Key Points
HERC4 is the first identified ubiquitin ligase that mediates c-Maf ubiquitination and degradation.
HERC4 suppresses MM cell proliferation and delays MM tumor growth.
Abstract
The transcription factor c-Maf is extensively involved in the pathophysiology of multiple myeloma (MM), a fatal malignancy of plasma cells. In the present study, affinity chromatography and mass spectrometry were used to identify c-Maf ubiquitination–associated proteins, from which the E3 ligase HERC4 was found to interact with c-Maf and catalyzed its polyubiquitination and subsequent proteasome-mediated degradation. HERC4 mediated polyubiquitination at K85 and K297 in c-Maf, and this polyubiquitination could be prevented by the isopeptidase USP5. Further analysis on the NCI-60 cell line collection revealed that RPMI 8226, a MM-derived cell line, expressed the lowest level of HERC4. Primary bone marrow analysis revealed HERC4 expression was high in normal bone marrow, but was steadily decreased during myelomagenesis. These findings suggested HERC4 played an important role in MM progression. Moreover, ectopic HERC4 expression decreased MM proliferation in vitro, and delayed xenograft tumor growth in vivo. Therefore, modulation of c-Maf ubiquitination by targeting HERC4 may represent a new therapeutic modality for MM.
Introduction
The Maf family proteins belong to the basic leucine zipper transcription factors, including c-Maf, MafA, MafB, MafF, MafG, MafK, and Nrl,1,2 of which c-Maf, MafA, and MafB belong to the large Maf subgroup containing the complete structure of a transcription factor, including a basic DNA-binding domain, a leucine zipper structure, and an activation domain.2 In the fetus, c-Maf is involved in the development of several organs and tissues, including islets.1 In the adult, c-Maf is associated with the differentiation of helper T cells, including Th2, and Th17.1,3 Overexpression of c-Maf, MafA, and MafB are frequently found in multiple myeloma (MM), a fatal hematologic disorder of plasma cells, and mantle cell lymphoma.4,5 c-Maf transgenic mice develop MM-like symptoms including hyperglobulinemia and associated kidney damage at 60 to 80 weeks.6 As a transcription factor, c-Maf modulates the expression of several key genes, including cyclin D2, a driver of cell-cycle progression and MM cell proliferation. Consistent with these findings, c-Maf knockdown by RNA interference leads to MM cell apoptosis.7
Previous studies demonstrated that c-Maf protein stability is modulated by the ubiquitin-proteasome system.8 c-Maf can be degraded in the proteasome after polyubiquitination,4,9 which is mediated by 3 types of sequential enzymes, including ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3). In humans there are 2 E1s, 35 known E2s, and ∼300 to 600 E3s; these enzymes coordinately modulate ubiquitination of a vast diversity of substrate proteins. In addition, attached ubiquitin molecules can be removed by ubiquitin-specific proteases, also known as deubiquitinases or Dubs. However, the enzymes mediating c-Maf ubiquitination and deubiquitination are unknown. In the present study, by using an affinity purification coupled with tandem mass spectrometry (AP-MS), HERC4 was identified as an E3 ubiquitin ligase mediating c-Maf ubiquitination in MM and inhibiting MM cell proliferation.
Materials and methods
Antibodies and plasmids
Rabbit anti-HERC4 antibody was obtained from Bethyl Laboratories, Inc. (Montgomery, TX). Rabbit anti–c-Maf and anti–cyclin D2 antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Mouse anti-HA, anti-Myc, anti-Flag, and anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies were obtained from MBL (Japan). An anti-USP5 antibody was purchased from Proteintech (Chicago, IL). c-Maf was cloned from myeloma cell line LP1, the MafB plasmid was purchased from Open Biosystems (Thermo Fisher), and MafA was cloned from HeLa genomic DNA as described previously.10 All these genes along with ubiquitin were subcloned into a pcDNA3.1 vector carrying an HA, Flag, or myc tag. The K to R c-Maf variants were generated by site-directed mutagenesis as reported previously.8 MG132 was purchased from Sigma-Aldrich.
