autosomal dominant | The mutation in this region may abrogate the function of the inhibitory JH2 domain, resulting in increased kinase activity. |
ATP, adenosine triphosphate; mRNA, messenger RNA; uORF, upstream open reading frame.
5q- syndrome.
Myelodisplastic syndromes (MDSs) are clonal preleukemic conditions characterized by ineffective hematopoiesis that frequently progress to AML. The 5q- syndrome is an MDS associated with isolated deletion on the long arm of chromosome 5 (del(5q)). Although ∼40% to 65% of MDS patients show thrombocytopenia, patients with the 5q- syndrome are characterized by normal or high platelet count and hypolobulated megakaryocytes in the bone marrow in addition to macrocytic anemia.49
Over the past few years, the pathogenic roles of specific genes located in the 5q- syndrome commonly deleted region, including the casein kinase 1A gene (CSNK1A1), the ribosomal gene RPS14, and the miRNA genes miR-145 and miR-146a (Figure 2) have been unveiled. In particular, the heterozygous inactivation of CSNK1A1 was recently demonstrated to drive hematopoietic stem cell expansion in mice, suggesting that CSNK1A1 haploinsufficiency may play a role in the initial clonal expansion in patients with the 5q- syndrome.50 In addition, loss of the ribosomal gene RPS14 is responsible for dyserythropoiesis and anemia, whereas deletions affecting miR-145 and miR-146a genes are associated with the thrombocytosis and megakaryocyte dysplasia observed in some patients with 5q- syndrome. Of note, miR-145 targets FLI1, whose expression levels are increased in patients with del(5q) MDS. However, the most striking results involving miR-145 and miR-146a in 5q- pathogenesis come from miR-145/miR-146a double-knockdown mice that show upregulation of the miR-145/miR-146a target TRAF6, and subsequent ectopic TRAF6-mediated induction of IL-6 expression. The increase in IL-6 levels in turn likely accounts for megakaryocyte dysplasia and thrombocytosis through a paracrine mechanism.51 These data collectively identify a cell-nonautonomous mechanism that underpins 5q- syndrome-related megakaryopoiesis and thrombopoiesis defects. They also underline the importance of extrinsic factors such as IL-6 in regulating megakaryocyte maturation and platelet production, with important implications for the optimization of ex vivo platelet production protocols.
RARS-T.
RARS-T is a rare condition with features of both MDSs and MPNs.20 It is characterized by anemia with dysplastic erythropoiesis, ring sideroblasts >15% of erythroid precursors, and thrombocytosis associated with the increased proliferation of large atypical MPN-like megakaryocytes52 (Figure 2). Even though associated with JAK2V617F at first,53 the recent identification of somatically acquired mutations in the RNA splicing protein SF3B1 observed in 70% to 90% of MDS or MDS-MPN patients with ring sideroblasts has been the major breakthrough in the field.54,55 SF3B1 encodes an essential U2snRNP spliceosomal complex component that participates in normal RNA splicing. Mutations in SF3B1 are considered the first hit in RARS-T patients and account for the myelodysplastic features, although a second hit to JAK2, MPL, or CALR is necessary to confer a myeloproliferative phenotype52 (Figure 2). Indeed, somatic CALR mutations have been reported in patients with RARS-T in addition to those with PMF and ET,32,56,57 further emphasizing the link between CALR mutations and abnormal megakaryopoiesis and thrombopoiesis. However, TET2, DNMT3A, and ASXL1, which encode for epigenetic modifiers, are the most frequently mutated genes outside of SF3B1 and JAK2 in RARS-T.
Overall these data further confirm the connection between the constitutive activation of MPL signaling through JAK2, MPL, or CALR mutations and the expansion of megakaryopoiesis and platelet production as a pathogenetic mechanism shared by MPNs and RARS-T.
AMKL.
