Key Points
Increased gene body methylation inhibits leukemia, and oncogenes require varying levels of DNA methylation for efficient leukemogenesis.
Dnmt3b-induced DNA methylation in mice targets stem cell–associated genes with prognostic association in acute myeloid leukemia patients.
Abstract
The de novo DNA methyltransferases Dnmt3a and Dnmt3b are of crucial importance in hematopoietic stem cells. Dnmt3b has recently been shown to play a role in genic methylation. To investigate how Dnmt3b-mediated DNA methylation affects leukemogenesis, we analyzed leukemia development under conditions of high and physiological methylation levels in a tetracycline-inducible knock-in mouse model. High expression of Dnmt3b slowed leukemia development in serial transplantations and impaired leukemia stem cell (LSC) function. Forced Dnmt3b expression induced widespread DNA hypermethylation in Myc-Bcl2–induced leukemias, preferentially at gene bodies. MLL-AF9–induced leukemogenesis showed much less pronounced DNA hypermethylation upon Dnmt3b expression. Nonetheless, leukemogenesis was delayed in both models with a shared core set of DNA hypermethylated regions and suppression of stem cell–related genes. Acute myeloid leukemia patients with high expression of Dnmt3b target genes showed inferior survival. Together, these findings indicate a critical role for Dnmt3b-mediated DNA methylation in leukemia development and maintenance of LSC function.
Introduction
DNA methylation of cytosine guanine dinucleotides (CpGs) is a key epigenetic modification that affects tissue- and time-dependent gene transcription and genome integrity.1 In mammals, CpG methylation is catalyzed by 3 DNA methyltransferases (DNMTs).2,3 The maintenance DNA methyltransferase Dnmt1 replicates preexisting methylation patterns,4 whereas Dnmt3a and Dnmt3b primarily act as de novo methyltransferases.5 Both enzyme classes cooperate to establish and maintain cellular methylation patterns.6,7 Dnmt deficiency induces embryonic lethality in case of Dnmt1 and Dnmt3b and death at roughly 3 weeks of age in Dnmt3a-knockout mice.5,8 Nonetheless, self-renewal potential is maintained in embryonic stem cells without Dnmt3a and Dnmt3b.6 DNA methylation influences hematopoietic differentiation and lineage decisions.9,10 In normal hematopoiesis, Dnmt1 is required for hematopoietic stem cell (HSC) self-renewal, differentiation, and niche retention.11 In HSC, loss of both Dnmt3a and Dnmt3b diminishes self-renewal potential, whereas the differentiation capacity into all lineages is maintained.12 Conditional deletion of Dnmt3a in the hematopoietic system severely impairs HSC differentiation, while simultaneously expanding HSC numbers in the bone marrow.13 Combined loss of Dnmt3a and Dnmt3b in HSC creates a synergistic effect resulting in a more severe differentiation block than in Dnmt3a-deficient cells alone. Nonetheless, residual Dnmt3b activity enabled differentiation of adult HSCs, especially upon loss of Dnmt3a.14
Aberrant DNA methylation constitutes a hallmark of cancer,15-17 and deregulated DNA methylation has been shown in a variety of hematologic malignancies.18 Recent findings in acute myeloid leukemia (AML) patient samples indicate that DNA methylation exhibits specific patterns in AML subtypes. These methylation profiles may reflect biological differences with therapeutic implications.19,20 Aberrant methylation patterns in cancer are characterized by global genome-wide hypomethylation,21 and simultaneously occurring regional hypermethylation of CpG islands.22-24 Both, hypo- and hypermethylation may facilitate tumorigenesis.10,25-28 Accordingly, the role of de novo methyltransferases in cancer has remained unclear. Forced expression of Dnmt3b in tumor-prone APCMin mice promoted development of gastrointestinal tumors by de novo methylation and transcriptional silencing of tumor suppressor genes.29 Conditional Dnmt3b knockout inhibited early intestinal tumor formation.30 Also, 2 independent studies identified Dnmt3b as a tumor suppressor in Eμ-Myc–driven lymphomas.31,32 Clinical data reflected these divergent roles of DNMTs in cancer. DNMT3A mutations have been identified in >20% of patients with AML33,34 and ∼10% of those with myelodysplastic syndromes.35 The identified mutations are invariably heterozygous and predicted to disrupt the catalytic enzyme activity.36 Of note, high expression levels of DNMT3B in leukemic bulk cells are associated with poor outcome in AML.37,38 These data suggest an important but tightly controlled role for DNA methylation in cancer initiation and progression. However, the precise function of increased DNA methylation is not fully understood.
Here, we show that forced expression of Dnmt3b severely impaired leukemia development. Dnmt3b expression induced DNA hypermethylation with subsequent downregulation of genes highly expressed in stem cells.
Materials and methods
Plasmids, retroviral supernatant production, and cell transduction
Murine stem cell virus (MSCV) retroviral construct MLL-AF9-IRES-GFP has been described.39 MSCV retroviral construct Myc-IRES-Bcl2-IRES-mCherry was generated by inserting an additional IRES and mCherry into the MSCV construct Myc-IRES-Bcl2.40 Retroviral supernatants were obtained by transfection of Plat-E cells. For transduction, bone marrow cells were MACS-lineage depleted (Miltenyi) and stimulated for 72 hours in Iscove modified Dulbecco medium (Invitrogen) containing 20% fetal calf serum (PAA), mouse interleukin (IL) 3 (10 ng/mL; Peprotech), human IL-6 (5 ng/mL; Peprotech), and mouse stem cell factor (50 ng/mL; Peprotech). Lineage-negative cells were infected using Retronectin (Takara) and sorted for GFP or mCherry expression after 3 days of spin infection.
Mice, genotyping, and transgene induction
Inducible Dnmt3b-knock-in mice were described previously.29 Inducible Dnmt3b-knock-in mice were purchased from the Jackson Laboratories, and C57Bl/6N mice from Janvier. All mice were kept in pathogen-free animal facilities at the University Hospital Münster. For genotyping, genomic tail-tip DNA was polymerase chain reaction (PCR) amplified using standard PCR. For transgene induction, mice were fed 0.5 mg/mL doxycycline in the drinking water. Drinking water was changed twice a week.
Transplantation experiments
All animal experiments were approved by the local authorities according to the German Federal Animal Protection Act. For bone marrow transplantation, 8 weeks-old C57Bl/6N recipient mice were sublethally irradiated with 7 Gy. Retrovirally transduced bone marrow cells were IV injected into the tail vein with 2 × 105 unfractionated nontransduced bone marrow cells. Expression of Dnmt3b was induced at time of transplantation, and recipient mice were kept on doxycycline until the end of the experiment. For serial transplantations, spleen cells from leukemic mice were sorted for GFP and c-Kit expression (MLL-AF9) or mCherry expression (Myc-Bcl2) and retransplanted into sublethally irradiated recipients. For competitive bone marrow transplantation, recipient mice were sublethally irradiated with 2 doses of 4.75 Gy. CD45.2+ bone marrow cells (105 or 106) were injected IV into the tail vein with 105 CD45.1+ competitor cells.
Microarray analysis
MLL-AF9 leukemic spleen cells were sorted for c-Kit expression, and gene expression was analyzed using the mouse Gene 2.0 ST Array (Affymetrix) according to the manufacturer’s instructions. Arrays were scanned at 1.56-μm resolution using the Affymetrix GeneChip Scanner 3000. Raw gene expression data were imported to the Affymetrix expression console and normalized using robust multiarray average (RMA). Differential gene expression was calculated using the R/Bioconductor package RankProd.41 Microarray data were deposited in the Gene Expression Omnibus database (accession number GSE71040).