Primary bone marrow cells
Primary bone marrow species were collected from healthy donors and MM patients who consented to donate a research sample from the First Affiliated Hospital of Soochow University. Mononuclear cells were isolated from the samples by Ficoll density centrifugation as described previously.11 The collection and use of human bone marrow for this study were approved by the institutional review board of Soochow University in accordance with the Declaration of Helsinki.
Affinity purification-coupled mass spectrometry
HEK293 cells were transfected with human c-Maf plasmid p-HA-c-Maf by using linear 25-Kd polyethyleimine (Sigma-Aldrich)8 ; 24 hours later, cells were treated with 20 µM of MG132 for 2 hours and then solubilized in a lysis buffer containing BenzoNuclease (Novus).12 After clarification by centrifugation and protein concentration determination, 10 mg of whole-cell lysates from each treatment were preincubated with protein A/G beads for 1 hour at 4°C, followed by incubation with anti–HA-conjugated agarose beads overnight at 4°C. After a gentle washing, proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), followed by EZ Blue Gel Staining (Sigma-Aldrich). The protein bands from the gel were excised and processed for mass spectrometry (MS) analysis as described previously.12 Tandem mass spectra were extracted by BioWorks version 3.3 followed by analyses using SEQUEST (Thermo Finnigan) and X! Tandem as described previously.12 After searching the human database (version 3.18), Scaffold software (Proteome Software Inc., Portland, OR; version 3) was used to evaluate MS/MS-based peptides and protein identifications. Proteins were included for analysis when identified by at least 2 unique peptides identified with a confidence interval >95%.
Luciferase-based screen
The cyclin D2 promoter containing a c-Maf recognition element was cloned into a luciferase report system (pGL4, Promega) as described previously.9 This reporter and c-Maf were cotransfected and stably expressed in NIH3T3 cells. These cells were then transfected with individual E3 ligase plasmids from a subset collection of E3 ligases (the SPARC BioCentre, Toronto, ON, Canada). Luciferase activity was analyzed with the Bright-Glo substrate (Promega) as described previously.8,9
Immunoblotting assay
Immunoblotting (IB) assay was performed against specific antibodies as described previously.11
Cycloheximide chase assay
After transfection with plasmids of interest for 24 hours, HEK293 cells were treated with cycloheximide (CHX) (100 μg/mL, Sigma-Aldrich) for 0 to 12 hours. Cell lysates were then prepared by using a lysis buffer containing 2% SDS, followed SDS-PAGE and IB analyses with specific antibodies.8
Confocal microscopy
NIH3T3 cells were transfected with HERC4 and c-MAF plasmids in pcDNA3.1 alone or together for 48 hours as described previously.8 Cells were then cultured on sterile glass coverslips in 6-well plates to 80% confluence in Dulbecco’s modified Eagle medium and then stained with an anti–c-Maf antibody and visualized on an Olympus FV1000MPE-share Multiphoton Laser Scanning Microscope.13
Interference analysis with shRNA of HERC4
HERC4 shRNAs (shHERC4) were obtained from Genechem Co. Ltd (Shanghai, China) and evaluated after transfection into HERC4-expressing HEK293 cells. The most effective shHERC4 was chosen to transfect HEK293 cells along with HERC4 and c-Maf. Forty-eight hours later, cells were prepared for IB assay.
c-Maf ubiquitination in vitro
This study was performed as described previously.14,15 Briefly, HA-c-Maf and Flag-HERC4 were separately introduced into HEK293 cells. Forty-eight hours later, c-Maf and HERC4 were purified with specific antibodies against HA and Flag, respectively. These proteins were then added to the reaction mixture containing adenosine triphosphate (ATP), HA-Ub, with or without E1 and E2 (Boston Biochem, Boston, MA). The reaction was then terminated and subjected to immunoprecipitation (IP) with Flag antibody and subsequent IB assay to measure co-IP of c-Maf.