AMKL is a subtype of AML characterized by the presence of numerous abnormal megakaryoblasts in the bone marrow as well as by considerable myelofibrosis. AMKL displays a bimodal age distribution and is more frequent in infants (3% to 14% of AML patients) than adults (1% of AML patients). Three main subtypes of AMKL can be distinguished: pediatric Down syndrome–related AMKL, pediatric non-Down syndrome AMKL, and adult AMKL. Pediatric patients often exhibit de novo AMKL, whereas adult AMKL frequently arises after chemotherapy or is secondary to MPNs or MDSs.58,59
AMKLs are commonly characterized by a higher incidence of complex karyotypes compared with other AML subtypes.58 There are several cytogenetic aberrations that are common in adult AMKL (t(9;22)(q34;q11), 3q21q26, −5/del(5q), −7/del(7q), and i(12)(p10)), but some pediatric mutations are also observed as described below.58 Of note, some of these lesions, such as del(5q) or del(7q), may also be found in MDS patients.
Although there are numerous cytogenetic aberrations that commonly cause adult AMKL, the most insightful mutations that inform us about normal megakaryopoiesis are found in children with Down syndrome who develop AMKL. These children often have a precedent indolent transient myeloproliferative disorder characterized by the accumulation of megakaryoblasts in the peripheral blood and by a moderate-to-high leukocytosis. Transient myeloproliferative disorder myeloid cells show just a few somatic lesions, with trisomy 21 (t21) and GATA1 mutations being the only recurrent alterations. The overexpression of some genes or microRNAs located in chromosome 21, such as DYRK1A,60 ERG,61 and miR-125b,62 has been shown to induce AMKL in mice. Recently, Ng et al63 have further characterized the role of the transcription factor ERG in a Down syndrome mouse model, suggesting that ERG is a key regulator of the erythroid-megakaryocytic cell fate in early progenitors, and its trisomy is sufficient to confer myeloproliferation in mice. In 2002, Wechsler and colleagues demonstrated that most of these patients have multiple, independent GATA1 mutations, resulting in a shorter GATA1 protein (GATA1s) with an N-terminal truncation.64 Interestingly, neither a mouse model carrying a complete copy of chromosome 21 nor GATA1s knockin mice develop any form of leukemia but show only an increased megakaryopoiesis. Surprisingly, the combination of these 2 hits further enhances the proliferation of megakaryocyte progenitors without leading to any overt leukemia phenotype.65,66
The pediatric non-Down syndrome AMKLs generally show copy number variations and numerical chromosomal abnormalities. Approximately 30% of these patients carry the OTT-MAL fusion gene.67,68 The conditional OTT-MAL knockin mouse model69 recapitulates an AMKL-like phenotype, although with a low penetrance. Functionally, the OTT-MAL fusion protein activates RBPJ-mediated transcriptional regulation, leading to increased proliferation and megakaryocyte lineage skewing. Because RBPJ is a key mediator of canonical Notch signaling, this work further highlights the importance of this pathway whose activation favors megakaryopoiesis in mice.70 However, Poirault-Chassac et al71 have reported a suppression effect of Notch signaling on human megakaryopoiesis. Although puzzling, these findings might pave the way to design better systems to obtain and/or culture megakaryocytes ex vivo. Of note, human CD34+ cells expanded in vitro in the presence of Notch ligands have already been used as transplantation units in clinical trials.72
Overall, the study of neoplastic megakaryopoiesis throughout the spectrum of malignancies affecting this lineage has allowed researchers to improve their understanding of how development of megakaryocytes can be driven by intrinsic and external stimuli. In particular, as discussed elsewhere in this review series, knowledge of cytokines (ie, TPO, IL-6), miRNAs (ie, miR-155, miR-28), and pathways (ie, JAK-STAT, Ca2+-dependent signaling, Notch pathway) can be exploited and applied to the production of megakaryocytes and platelets for therapy.