Reduced representation bisulfite sequencing (RRBS)
A total of 0.3 to 1 µg of DNA was used for RRBS library preparation using published protocols with minor modifications.42 Briefly, genomic DNA was digested with MspI (NEB), end repaired, A-tailed, and ligated to Illumina TruSeq adapters (Illumina). Fragments in the range of 50- to 220-bp size were gel purified (NuSieve 3:1 agarose; Lonza). Libraries were bisulfite converted and sequenced on a HiScanSQ instrument (Illumina). See supplemental Methods for further details (available on the Blood Web site). RRBS data were deposited in the Gene Expression Omnibus database (accession number GSE71039).
Statistical analysis
All data are shown as mean ± standard deviation (SD) if not indicated otherwise. Statistical analyses were done in SPSS 22 (IBM). Student t tests were used to determine statistical significance. Survival was analyzed using Kaplan-Meier curves and log-rank test. A P value ≤.05 was considered significant.
Results
High levels of Dnmt3b expression prolong leukemia latency
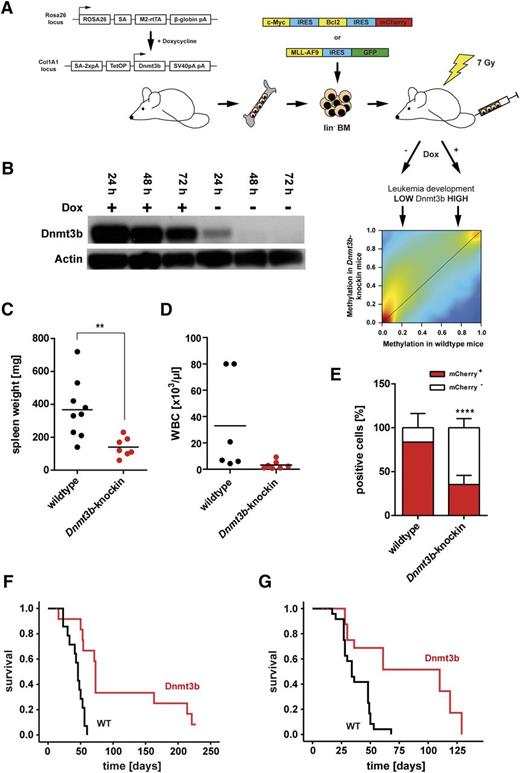
Several studies described higher expression of Dnmt3b in AML samples when compared with healthy controls,43,44 whereas others did not show a significant difference.45,46 Compared with HSCs, DNMT3B expression was decreased in primary AML blasts (supplemental Figure 1A). To functionally investigate the role of Dnmt3b in leukemogenesis, we generated murine leukemias in a tetracycline-inducible Dnmt3b-knock-in mouse model by retroviral expression of an Myc-Bcl2-mCherry construct in lineage-negative bone marrow cells. The Myc-Bcl2 combination induces a biphenotypic leukemia with myeloid and B-lymphoid blasts in mice.40 Cells from Dnmt3b-wild-type and heterozygous Dnmt3b-knock-in cells (supplemental Figure 1B) were transduced with similar efficiency (supplemental Figure 1C). Cells were transplanted into sublethally irradiated C57Bl/6N recipient mice (Figure 1A). Dnmt3b expression was detected in Myc-Bcl2 Dnmt3b-knock-in cells (Figure 1B). All recipients of Dnmt3b-wild-type Myc-Bcl2-mCherry cells rapidly died of lethal leukemia characterized by splenomegaly (Figure 1C) and increased numbers of leukemic cells in bone marrow (Figure 1D) and spleen (data not shown). Recipients of Dnmt3b-knock-in Myc-Bcl2 cells showed less pronounced splenomegaly (Figure 1C) and only slightly increased white blood cell counts in peripheral blood (Figure 1D). Upon cotransplantation of leukemic mCherry+ and nonleukemic mCherry− cells, Dnmt3b-knock-in were inferior in replacing nonleukemic bone marrow cells indicating an intrinsic defect in leukemia-initiating cells (LICs) of Dnmt3b-knock-in mice (Figure 1E). Recipients of Dnmt3b-knock-in Myc-Bcl2 cells developed leukemia with substantially prolonged latency with mean latency times of 43.4 ± 3.2 days posttransplantation (wild-type cells) and 107.3 ± 21.0 days (Dnmt3b-knock-in cells) (P < .001; Figure 1F). Transformation of Myc-Bcl2–transduced cells was confirmed by secondary transplantation of mCherry-sorted spleen cells (Figure 1G). Dnmt3b-knock-in leukemias were characterized by high expression of myeloid surface markers, whereas the bilinear phenotype of Myc-Bcl2 leukemias40 was only observed in recipients of wild-type cells (supplemental Figure 1D). These data indicate that deviation from physiological methylation precluded development of lymphoid neoplasia.
Dnmt3b expression prolongs leukemia latency. (A) Bone marrow from tetracycline-inducible Dnmt3b overexpressing mice was retrovirally transduced with MSCV-Myc-IRES-Bcl2-IRES-mCherry or MSCV-MLL-AF9-IRES-GFP oncogene vectors. Transduced cells were sorted for mCherry or GFP, respectively, and transplanted into sublethally irradiated recipient mice. Leukemia development was analyzed under conditions of high vs low DNA methylation. (B) Doxycycline-regulated Dnmt3b expression in sorted Myc-Bcl2 leukemic spleen cells from primary recipient mice of Dnmt3b-knock-in cells. Spleen cells were isolated from diseased mice and cultured in the presence of 1 µg/mL doxycycline, and protein expression was analyzed after different time points of addition and subsequent withdrawal of doxycycline by western blot. Spleen weight (C) and white blood cell (WBC) count (D) in primary recipients of Myc-Bcl2 leukemic Dnmt3b-knock-in cells at end of experiment. Each dot represents 1 leukemic mouse. **P < .01. (E) Percentage of leukemic (mCherry+) and nonleukemic (mCherry−) cells in bone marrow of primary recipient mice at end of experiment. Mean ± SD are shown (n = 5-9 mice). ****P < .0001. (F) Survival of primary recipients of Myc-Bcl2 leukemic wild-type and Dnmt3b-knock-in cells (n = 15 mice for each group). Dnmt3b expression was induced at time of transplantation. P < .001. (G) Survival of secondary recipients of Myc-Bcl2 leukemias (n = 15 mice for each group). P < .01.
Dnmt3b expression prolongs leukemia latency. (A) Bone marrow from tetracycline-inducible Dnmt3b overexpressing mice was retrovirally transduced with MSCV-Myc-IRES-Bcl2-IRES-mCherry or MSCV-MLL-AF9-IRES-GFP oncogene vectors. Transduced cells were sorted for mCherry or GFP, respectively, and transplanted into sublethally irradiated recipient mice. Leukemia development was analyzed under conditions of high vs low DNA methylation. (B) Doxycycline-regulated Dnmt3b expression in sorted Myc-Bcl2 leukemic spleen cells from primary recipient mice of Dnmt3b-knock-in cells. Spleen cells were isolated from diseased mice and cultured in the presence of 1 µg/mL doxycycline, and protein expression was analyzed after different time points of addition and subsequent withdrawal of doxycycline by western blot. Spleen weight (C) and white blood cell (WBC) count (D) in primary recipients of Myc-Bcl2 leukemic Dnmt3b-knock-in cells at end of experiment. Each dot represents 1 leukemic mouse. **P < .01. (E) Percentage of leukemic (mCherry+) and nonleukemic (mCherry−) cells in bone marrow of primary recipient mice at end of experiment. Mean ± SD are shown (n = 5-9 mice). ****P < .0001. (F) Survival of primary recipients of Myc-Bcl2 leukemic wild-type and Dnmt3b-knock-in cells (n = 15 mice for each group). Dnmt3b expression was induced at time of transplantation. P < .001. (G) Survival of secondary recipients of Myc-Bcl2 leukemias (n = 15 mice for each group). P < .01.