GEO dataset analyses
A DNA microarray dataset generated from healthy donors and MM patients with monoclonal gammopathy of undetermined significance (MGUS), smoldering MM (SMM), and MM,16 and a DNA microarray dataset from NCI 60 cell lines17 were retrieved from Gene Expression Omnibus (GEO) databases (http://www.ncbi.nlm.nih.gov/gds). These datasets were used to analyze the expression of HERC4, followed by verification by IB assay with lysates from a panel of MM and solid tumor cell lines. Statistical difference between the control and each patient group was analyzed by Student t test.
Reverse-transcription polymerase chain reaction
Total RNA was extracted using Trizol (Transgene, Beijing, China). RNA (2.5 μg) was reverse-transcribed using an EasyScipt First-strand cDNA Synthesis SuperMix (Transgene) according to the manufacturer’s instruction. Polymerase chain reaction (PCR) amplification was carried out using the following primers: for HERC4, 5′-GAGATAGCCAATGAGATAGATG-3′ (Forward) and 5′-TTGAGGAATAGTGGAGGT-3′ (Reverse); for GAPDH, 5′-AATCCCATCACCATCTTCC-3′ (Forward) and 5′-CATCACGCCACAGTTTCC-3′ (Reverse). The PCR products were visualized by Goldview staining, following electrophoresis on 1.5% agarose gels. The optical densities of the bands were quantified using an image analysis system and ImageJ software (National Institutes of Health, Bethesda, MD). For real-time PCR, Reactions were processed and analyzed on an ABI 7700 Sequence Detection System (Applied Biosystems). Relative mRNA expression was determined using the ΔΔCT model as described previously.4
HERC4 lentivirus construction
HERC4 cDNA was generated by using the following primers: 5′-CCGCTCGAGATGTTGTGCTGGGGAAA-3′ (Forward) and 5′-GCCGGAATTCTTATATTAAACTGAAGCC-3′ (Reverse), and inserted into pLVX-AcGFP lentiviral vector (Clontech) within the XhoI and EcoRI sites. To generate lentiviral particles, HEK293T cells at 80% confluence were transfected with 10 μg of pLVX-AcGFP-HERC4, 3.5 μg of VSV-G envelope glycoprotein, 2.5 μg of packaging proteins (Rev), and 6.5 μg of packaging proteins (ΔR8.74) using Lipofectamine 2000 (Invitrogen). Cells were washed and refreshed with Dulbecco’s modified Eagle medium 12 hours later. The lentiviral particle-enriched supernatant was harvested 48 hours later, filtered, and stored frozen at −80°C. The titration of viral particles was determined by flow cytometry. The pLVX-AcGFP lentiviral particle was used as a mock control in the infection assay.
MM xenografts
RPMI 8226 cells infected with HERC4-carrying mock lentivirus were sorted by flow cytometry and suspended in 100 μL of phosphate-buffered saline and inoculated subcutaneously into the flanks of SCID mice (n = 6 for each group, 5-6 weeks old) from Shanghai Slac Laboratory Animal Co. Ltd (Shanghai, China). The tumor size was measured every other day for 14 days and the tumor sizes were calculated as described previously.13 On the 14th day after inoculation, all mice were sacrificed and tumor tissues were removed, weighed, and stored frozen. Proteins from the tumor tissues were subjected to IB analyses for the expression levels of c-Maf, HERC4, and cyclin D2. This experiment was approved by the Review Board for Animal Welfare and Ethics of Soochow University.