Genomic lesions in inherited platelet function disorders
The transition of megakaryocytes to platelets for therapeutic purposes ideally requires optimal platelet function. Inherited platelet function disorders frequently entail alterations in platelet number and size.73 The characterization of their genomic landscape has provided much insight into the molecular mechanisms crucial for proper platelet production and function. Here we discuss the genomic landscape of the inherited platelet function disorders (Table 2), because it could provide important clues into how to improve and preserve the production of functional platelets for therapeutic purposes. Defects primarily responsible for thrombocytopenia are covered in the review by Eto and Kunishima.
Principal inherited platelet function disorders and underlying genomic defects
| Disorder . | Gene . | Chromosome location . | Main mode of inheritance . |
|---|---|---|---|
| Adhesion defects | |||
| Bernard-Soulier syndrome | GP1BA | 17p13.2 | Autosomal recessive |
| GP1BB | 22q11.21 | Autosomal recessive | |
| GP9 | 3q21.3 | Autosomal recessive | |
| Platelet-type von Willebrand disease | GP1BA | 17p13.2 | Autosomal dominant |
| GP6 deficiency | GP6 | 19q13.4 | Autosomal recessive |
| Receptors and signaling defects | |||
| ADP receptor deficiency | P2RY12 | 3q25.1 | Autosomal recessive |
| Thromboxane A2 receptor deficiency | TBXA2R | 19p13.3 | Autosomal recessive |
| PAR-4 receptor defects | F2RL3 | 19p13.11 | Autosomal dominant |
| Storage pool disease | |||
| Gray platelet syndrome | NBEAL2 | 3p21.31 | Autosomal recessive |
| GFI1B | 9q34.13 | Autosomal dominant | |
| Quebec platelet syndrome | PLAU | 10q22.2 | Autosomal dominant |
| Hermansky-Pudlak syndrome | HPS1 | 10q23.1-q23.3 | Autosomal recessive |
| AP3B1 | 5q14.1 | Autosomal recessive | |
| HPS3 | 3q24 | Autosomal recessive | |
| HPS4 | 22cen-q12.3 | Autosomal recessive | |
| HPS5 | 11p14 | Autosomal recessive | |
| HPS6 | 10q24.32 | Autosomal recessive | |
| HPS7 | 6p22.3 | Autosomal recessive | |
| HPS8 | 19q13.32 | Autosomal recessive | |
| HPS9 | 15q21.1 | Autosomal recessive | |
| Chediak-Higashi syndrome | LYST | 1q41.3 | Autosomal recessive |
| Aggregation defects | |||
| Glanzmann thrombasthenia | ITGA2B | 17q21.31 | Autosomal recessive |
| ITGB3 | 17q21.32 | Autosomal recessive | |
| Glanzmann thrombasthenia-like disorder | RASGRP2 | 11q13 | Autosomal recessive |
| Leukocyte adhesion deficiency-III syndrome | FERMT3 | 11q13.1 | Autosomal recessive |
| Procoagulant activity defects | |||
| Scott syndrome | TMEM16F | 12q12 | Autosomal dominant |
| Disorder . | Gene . | Chromosome location . | Main mode of inheritance . |
|---|---|---|---|
| Adhesion defects | |||
| Bernard-Soulier syndrome | GP1BA | 17p13.2 | Autosomal recessive |
| GP1BB | 22q11.21 | Autosomal recessive | |
| GP9 | 3q21.3 | Autosomal recessive | |