The increased latency of Myc-Bcl2 Dnmt3b-knock-in leukemias and reduced numbers of leukemic cells in competitive transplantation assays suggested that constitutive methylation is required for leukemia stem cell (LSC) function. In murine MLL-AF9 leukemia, c-Kit+ cells are highly enriched for LICs and LSCs.39,47 We analyzed effects of enhanced DNA methylation on LSC function in MLL-AF9 leukemia (supplemental Figure 2A). Transplantation of MLL-AF9–expressing Dnmt3b-knock-in cells led to leukemia development with prolonged latency (mean survival: MLL-AF9 wild-type, 85.08 ± 22.44 days; MLL-AF9 Dnmt3b-knock-in, 153.55 ± 30.88 days) (supplemental Figure 2B). Spleen weights did not differ between recipients of wild-type and Dnmt3b-knock-in cells (supplemental Figure 2C). We sorted primary leukemias for high expression of c-Kit (gated for top 20%) (supplemental Figure 2D) and performed serial transplantations. Superior colony formation potential of c-Kithigh cells (compared with Kitlow cells) was confirmed by colony-forming unit assays (supplemental Figure 2E). Upon transplantation of sorted c-Kit+ cells, leukemia latency was significantly prolonged in secondary (Figure 2A) and tertiary recipients (Figure 2B). Similar to Dnmt3b-expressing Myc-Bcl2 leukemias, diseased recipients of Dnmt3b-knock-in MLL-AF9 LSCs showed reduced numbers of white blood cells in peripheral blood at time of analysis (Figure 2C). MLL-AF9–induced leukemias were exclusively of myeloid lineage (Figure 2D). Of note, when secondary recipients died of leukemia, recipients of MLL-AF9 Dnmt3b-knock-in cells showed increased numbers of c-Kit+ cells (Figure 2D). The c-Kit+Dnmt3b-knock-in LSCs did not show altered cell surface expression of other HSC markers (Figure 2E), which indicates that high Dnmt3b expression did not affect the cellular composition of c-Kit+MLL-AF9 LSCs. The colony formation potential of c-Kit+Dnmt3b-knock-in LSCs was reduced when compared with recipients of MLL-AF9 wild-type cells (Figure 2F). Colony numbers of total spleen cells were similar (Figure 2G), pointing toward impaired leukemic potential of c-Kit+ leukemia cells with high Dnmt3b expression.
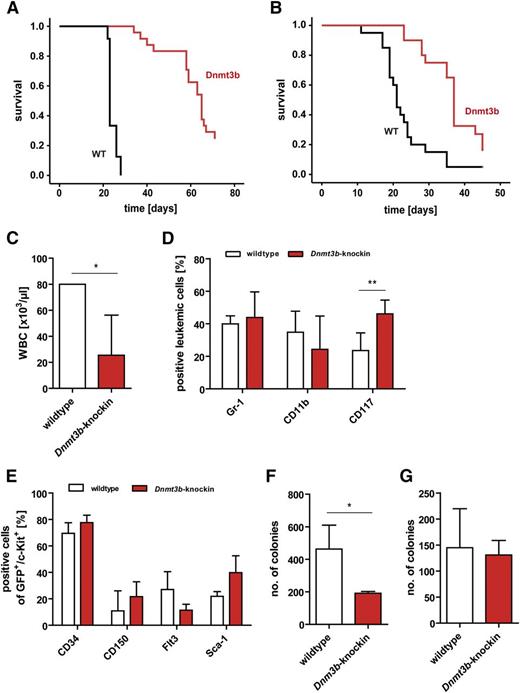
Effects of Dnmt3b-knock-in in MLL-AF9 leukemia model. (A-B) Survival of secondary (P < .001) (A) and tertiary (P < .001) (B) recipients of MLL-AF9–transduced wild-type and Dnmt3b-knock-in c-Kit+ LSCs. Dnmt3b expression was induced in all Dnmt3b-knock-in mice (n = 18-24 per group). (C) White blood cell counts of secondary recipients of wild-type and Dnmt3b-knock-in MLL-AF9–transduced cells at end of experiment; n = 3 for wild-type, n = 6 for Dnmt3b-knock-in. (D) Percentage of Gr-1+, CD11b+, and c-Kit+ cells in bone marrow of secondary recipients of wild-type (n = 11) and Dnmt3b-knock-in (n = 4) MLL-AF9 LSCs as determined by fluorescence-activated cell sorter analysis. (E) Expression of HSC markers CD34, CD150, Flt3, and Sca-1 on GFP+/c-Kit+MLL-AF9 LSCs from bone marrow of recipients of wild-type and Dnmt3b-knock-in leukemias (n = 3 for each genotype). (F-G) Dnmt3b expression suppresses colony growth of c-Kit+MLL-AF9 LSCs (top 20% of c-Kit–expressing cells were sorted for wild-type and knock-in mice). C-Kit+ cells (F) and total spleen cells (G) from secondary recipients of wild-type and Dnmt3b-knock-in MLL-AF9 LSCs were plated in triplicates in Methocult (n = 3-11 mice). Values in panels C-G are mean ± SD. *P < .05; **P < .01
Effects of Dnmt3b-knock-in in MLL-AF9 leukemia model. (A-B) Survival of secondary (P < .001) (A) and tertiary (P < .001) (B) recipients of MLL-AF9–transduced wild-type and Dnmt3b-knock-in c-Kit+ LSCs. Dnmt3b expression was induced in all Dnmt3b-knock-in mice (n = 18-24 per group). (C) White blood cell counts of secondary recipients of wild-type and Dnmt3b-knock-in MLL-AF9–transduced cells at end of experiment; n = 3 for wild-type, n = 6 for Dnmt3b-knock-in. (D) Percentage of Gr-1+, CD11b+, and c-Kit+ cells in bone marrow of secondary recipients of wild-type (n = 11) and Dnmt3b-knock-in (n = 4) MLL-AF9 LSCs as determined by fluorescence-activated cell sorter analysis. (E) Expression of HSC markers CD34, CD150, Flt3, and Sca-1 on GFP+/c-Kit+MLL-AF9 LSCs from bone marrow of recipients of wild-type and Dnmt3b-knock-in leukemias (n = 3 for each genotype). (F-G) Dnmt3b expression suppresses colony growth of c-Kit+MLL-AF9 LSCs (top 20% of c-Kit–expressing cells were sorted for wild-type and knock-in mice). C-Kit+ cells (F) and total spleen cells (G) from secondary recipients of wild-type and Dnmt3b-knock-in MLL-AF9 LSCs were plated in triplicates in Methocult (n = 3-11 mice). Values in panels C-G are mean ± SD. *P < .05; **P < .01
Limited effects of Dnmt3b in hematopoiesis
We analyzed whether forced Dnmt3b expression affected hematopoietic differentiation under nonleukemic conditions. Dnmt3b expression was induced in vivo for 4 to 44 weeks before analysis. Dnmt3b-knock-in mice showed a 1.5-fold increase in Lin−Sca-1+c-Kit+ (LSK) cell numbers after 4 weeks of induction (P < .05), mainly because of an increase in the multipotent progenitor cell compartment (LMPP) (P < .01; Figure 3A). The dominance of the LMPP pool in Dnmt3b-knock-in mice was not matched by a similar contribution of these LMPPs to the downstream progenitor populations of granulocyte-macrophage progenitors (supplemental Figure 3A) and megakaryocyte-erythroid progenitors (supplemental Figure 3B), neither were cell numbers of neutrophils altered in bone marrow of Dnmt3b-knock-in mice (supplemental Figure 3C). Instead, Dnmt3b-knock-in mice showed ∼1.7-fold reduced white blood cell counts in peripheral blood (Figure 3B) and ∼2.3-fold reduced lymphocyte counts in peripheral blood (Figure 3C). Fluorescence-activated cell sorter analysis of erythroid differentiation revealed slightly increased numbers of erythroblastic cells originating from Dnmt3b-knock-in HSCs (P < .05) (supplemental Figure 3D), whereas other blood parameters did not differ (supplemental Figure 3E-G). Expansion of Dnmt3b-knock-in LMPPs in vivo could not be attributed to altered apoptosis rates (supplemental Figure 4A). Dnmt3b-knock-in mice and wild-type mice showed similar survival times (Figure 3D). Both wild-type and Dnmt3b-knock-in animals showed comparable spleen weights (supplemental Figure 4B), and total bone marrow cells gave rise to equal colony numbers (supplemental Figure 4C). Competitive transplantation of 105 or 106 CD45.2+ donor-derived bone marrow cells with 105 CD45.1+ competitor cells did not yield differences between wild-type and Dnmt3b-knock-in donor-derived cells (Figure 3E), nor was multilineage engraftment altered by high Dnmt3b expression (supplemental Figure 4D). But Dnmt3b-knock-in bone marrow cells showed reduced growth in in vitro culture (Figure 3F). High Dnmt3b expression has been shown in mouse wild-type HSCs and different multipotent progenitor cell stages by RNA sequencing.48 In our model, Dnmt3b was also highly expressed in LSK, myeloid, lymphoid, and erythroid cells upon induction. Although Dnmt3a proteins were slightly elevated in 2 out of 4 mice after Dnmt3b induction, neither Dnmt3a nor Dnmt1 showed consistent deregulation upon overexpression of Dnmt3b (supplemental Figure 4F).
Dnmt3b expression increases the number of multipotent progenitor cells in nonleukemic mice. (A) Induction of Dnmt3b expression in nonleukemic Dnmt3b-knock-in mice increased the numbers of multipotent progenitor cells after 4 weeks (n = 3 per group). LT-HSC gated for LSK CD34−Flt3−; ST-HSC gated for LSK CD34+Flt3−; LMPP gated for LSK CD34+Flt3+. (B) White blood cell counts of wild-type (black circles) and Dnmt3b-knock-in (red circles) mice after 16, 28, and 44 weeks of in vivo induction of Dnmt3b expression. (C) Total cell counts for lymphocytes in peripheral blood of wild-type and Dnmt3b-knock-in mice at the indicated time points after induction of Dnmt3b. Each dot represents 1 individual mouse. (D) Survival of wild-type and Dnmt3b-knock-in mice after induction of Dnmt3b expression for up to 1 year (n = 6 for wild-type [WT] and n = 8 for Dnmt3b-knock-in mice). (E) Percentage of CD45.2+ donor-derived peripheral blood cells in primary recipients for up to 24 weeks posttransplantation. Transplanted cell doses are 106 and 105 cells mixed with 105 competitor cells (n = 4 mice each). (F) Dnmt3b-knock-in total bone marrow cells were impaired in growth upon culture in the presence of IL-3, IL-6, and murine stem cell factor; n = 3 per group. Data in panels A-C and E-F are mean ± SD values. *P < .05; **P < .01; ***P < .001.
Dnmt3b expression increases the number of multipotent progenitor cells in nonleukemic mice. (A) Induction of Dnmt3b expression in nonleukemic Dnmt3b-knock-in mice increased the numbers of multipotent progenitor cells after 4 weeks (n = 3 per group). LT-HSC gated for LSK CD34−Flt3−; ST-HSC gated for LSK CD34+Flt3−; LMPP gated for LSK CD34+Flt3+. (B) White blood cell counts of wild-type (black circles) and Dnmt3b-knock-in (red circles) mice after 16, 28, and 44 weeks of in vivo induction of Dnmt3b expression. (C) Total cell counts for lymphocytes in peripheral blood of wild-type and Dnmt3b-knock-in mice at the indicated time points after induction of Dnmt3b. Each dot represents 1 individual mouse. (D) Survival of wild-type and Dnmt3b-knock-in mice after induction of Dnmt3b expression for up to 1 year (n = 6 for wild-type [WT] and n = 8 for Dnmt3b-knock-in mice). (E) Percentage of CD45.2+ donor-derived peripheral blood cells in primary recipients for up to 24 weeks posttransplantation. Transplanted cell doses are 106 and 105 cells mixed with 105 competitor cells (n = 4 mice each). (F) Dnmt3b-knock-in total bone marrow cells were impaired in growth upon culture in the presence of IL-3, IL-6, and murine stem cell factor; n = 3 per group. Data in panels A-C and E-F are mean ± SD values. *P < .05; **P < .01; ***P < .001.
Dnmt3b expression induces widespread DNA methylation changes
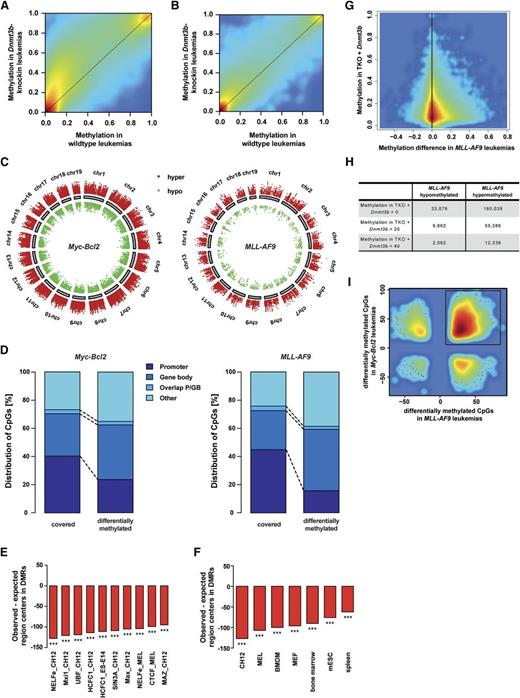
We used RRBS to map genome-wide DNA methylation in Dnmt3b-knock-in leukemias. For each genotype, RRBS data from 2 mice with Myc-Bcl2 leukemias and from MLL-AF9 LSCs (GFP+/c-Kit+) from 3 mice were analyzed. Raw methylation data from all covered CpG sites were used to generate an unsupervised hierarchical clustering (supplemental Figure 5A). Overall, Dnmt3b-knock-in Myc-Bcl2 leukemias exhibited a larger fraction of intermediately methylated (20% to 80%) and highly methylated CpGs (80% to 100%) than wild-type Myc-Bcl2 leukemias (supplemental Figure 5B). A similar pattern was observed in MLL-AF9 leukemias, yet it was far less pronounced (supplemental Figure 5B-C). Smoothed methylation values confirmed genome-wide hypermethylation in both Myc-Bcl2 and MLL-AF9 Dnmt3b-knock-in leukemias (Figure 4A-B). Of note, Dnmt3b-induced hypermethylation in MLL-AF9 leukemias was less pronounced than in Myc-Bcl2 leukemias (Figure 4B). Differential methylation analysis in Dnmt3b-knock-in samples yielded 2092 differentially methylated regions (DMRs) in Myc-Bcl2 leukemias (supplemental Figure 5D) and 105 DMRs in MLL-AF9 LSCs (supplemental Figure 5E). Gene promoters and exons were underrepresented in hypermethylated DMRs, but transcribed regions were overrepresented in hypermethylated DMRs (supplemental Figure 5F).49
Dnmt3b induces DNA methylation changes detected by RRBS. (A-B) Smoothened scatter plots of methylation values for Dnmt3b-knock-in vs wild-type control samples in Myc-Bcl2 (A) and MLL-AF9 (B) leukemias, respectively. Colors represent the density of points ranging from red (high density) to blue (low density). (C) Chromosomal distribution of DMCs in Myc-Bcl2 (left) and MLL-AF9 (right) leukemias. Every dot represents 1 DMC. Red indicates DNA hypermethylated DMCs in Dnmt3b-knock-in samples, green indicates hypomethylated DMCs. (D) Region presence of covered and DMC sites across different genomic regions. Promoter regions were defined as 1000 bp upstream of transcriptional start site (TSS) to 500 bp downstream of TSS; gene bodies were defined as 500 bp downstream of TSS to end of gene. GB, gene bodies; P, promoter. (E-F) Presence of transcription factor binding sites (E), and RNA polymerase II binding sites (F) among