| Platelet-type von Willebrand disease | GP1BA | 17p13.2 | Autosomal dominant |
| GP6 deficiency | GP6 | 19q13.4 | Autosomal recessive |
| Receptors and signaling defects | |||
| ADP receptor deficiency | P2RY12 | 3q25.1 | Autosomal recessive |
| Thromboxane A2 receptor deficiency | TBXA2R | 19p13.3 | Autosomal recessive |
| PAR-4 receptor defects | F2RL3 | 19p13.11 | Autosomal dominant |
| Storage pool disease | |||
| Gray platelet syndrome | NBEAL2 | 3p21.31 | Autosomal recessive |
| GFI1B | 9q34.13 | Autosomal dominant | |
| Quebec platelet syndrome | PLAU | 10q22.2 | Autosomal dominant |
| Hermansky-Pudlak syndrome | HPS1 | 10q23.1-q23.3 | Autosomal recessive |
| AP3B1 | 5q14.1 | Autosomal recessive | |
| HPS3 | 3q24 | Autosomal recessive | |
| HPS4 | 22cen-q12.3 | Autosomal recessive | |
| HPS5 | 11p14 | Autosomal recessive | |
| HPS6 | 10q24.32 | Autosomal recessive | |
| HPS7 | 6p22.3 | Autosomal recessive | |
| HPS8 | 19q13.32 | Autosomal recessive | |
| HPS9 | 15q21.1 | Autosomal recessive | |
| Chediak-Higashi syndrome | LYST | 1q41.3 | Autosomal recessive |
| Aggregation defects | |||
| Glanzmann thrombasthenia | ITGA2B | 17q21.31 | Autosomal recessive |
| ITGB3 | 17q21.32 | Autosomal recessive | |
| Glanzmann thrombasthenia-like disorder | RASGRP2 | 11q13 | Autosomal recessive |
| Leukocyte adhesion deficiency-III syndrome | FERMT3 | 11q13.1 | Autosomal recessive |
| Procoagulant activity defects | |||
| Scott syndrome | TMEM16F | 12q12 | Autosomal dominant |
Adhesion defects
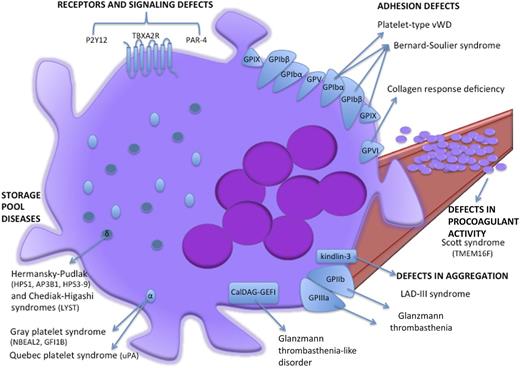
Bernard-Soulier syndrome (BSS) is a rare disorder characterized by thrombocytopenia with giant platelets and bleeding tendency owing to a platelet adhesion deficiency. BSS is caused by qualitative or quantitative defects in the platelet GPIb-IX-V complex that includes GPIbA, GPIbB, GPIX, and GPV proteins (Figure 3) and binds the von Willebrand factor (VWF). The most comprehensive characterization of the genomic defects underlying BSS led to the identification of 112 different mutations in the GP1BA, GP1BB, or GP9 genes.74,75 With a few exceptions of monoallelic BSS resulting from dominant mutations (eg, the Bolzano mutation affecting GP1BA), biallelic GP1BA, GP1BB, or GP9 lesions are usually found in BSS patients. Most of them impair GPIb-IX-V complex assembly and/or trafficking but more rarely affect VWF binding ability.75-77
Schematic cartoon representing the proteins mutated in inherited platelet function disorders. Light blue ovoids represent α-granules, dark blue spheres represent δ-granules. LAD-III, leukocyte adhesion deficiency-III syndrome; VWD, von Willebrand disease.
Schematic cartoon representing the proteins mutated in inherited platelet function disorders. Light blue ovoids represent α-granules, dark blue spheres represent δ-granules. LAD-III, leukocyte adhesion deficiency-III syndrome; VWD, von Willebrand disease.
Megakaryocytes from both BSS patients78 and GP1BB−/− BSS mouse models79 show that GPIb-IX-V deficiency underpins an impaired proplatelet formation that in turn accounts for macrothrombocytopenia. Notably, the proplatelet formation defects are independent from VWF binding. Indeed, megakaryocytes from BSS patients display an impaired proplatelet formation in vitro not only when megakaryocytes are cultured in adhesion on VWF or fibrinogen-coated surfaces but also when megakaryocytes are cultured in suspension. These observations suggest that the proplatelet formation defects cannot be exclusively ascribed to the deficient interaction of GPIb-IX-V with extracellular ligands (eg, P-selectin in addition to VWF) but rather are intrinsic to megakaryocytes. Additional GPIb-IX-V–mediated actomyosin-to-plasma membrane connections might be involved.77,79
In contrast to the loss-of-function mutations in GPIb-IX-V components that cause BSS, a few gain-of-function GP1BA mutations underlie the platelet hyperresponsiveness observed in platelet-type von Willibrand disease, a rare autosomal dominant disorder. Platelet-type von Willebrand disease patients display an excessive platelet-VWF interaction that exhausts the ability of platelets to bind to fibrinogen upon stimulation and leads to mild to moderate bleeding.73,80,81
Receptors and signaling defects
Defects in signaling pathways that control platelet secretion and aggregation are the most common cause of mild to moderate bleeding in inherited platelet function disorders. Among them, mutations in P2RY12 and TBXA2R genes coding for G-protein–coupled receptors (P2Y12 and TBXA2R, respectively) for soluble agonists adenosine 5′-diphosphate (ADP) and thromboxane A2 have provided important insights into the molecular mechanisms underlying platelet activation.
Multiple P2RY12 genomic lesions have been described as causing decreased and reversible ADP-induced platelet aggregation. The majority of lesions are nonsense and missense mutations, the latter affecting ADP binding to P2Y12 or P2Y12 receptor trafficking and function.73,82 Most of the qualitative defects identified (eg, the recently described P2RY12H187Q mutation that impairs ADP binding83 ) have shed new light on P2Y12 structure-function relationships.
Although extremely rare, genomic lesions in TBXA2R similarly account for defects in TXA2-mediated platelet aggregation and underpin a mild bleeding phenotype.76 Genomic lesions that lead to the loss of TBXA2R expression or disrupt the TBXA2R cell surface expression, ligand binding affinity, or G-protein coupling have not been reported till now.73,82
Storage pool diseases
Storage pool diseases are inherited defects of secretion from storage organelles (α-granules and δ-granules; Figure 3). α-granules are the most abundant platelet organelles. α-granule protein release triggers platelet adhesion, hemostasis, and wound healing.73 Gray platelet syndrome is the main inherited platelet function defect resulting from defective α-granule biogenesis, protein packaging, and storage.76 Patients with gray platelet syndrome display mild to moderate bleeding, myelofibrosis, and splenomegaly. In 2011, exome and RNA sequencing studies from 3 independent groups led to the identification of NBEAL2 hypomorphic mutations as the genetic lesion underlying autosomal recessive gray platelet syndrome.85-87 By an as yet unclarified mechanism, NBEAL2 is involved in α-granule formation and trafficking. It is noteworthy that NBEAL2−/− gray platelet syndrome mouse models reveal the role of α-granule protein deficiency and megakaryocyte proinflammatory phenotypes that drive the development of bone marrow fibrosis.88 However, NBEAL2 mutations cannot explain all gray platelet syndrome cases. Interestingly, Monteferraio et al89 recently identified a nonsense mutation in the GFI1B gene that codes for a transcriptional regulator of erythroid and megakaryocyte development as being responsible for an autosomal dominant form of gray platelet syndrome coupled with red cells defects.77,84,89