DMRs. The number of centers of a particular region of interest that could be expected in DMRs under the assumption of a uniform distribution in RRBS-covered regions (expected region centers) was subtracted from the number of region centers actually found in DMRs (observed region centers). The bars visualize the differences of observed and expected region centers in hypermethylated DMRs. ***P < .001. (G) Smoothed scatter plot shows CpG sites that gained DNA methylation in previously published Dnmt triple-knockout mice (TKO) with reintroduced Dnmt3b50 and the corresponding DNA methylation difference in MLL-AF9 leukemias (Dnmt3b-knock-in − wild-type). Methylation in TKO + Dnmt3b indicates CpGs that gain DNA methylation after eradication of existing DNA methylation patterns in TKO cells following reintroduction of Dnmt3b. (H) Number of CpG sites that show methylation gain in TKO murine embryonic stem cells >0, >20, or >40, and hypo- or hypermethylation in Dnmt3b-knock-in MLL-AF9 leukemias. (I) Smoothed scatter plot showing all CpGs that are differentially methylated in Myc-Bcl2 and MLL-AF9 leukemias, respectively. The majority of DMCs showed hypermethylation in both leukemias.
Dnmt3b induces DNA methylation changes detected by RRBS. (A-B) Smoothened scatter plots of methylation values for Dnmt3b-knock-in vs wild-type control samples in Myc-Bcl2 (A) and MLL-AF9 (B) leukemias, respectively. Colors represent the density of points ranging from red (high density) to blue (low density). (C) Chromosomal distribution of DMCs in Myc-Bcl2 (left) and MLL-AF9 (right) leukemias. Every dot represents 1 DMC. Red indicates DNA hypermethylated DMCs in Dnmt3b-knock-in samples, green indicates hypomethylated DMCs. (D) Region presence of covered and DMC sites across different genomic regions. Promoter regions were defined as 1000 bp upstream of transcriptional start site (TSS) to 500 bp downstream of TSS; gene bodies were defined as 500 bp downstream of TSS to end of gene. GB, gene bodies; P, promoter. (E-F) Presence of transcription factor binding sites (E), and RNA polymerase II binding sites (F) among DMRs. The number of centers of a particular region of interest that could be expected in DMRs under the assumption of a uniform distribution in RRBS-covered regions (expected region centers) was subtracted from the number of region centers actually found in DMRs (observed region centers). The bars visualize the differences of observed and expected region centers in hypermethylated DMRs. ***P < .001. (G) Smoothed scatter plot shows CpG sites that gained DNA methylation in previously published Dnmt triple-knockout mice (TKO) with reintroduced Dnmt3b50 and the corresponding DNA methylation difference in MLL-AF9 leukemias (Dnmt3b-knock-in − wild-type). Methylation in TKO + Dnmt3b indicates CpGs that gain DNA methylation after eradication of existing DNA methylation patterns in TKO cells following reintroduction of Dnmt3b. (H) Number of CpG sites that show methylation gain in TKO murine embryonic stem cells >0, >20, or >40, and hypo- or hypermethylation in Dnmt3b-knock-in MLL-AF9 leukemias. (I) Smoothed scatter plot showing all CpGs that are differentially methylated in Myc-Bcl2 and MLL-AF9 leukemias, respectively. The majority of DMCs showed hypermethylation in both leukemias.
Analysis of single CpGs revealed 89 533 and 16 572 differentially methylated CpGs (DMCs) in Myc-Bcl2 and MLL-AF9 murine leukemias, respectively (supplemental Table 1). The vast majority of DMCs were hypermethylated upon Dnmt3b expression (supplemental Table 1). Aberrant DNA methylation was observed across all chromosomal regions, independently of the oncogene (Figure 4C). However, CpG-rich and CpG-poor regions were affected differently. DMCs more frequently mapped to CpG islands in Myc-Bcl2 leukemias (45% vs 26%). In contrast, 60% of DMCs in MLL-AF9 leukemias were found neither in CpG islands nor CpG shores (supplemental Figure 6A), thereby reflecting the high abundance of DMCs in regions beyond CpG shores in MLL-AF9 leukemias. Because Dnmt3b has recently been identified as the main de novo methyltransferase of gene bodies,50 we analyzed gene body methylation in murine leukemias. Gene bodies were overrepresented in hyper- and hypomethylated CpG sites in Myc-Bcl2 and MLL-AF9 leukemias (Figure 4D; supplemental Figure 6B-E). We also analyzed the overlap of hypermethylated regions in Dnmt3b-knock-in leukemias with published mouse ENCODE51 chromatin immunoprecipitation–sequencing data. Known transcription factor binding sites were heavily underrepresented in DMRs (Figure 4E). This also held true for the transcription factors MAX and the MAX interacting protein MXI1, which have been linked to human cancers.52 Most commonly affected by DMRs were binding sites of transcription factors that have previously been associated with leukemia (supplemental Table 4). The chromatin organizing factor CTCF was recently found to be mutated in AML patient samples53 and in pre-LSCs.54 Other transcription factors (eg, HCFC1 and P300) have been described to play a role in AML development and maintenance,55 especially in MLL-rearranged leukemias.56-58 In addition, sites of H3K4 trimethylation (supplemental Figure 6F) and polymerase II binding sites (Figure 4F) also showed reduced representation in DMRs. CpG sites that showed altered methylation levels in Dnmt3b-knock-in leukemias were compared with CpG sites that gained methylation in murine Dnmt-triple-knockout embryonic stem (ES) cells after reintroduction of Dnmt3b.50 The number of CpG sites with methylation gain in murine ES cells and hypermethylation in Dnmt3b-expressing Myc-Bcl2 and MLL-AF9 leukemias was higher than the number of those CpG sites affected by hypomethylation in Dnmt3b-expressing leukemias (Figure 4G-H; supplemental Figure 7A-B). Interestingly, this phenomenon was also shown for Dnmt3a target sites (supplemental Figure 7C-F). In addition, the majority of DMCs identified in both Myc-Bcl2 and MLL-AF9 Dnmt3b-knock-in samples showed DNA hypermethylation (Figure 4I), which elucidated a shared set of commonly hypermethylated CpG sites.