Storage pool diseases affecting δ-granule secretion lead to platelet aggregation defects and bleeding diatheses.73 When associated with defects in other lysosome-related organelles, they entail clear phenotypic features. Notably, melanosomal defects account for the lack of skin and hair pigmentation in patients with Hermansky-Pudlak syndrome and Chediak-Higashi syndrome.76,80 Defects in formation, shuttle, and release of platelet δ-granules account for the bleeding tendency in patients with Hermansky-Pudlak syndrome. Genomic lesions in 9 genes (HPS1, AP3B1, HPS3-9) that code for biogenesis of lysosome-related organelle complexes 1, 2, and 3 (BLOC-1, BLOC-2, and BLOC-3) and AP-3 complex subunits have been linked to distinct Hermansky-Pudlak syndrome subtypes,73 with relatively mild clinical features for those linked to defects in BLOC-2 components (HPS-3, HPS-5, and HPS-6 subtypes) and more severe forms associated with mutations in BLOC-3 components (HPS-1 and HPS-4 subtypes).90 In Chediak-Higashi syndrome, δ-granule deficiency and hypopigmentation are associated with recurrent infections, severe immunologic defects, and progressive neurologic dysfunction.73,76 Frameshift and nonsense mutations in the LYST gene (coding for LYST, lysosomal-trafficking regulator) underlie the defective exocytosis of secretory lysosomes and lysosome-related organelles.73
Defects in aggregation
Integrin αIIbβ3 on activated platelets enables the binding of fibrinogen and other soluble adhesive proteins involved in platelet aggregation.
Glanzmann thrombasthenia is caused by biallelic mutations in ITGA2B or ITGB3 genes encoding αIIb (ie, GPIIb) and β3 (ie, GPIIIa), respectively, which are usually unique to each family. Despite their normal platelet count and morphology, Glanzmann thrombasthenia patients have a severe bleeding diathesis due to αIIbβ3 qualitative or quantitative defects that impair platelet binding to fibrinogen during platelet activation.91 Genomic lesions underlying Glanzmann thrombasthenia are mainly represented by nonsense and missense mutations in ITGA2B or ITGB3 genes that impair either the αIIb and β3 subunit biosynthesis or the pro-αIIbβ3 complex formation, maturation, and translocation to the plasma membrane in megakaryocytes (Figure 3).73,84,92 Nurden et al93 recently screened a large cohort (76 families) of Glanzmann thrombasthenia patients by Sanger sequencing analysis on exon and splice sites and identified 55 novel mutations in ITGA2B and ITGB3 genes.
During platelet activation, integrin αIIbβ3 acts as a high-affinity binding site for fibrinogen. Proper αIIbβ3-mediated platelet activation requires interaction between β3 and the cytoplasmic proteins talin and kindlin-3. Indeed, β3 mutations that disrupt binding to talin/kindlin-3 cause platelet aggregation defects. Similarly, mutations affecting the kindlin-3–coding FERMT3 gene underlie leukocyte adhesion deficiency-III syndrome, whose features are a Glanzmann thrombasthenia-like phenotype because of defective β1 and β3 function in platelets, coupled with recurrent infections owing to the impaired β2 function in leukocytes.73,80,84,94,95
The small GTPase RAP1 is required for full αIIbβ3 activation. RAP1 activity, in turn, is tuned by the guanine nucleotide exchange factor CalDAG-GEFI. Indeed, mutations affecting the CalDAG-GEFI–coding RASGRP2 gene have recently been described as causative for a variant Glanzmann thrombasthenia-like phenotype with normal αIIbβ3 expression but defective platelet aggregation and severe bleeding.84,96
Defects in procoagulant activity
Phosphatidylserine flipping to the outer plasma membrane is essential for both proper platelet-mediated procoagulant activity and shedding of microparticles from activated, apoptotic, or necrotic cells. In fact, phosphatidylserine exposure upon vascular injury triggers the binding of coagulation factors and assembly of the prothrombinase complex, ultimately leading to platelet-mediated thrombin generation and fibrin clot formation. Platelet microparticles further enhance the hemostatic response through the exposure of phosphatidylserine and coagulation factor binding sites.