Dnmt3b-induced DNA methylation alters LSC gene expression profile
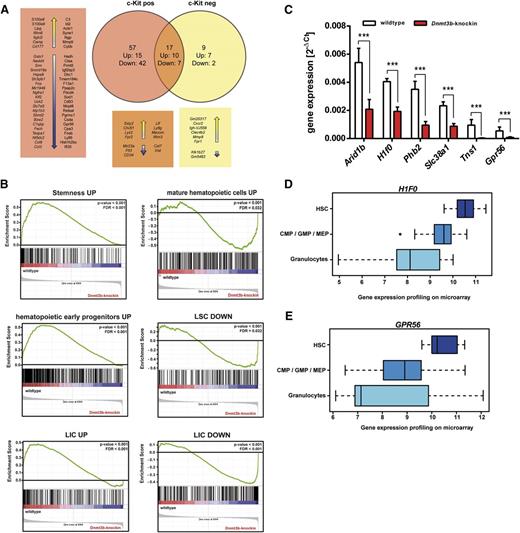
We generated genome-wide messenger RNA expression profiles of GFP+/c-Kit+ wild-type and Dnmt3b-knock-in MLL-AF9 LSCs from the exact same donor mice that were used for DNA methylation profiling. We identified 74 differentially expressed genes in c-Kit+Dnmt3b-knock-in LSCs (Figure 5A). The group of genes downregulated in Dnmt3b-knock-in LSCs was enriched for genes highly expressed in normal HSCs, including the tyrosine kinase receptor Flt3 and the early hematopoietic marker CD34. Conversely, Dnmt3b-knock-in LSCs showed higher expression of HSC differentiation factors. Ingenuity pathway analysis revealed significant enrichment for genes downregulated in Dnmt3-knock-in LSCs in the categories of cancer (25 genes), cellular growth and proliferation (18 genes), and hematologic system development and function (12 genes) (P < .05). Upstream regulators of differentially expressed genes in Dnmt3b-knock-in leukemias included transcription factors (eg, Myc and Srf), as well as the epigenetic factors Kdm6b and Asxl1. Gene set enrichment analysis revealed a strong enrichment of “stemness” genes and of genes expressed in early hematopoietic progenitors and LICs in wild-type LSCs (Figure 5B, left). Vice versa, Dnmt3b-knock-in hypermethylated LSCs were enriched for genes expressed in mature hematopoietic cells and genes downregulated in normal LSCs and LICs (Figure 5B, right). The concomitant downregulation of HSC and LSC genes and upregulation of differentiation genes may contribute to increased leukemia latencies upon high Dnmt3b expression.
Dnmt3b alters the expression profile of LSCs and leads to downregulation of stem cell genes. (A) Genome-wide mRNA expression profiles of c-Kit+MLL-AF9 LSCs and c-Kit−MLL-AF9 leukemic bulk cells were generated using Affymetrix Mouse Gene ST 2.0 arrays. Numbers of up- and downregulated genes in c-Kit+ and c-Kit− bone marrow cells from primary recipients of wild-type and Dnmt3b-knock-in MLL-AF9 leukemic cells were compared. (B) Gene set enrichment analysis of expression profiles of c-Kit+ wild-type and Dnmt3b-knock-in MLL-AF9 LSCs. Stemness genes and genes expressed in LICs were found in wild-type LSCs (left), whereas the expression profile of Dnmt3b-knock-in LSCs was enriched for genes expressed in mature hematopoietic cells or genes that are downregulated in LSCs and LICs (right). (C) Differentially expressed genes were filtered for those with reduced expression levels with DNA hypermethylation in both MLL-AF9 and Myc-Bcl2 leukemias at the same time. Relative expression levels of representative candidate genes were measured by real-time PCR analysis. Mean ± SD values are from 3 replicates normalized to expression levels of GAPDH. (D-E) Gene expression profiles of representative candidate genes in HSCs, granulocyte-macrophage progenitors, and mature granulocytes. Microarray data sets were analyzed using the Leukemia Gene Atlas (http://www. leukemia-gene-atlas.org). ***P < .001
Dnmt3b alters the expression profile of LSCs and leads to downregulation of stem cell genes. (A) Genome-wide mRNA expression profiles of c-Kit+MLL-AF9 LSCs and c-Kit−MLL-AF9 leukemic bulk cells were generated using Affymetrix Mouse Gene ST 2.0 arrays. Numbers of up- and downregulated genes in c-Kit+ and c-Kit− bone marrow cells from primary recipients of wild-type and Dnmt3b-knock-in MLL-AF9 leukemic cells were compared. (B) Gene set enrichment analysis of expression profiles of c-Kit+ wild-type and Dnmt3b-knock-in MLL-AF9 LSCs. Stemness genes and genes expressed in LICs were found in wild-type LSCs (left), whereas the expression profile of Dnmt3b-knock-in LSCs was enriched for genes expressed in mature hematopoietic cells or genes that are downregulated in LSCs and LICs (right). (C) Differentially expressed genes were filtered for those with reduced expression levels with DNA hypermethylation in both MLL-AF9 and Myc-Bcl2 leukemias at the same time. Relative expression levels of representative candidate genes were measured by real-time PCR analysis. Mean ± SD values are from 3 replicates normalized to expression levels of GAPDH. (D-E) Gene expression profiles of representative candidate genes in HSCs, granulocyte-macrophage progenitors, and mature granulocytes. Microarray data sets were analyzed using the Leukemia Gene Atlas (http://www. leukemia-gene-atlas.org). ***P < .001
We next investigated a potential connection between DNA hypermethylation, deregulated gene expression, and altered functional properties of Dnmt3b-knock-in LSCs. We focused on genes with extensive hypermethylation in both MLL-AF9 and Myc-Bcl2 leukemias and reduced expression levels in Dnmt3b-knock-in MLL-AF9 LSCs. We identified 41 candidate genes, which showed at least 20% hypermethylation in murine Dnmt3b-knock-in leukemias and an expression log ratio of −0.5 in Dnmt3b-knock-in MLL-AF9 LSCs. Expression changes for 19 of these genes were confirmed by real-time PCR, exemplarily depicted for 6 candidates (Figure 5C). Comparison of the obtained candidate genes with published expression data sets using the Leukemia Gene Atlas59 and Gene Expression Commons60 revealed high expression of 12 out of 19 genes in HSCs and decreasing expression levels in more differentiated cells (Figure 5D-E; supplemental Figure 8). The histone protein (H1F0) (Figure 5D) and the G-protein coupled receptor 56 (GPR56) (Figure 5E) exhibited the strongest expression differences. These data suggest that Dnmt3b-induced hypermethylation may have altered the gene expression profile of LSCs. Dnmt3b-knock-in MLL-AF9 LSCs may have lost the characteristic stem cell expression profile and possibly even their functional stem cell properties. Of note, differential gene expression did not correlate with DNA methylation changes in gene bodies (Spearman’s ρ = −0.073, P < .001) (supplemental Figure 9A) or promoter regions (Spearman’s ρ = −0.021, P = .280) (supplemental Figure 9B). Other studies have similarly observed no correlation between gene expression and promoter methylation in human disease.33,61 High gene body methylation has been associated with gene activation rather than transcriptional silencing.62,63 These data support the view that Dnmt3b affects nonpromoter methylation and that Dnmt3b-mediated enhanced gene body methylation is, at least in AML, not necessarily associated with activation of gene expression.
Deregulated Dnmt3b targets have prognostic impact in AML patients
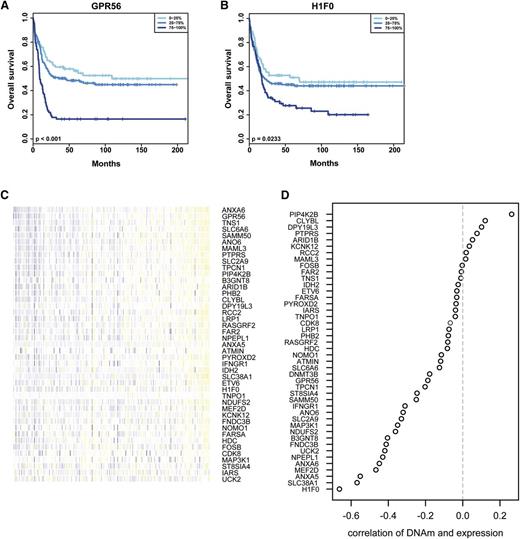
Candidate genes that showed hypermethylation and reduced gene expression in Dnmt3b-knock-in leukemias were analyzed for a potential association with prognosis in AML patients.59 Overall, 5 of 19 candidate genes were associated with better overall survival when expressed at low levels in AML patients (Figure 6A-B; supplemental Figure 10A-C).64 The methylation status of candidate genes with putative functional importance was determined in 194 AML patient samples available from the TCGA database (http://cancergenome.nih.gov/). All of the candidate genes identified in the Dnmt3b-knock-in mouse model showed low methylation levels across the analyzed patient cohort (Figure 6C). For 34 of 41 candidates, gene expression negatively correlated with DNA methylation levels (Figure 6D). These data indicate that the identified target genes that are deregulated by Dnmt3b-mediated DNA methylation in mice may also be regulated by DNA methylation in human AML.
Analysis of Dnmt3b-regulated genes in human AML data sets. (A-B) Candidate genes were analyzed for their prognostic impact in human leukemia using published patient data and the Leukemia Gene Atlas. Gene expression data were grouped into quartiles, and survival outcome was analyzed in a patient data set published in 2009 by Verhaak et al.64 Shown is overall survival in AML patients for low (0% to 25%), intermediate (25% to 75%), and high (75% to 100%) expression levels of the candidate genes GPR56 (P < .001) (A) and H1F0 (P = .023) (B). (C) DNA methylation (DNAm) levels of candidate genes were analyzed in 194 AML patients using Illumina bead array data available from the TCGA database. Each column represents 1 patient, and methylation levels are color coded with blue representing low methylation and yellow representing high methylation. (D) Correlation between DNA methylation levels and gene expression for the identified target genes from AML patients in the TCGA database. Depicted is the correlation coefficient for all candidate genes. Overall, 34 genes showed significant negative correlation between methylation and expression.
Analysis of Dnmt3b-regulated genes in human AML data sets. (A-B) Candidate genes were analyzed for their prognostic impact in human leukemia using published patient data and the Leukemia Gene Atlas. Gene expression data were grouped into quartiles, and survival outcome was analyzed in a patient data set published in 2009 by Verhaak et al.64 Shown is overall survival in AML patients for low (0% to 25%), intermediate (25% to 75%), and high (75% to 100%) expression levels of the candidate genes GPR56 (P < .001) (A) and H1F0 (P = .023) (B). (C) DNA methylation (DNAm) levels of candidate genes were analyzed in 194 AML patients using Illumina bead array data available from the TCGA database. Each column represents 1 patient, and methylation levels are color coded with blue representing low methylation and yellow representing high methylation. (D) Correlation between DNA methylation levels and gene expression for the identified target genes from AML patients in the TCGA database. Depicted is the correlation coefficient for all candidate genes. Overall, 34 genes showed significant negative correlation between methylation and expression.
Discussion
DNA methylation patterns are altered during leukemogenesis. But the mechanistic relevance of altered DNA methylation for leukemogenesis has remained unclear. In the current study, we demonstrate that increased activity of de novo DNA methyltransferase Dnmt3b inhibits leukemogenesis. Myc-Bcl2–induced leukemias with high levels of DNA methylation were affected as well as MLL-AF9–induced leukemias with much lower levels of altered DNA methylation.
DNA methylation plays an important role in HSC self-renewal as well as in differentiation and lineage commitment.10,13,14 Multiple tumor suppressor genes have been identified to be epigenetically silenced presumably by DNA methylation in AML.65-67 Oncogenes such as AML1-ETO were described to directly recruit DNMT activity to their target genes.68 On the other hand, PML-RARα by itself did induce a differentiation block in promyelocytes without causing increased DNA methylation.49 The most common mutations in the DNA methylation machinery affect DNMT3A and appear to induce DNA hypomethylation. Loss of Dnmt3a either alone or in combination with Dnmt3b induces stem cell expansion as an important prerequisite for leukemogenesis.14 DNMT3A mutations are also the most common finding in clonal hematopoiesis in older adults.69 These seemingly contradictory findings prompted us to evaluate the functional consequences of high de novo Dnmt activity in leukemogenesis. We chose Dnmt3b, because its expression is highest in HSCs.12 High DNMT3B expression in leukemic bulk cells was associated with poor prognosis.37
The inducible Dnmt3b model has previously been used to analyze colon adenoma formation. Forced Dnmt3b expression in colon was not toxic but increased the number of colon adenomas.29 In our analyses, we did not observe toxic effects in healthy hematopoiesis in vivo or in vitro.
Dnmt3b was previously identified as a haploinsufficient tumor suppressor in Eμ-myc mice.32 Loss of Dnmt3b led to the upregulation of the tumor modifier Ment and accelerated mouse lymphomagenesis. In our analyses, leukemias with high Dnmt3b levels showed increased disease latency in serial transplantations. Accordingly, Dnmt3b might exhibit tumor suppressor functions in myeloid disease.
DNA methylation is essential for lymphoid development.10 The Myc-Bcl2 leukemia model can induce lymphoid as well as myeloid leukemias.40 Upon forced Dnmt3b expression, we observed a shift toward myeloid leukemia with stronger suppression of lymphoid leukemias. These findings may reemphasize the notion that the myeloid lineage as the phylogenetically older system is the default pathway. DNA methylation changes might preferentially induce myeloid leukemia. Lymphoid leukemogenesis was also inhibited by global DNA hypomethylation (eg, by loss of Dnmt1 [hypomorph]), whereas myeloid leukemias could still be formed.10 In line with our new findings, these data suggest that lymphoid leukemias depend on a tightly controlled DNA methylation machinery whereas myeloid leukemias are more permissive for global changes in DNA methylation levels.
Leukemogenesis depends on LSC activity. In MLL-AF9 leukemia, the population of c-Kit+ cells is highly enriched for LICs.39,47 Forced Dnmt3b expression in the present study increased numbers of c-Kit+ leukemic cells but reduced colony formation potential. In accordance, transplantation of c-Kithigh cells resulted in delayed leukemia development in recipients of Dnmt3b-knock-in cells. Still, secondary and tertiary recipients of Dnmt3b-expressing c-Kit+ LSCs developed a leukemia-like disease, indicating that LSCs were still present in the population of c-Kit–expressing cells, yet they induced disease with increased latency. Colony formation potential was not impaired in leukemic cells from spleen. Accordingly, DNA methylation might be most relevant at the stem cell level.