Notably, phosphatidylserine externalization is a Ca2+-dependent process regulated by TMEM16F activity. TMEM16F is a Ca2+-activated cation channel that tunes Ca2+ levels, therefore affecting phosphatidylserine scrambling in platelets during coagulation. Indeed, both homozygous nonsense and compound heterozygous in-frame deletion or nonsense mutations in the TMEM16F gene97,98 were identified in Scott syndrome patients. Scott syndrome is an extremely rare bleeding disorder characterized by defects in activation-induced phosphatidylserine externalization on platelets and subsequent impairment of tissue factor–induced thrombin generation and procoagulant microparticle shedding by platelets (Figure 3).76,84 Indeed, platelet-specific TMEM16F-null mice recapitulate all of these major platelet defects99 further emphasizing the crucial role of TMEM16F in platelet function.
Conclusions and future directions
Over the past decade, approaches based on next-generation sequencing have prompted the description of the mutational landscape for megakaryopoiesis and platelet function disorders. For some disorders, such as MPNs, genomics has reached the bedside to help patients, but unraveling these disorders is also providing us with a better knowledge of the mechanisms that underpin normal megakaryocyte-to-platelet transition. From this point of view, some of the newly uncovered molecular targets might be exploitable for the development of megakaryocyte precursor–related therapeutic strategies based on gene therapy for inherited platelet disorders as illustrated in the review by Wilcox in this series, but also in the quest to improve the megakaryocyte-to-platelet transition for ex vivo production of functional platelets as described in the review by Sim et al. Indeed, one of the major challenges for therapeutic platelet production is the requirement to produce functional platelets (ie, platelets that fulfill their proper physiological role in hemostasis). The genomic landscape of inherited platelet function defects further emphasizes which signaling pathways and molecular mechanisms are essential for proper platelet function.
With regard to the genomic landscape of malignant megakaryopoiesis, the recent development of methods for genome-wide analysis at a single-cell level will enable us to further investigate how clonal architecture can influence the disease phenotype in MPNs and further dissect the role of every single mutational event in the skewing of megakaryopoiesis and thrombopoiesis. In addition, clinical studies already underway that evaluate how mutations impact the clinical outcome of MPNs should be implemented through the assessment of global accuracy, sensitivity, and specificity of next-generation–based platforms compared with conventional assays.
Despite the effort to characterize the genomic landscape of AMKL, there are still gaps in our understanding. Thus, future studies should be focused on the development of targeted therapies toward recently identified fusion genes or deregulated downstream pathways.
Next-generation sequencing–based techniques have had a significant impact on the characterization of the mutational landscape of platelet function disorders.86,100 The ThromboGenomics Consortium of the International Society on Thrombosis and Haemostasis (https://haemgen.haem.cam.ac.uk/thrombogenomics) developed a platform for diagnosis of rare inherited bleeding and platelet disorders based on next-generation sequencing that has been available for clinical use since July 2015. Major efforts for characterizing platelet function disorders should be directed toward the assessment of tools for genotype-based prediction of bleeding severity to improve patient care.
Acknowledgments
This work was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC) project No. 10005 “Special Program Molecular Clinical Oncology 5 × 1000” (AIRC-Gruppo Italiano Malattie Mieloproliferative; http://www.progettoagimm.it); AIRC project No. 15337; Italian Ministry of University and Research (Fondo per gli Investimenti della Ricerca di Base Project 2011 [project No. RBAP11CZLK], and Progetti di Rilevante Interesse Nazionale 2010-11 [project No. 2010NYKNS7]).
Authorship
Contribution: E.B., R.N., V.P., R.Z., and R.M. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rossella Manfredini, Centre for Regenerative Medicine Stefano Ferrari, University of Modena and Reggio Emilia, via Gottardi 100, 41125 Modena, Italy; e-mail: rossella.manfredini@unimore.it.
References
Author notes
E.B. and R.N. contributed equally to this work.