Increased numbers of c-Kit+ LSCs were found in recipients of Dnmt3b-knock-in cells, although leukemia development was delayed. In nonleukemic Dnmt3b-knock-in mice, Dnmt3b overexpression led to a block at the multipotent progenitor (LMPP) stage. Multilineage repopulation potential of Dnmt3b-knock-in cells remained unaltered in competitive transplantation assays. In line with previous findings, higher Dnmt3b levels were not sufficient to ultimately drive differentiation of healthy HSCs. But altered Dnmt3b levels had a crucial effect on LSCs in AML. Nonetheless, an involvement of Dnmt3a cannot be completely excluded, because high levels of Dnmt3b might stabilize protein complexes formed by Dnmt3b, Dnmt3a, and Dnmt3L.70,71
Studies concerning DNA methylation patterns in human AML have found that a small common set of genes exhibited consistent aberrant methylation across several hundred cases.19,20 Dnmt3b-knock-in leukemias showed increased levels of DNA methylation, as analyzed by RRBS. Dnmt3b-knock-in Myc-Bcl2 leukemia samples showed a much higher number of hypermethylated CpGs than the respective MLL-AF9 leukemic samples. This is in line with the finding that MLL-rearranged leukemias in AML patients are characterized by DNA hypomethylation when compared with IDH-mutated leukemias.20 The extent of hypermethylation induced by forced Dnmt3b expression possibly depends on the basal DNA methylation level that is induced by the specific oncogene. Less pronounced DNA hypermethylation in MLL-AF9 leukemias further suggests that these may be more susceptible to methylation-associated delay in leukemogenesis. Detailed analysis of MLL-AF9 patient samples with high and low DNMT3B expression would improve our understanding of the role DNMT3B-mediated DNA methylation in human leukemogenesis. Of note, the majority of DMCs in Dnmt3b-knock-in leukemias mapped to gene bodies. This preferential targeting of Dnmt3b-mediated DNA methylation to nonpromoter CpGs has been described (eg, in ES cells),50,72 but the role of gene body methylation in leukemia has not been investigated so far. An inverse relationship between transcription factor binding and DNA methylation was found in several studies involving diverse human cell and tissue types73 and acute promyelocytic leukemia.49 Protection of actively transcribed genes from de novo methylation has been attributed to loss of nucleosomes at sites of transcription.70
Apart from unique DNA methylation patterns in different leukemia subtypes, both, Myc-Bcl2 and MLL-AF9 leukemias shared a significant set of commonly hypermethylated CpGs. These CpGs were possibly instrumental for the consistently observed effect of delayed leukemogenesis. Although overexpression of Dnmt3b induced thousands of DNA methylation alterations, only few changes in gene expression were observed. We and others have shown that promoter and gene body methylation does not necessarily correlate with altered gene expression.33,61 Low numbers of differentially expressed genes in Dnmt3b-knock-in leukemias could also be because of heavy underrepresentation of transcription factor binding sites in DMRs. In addition, predicted upstream regulators can affect the expression of target genes independently of DNA methylation levels. An essential role in initiation and maintenance of T-ALL was recently demonstrated for the H3K27-demethylase Kdm6b.74
Hypermethylated LSCs showed reduced expression of genes related to cancer and proliferation, which is in accordance with increased disease latency of Dnmt3b-knock-in leukemias. Furthermore, hypermethylated LSCs were characterized by loss of a stemness-associated gene expression signature. Of note, high expression of LSC- and HSC-related genes in leukemic blast cells was associated with worse survival in AML patients.75,76 In our analyses, genes with DNA hypermethylation and reduced gene expression marked several candidate genes that were associated with poor outcome upon high expression in AML patients. Thus, suppression of selected stem cell genes by DNMT3B could be a protective mechanism that prevents LSC functions. The correlation between overall DNMT3B expression in leukemic bulk cells with poor outcome might reflect the stem cell phenotype of the leukemia. Studies in AML patients have only addressed the effect of DNMT3B expression in unsorted leukemic bulk cells from various leukemia subtypes on patient survival.37,38 Here, we show for the first time the role of high Dnmt3b expression in murine LSCs and a potential relevance of DNMT3B-target genes in AML patients. Additional studies in patients with different AML subtypes would help to further clarify the function of DNMT3B in LSCs.
Taken together, increased activity of the de novo DNA methyltransferase Dnmt3b impairs leukemia development. Protection from DNA methylation and/or specific loss of DNA methylation at regions that are critical for LSC formation may constitute an important step in leukemogenesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from Deutsche Forschungsgemeinschaft (MU 1328/15-1 and MU 1328/9-2). W.E.B.’s laboratory is supported by Deutsche Forschungsgemeinschaft (EXC1003, Cells in Motion).
Authorship
Contribution: I.S. and C.M.-T. designed the research and wrote the manuscript; I.S., M.S.-W., N.B., A.K., P.R., H.G., F.H., and P.T. performed experiments; C.R. performed all bioinformatics analyses on single CpG level; K.H. performed BiSeq analysis; Q.L. and W.W. performed bioinformatics analyses of TCGA data; H.L. and L.A.G. provided mice; and all authors contributed to data interpretation and the manuscript writing process.
Conflict of interest disclosure: The authors declare no competing financial interests.
Correspondence: Carsten Müller-Tidow, Department of Hematology and Oncology, University of Halle, Ernst-Grube-Strasse 40, 06108 Halle, Germany; e-mail: carsten.mueller-tidow@uk-halle.de.

![Figure 3. Dnmt3b expression increases the number of multipotent progenitor cells in nonleukemic mice. (A) Induction of Dnmt3b expression in nonleukemic Dnmt3b-knock-in mice increased the numbers of multipotent progenitor cells after 4 weeks (n = 3 per group). LT-HSC gated for LSK CD34−Flt3−; ST-HSC gated for LSK CD34+Flt3−; LMPP gated for LSK CD34+Flt3+. (B) White blood cell counts of wild-type (black circles) and Dnmt3b-knock-in (red circles) mice after 16, 28, and 44 weeks of in vivo induction of Dnmt3b expression. (C) Total cell counts for lymphocytes in peripheral blood of wild-type and Dnmt3b-knock-in mice at the indicated time points after induction of Dnmt3b. Each dot represents 1 individual mouse. (D) Survival of wild-type and Dnmt3b-knock-in mice after induction of Dnmt3b expression for up to 1 year (n = 6 for wild-type [WT] and n = 8 for Dnmt3b-knock-in mice). (E) Percentage of CD45.2+ donor-derived peripheral blood cells in primary recipients for up to 24 weeks posttransplantation. Transplanted cell doses are 106 and 105 cells mixed with 105 competitor cells (n = 4 mice each). (F) Dnmt3b-knock-in total bone marrow cells were impaired in growth upon culture in the presence of IL-3, IL-6, and murine stem cell factor; n = 3 per group. Data in panels A-C and E-F are mean ± SD values. *P < .05; **P < .01; ***P < .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/12/10.1182_blood-2015-07-655928/5/m_1575f3.jpeg?Expires=1770077497&Signature=B9OvtL~xfGRTFGc4MaqQoO9aGZaQBw3nOAa~TO6P5buJk2ZUI5AQUWygxx9rKKgFa2DTRSkd5KpV6~UtRUiEBKMcWjKIPuRJEXdXj1gLu3OcFW9kh6SfFO4aZfosDq8-5DkBEe3u5NNwbMK3O2xC-fdOoaN0FpcUDA-r1vObsytmIfyPNINGx3~86~cKU9R9zT4xN0OxDcimww9Br2pJBscENurhPg~qFLv9BLGQnipxHfaNM1dVWPhM4a8skf6C4Ofqo-sLMJbSuqiriPQ1bzsxOjpbOEKHPlPVhY2vgY~DIw~9cjZc2O2OFE58Vfp6~KROmW2RvNsJgcjOdqnoFg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)