Results
HERC4 interacts with c-Maf
To find specific enzymes mediating c-Maf ubiquitination, an AP-MS strategy was performed. After co-IP with HA-conjugated agarose beads, c-Maf immune complexes were resolved by SDS-PAGE and visualized by using Coomassie brilliant blue stain. As shown in Figure 1A, there were several proteins specifically isolated from the HA-c-Maf–expressing cells compared with the background of proteins recovered with the anti-HA control from the cells not expressing HA-c-Maf. These protein bands were isolated, digested, purified, and subjected to liquid chromatography-MS/MS. One of these bands (as indicated in Figure 1A) was identified as c-Maf by MS, confirming its expression and recovery by the anti-HA IP. The MS analysis revealed a group of ubiquitination-associated enzymes recovered in the c-Maf immune complex, including the ubiquitin ligases (RNF114, HERC4, UBR5, and HUWE1) and the Dub USP5 (Figure 1A). Of these E3 ligases, RNF114 has been well studied, and HUWE1 and UBR5 were difficult to clone owing to their big sizes with molecular weights of 350 and 480 kD, respectively. In contrast, HERC4 was less reported and its substrate was not reported and thus was chosen for further studies. The unique peptides of HERC4 identified by MS are highlighted in Figure 1B. To verify this finding of an apparent HERC4–c-Maf interaction, their respective expression plasmids were cotransfected into HEK293 cells followed by IP with a HERC4-specific antibody. As shown in Figure 1C, ectopically expressed HA-c-Maf was identified by co-IP with an anti-HERC4 antibody (anti-Flag). Moreover, when the MM cell line MM1.S expressing constitutive c-Maf was infected with a HERC4 lentivirus and the cell lysates subjected to IP with a c-Maf antibody, subsequent IB assays confirmed the association of HERC4 with c-Maf (Figure 1D). This reciprocal IP/IB analysis thus demonstrated that HERC4 interacts with c-Maf. To further characterize this interaction, NIH3T3 cells were transfected with c-Maf and HERC4 alone or together. Immunofluorescent confocal microscopic analysis confirmed that the c-Maf transcription factor was localized to nuclei, whereas HERC4 was mainly found in the cytoplasm (Figure 1E). Interestingly, when the 2 genes were cotransfected, HERC4 became localized mainly in nuclei (Figure 1E). These results collectively showed that the HERC4 and c-Maf proteins interact, and that HERC4 becomes conditionally localized to the nucleus when coexpressed with c-Maf.
HERC4 interacts with the c-Maf protein. (A) Affinity-purification assay was performed using an anti-HA–specific antibody. c-Maf, HERC4, and USP5 were indicated by the arrows. (B) The HERC4-unique peptides identified by MS/MS are highlighted in yellow. (C) HEK293 cells were transiently transfected with HA-c-Maf and or HERC4 for 48 hours. Cell lysates were subjected to IP with a HERC4 antibody and subsequent IB with c-Maf or HERC4 antibodies. (D) MM1.S cells were infected with HERC4 lentivirus. Ninety-six hours later, cell lysates were prepared for co-IP with a c-Maf–specific antibody, followed by IB with a HERC4-specific antibody. (E) NIH3T3 cells were transfected with c-Maf, HERC4, or both plasmids; 48 hours later, cells were subjected to immunofluorescent staining and confocal analysis.
HERC4 interacts with the c-Maf protein. (A) Affinity-purification assay was performed using an anti-HA–specific antibody. c-Maf, HERC4, and USP5 were indicated by the arrows. (B) The HERC4-unique peptides identified by MS/MS are highlighted in yellow. (C) HEK293 cells were transiently transfected with HA-c-Maf and or HERC4 for 48 hours. Cell lysates were subjected to IP with a HERC4 antibody and subsequent IB with c-Maf or HERC4 antibodies. (D) MM1.S cells were infected with HERC4 lentivirus. Ninety-six hours later, cell lysates were prepared for co-IP with a c-Maf–specific antibody, followed by IB with a HERC4-specific antibody. (E) NIH3T3 cells were transfected with c-Maf, HERC4, or both plasmids; 48 hours later, cells were subjected to immunofluorescent staining and confocal analysis.
HERC4 is required for c-Maf ubiquitination
Because HERC4 is a ubiquitin ligase, we wondered whether it mediates c-Maf ubiquitination. HA-c-Maf and Flag-HERC4 plasmids were cotransfected into HEK293 cells followed by IP/IB analysis. As shown in Figure 2A, polyubiquitinated c-Maf was identified in the presence of HERC4. To find out whether HERC4 was able to induce ubiquitination of endogenous c-Maf, HERC4-lentivirus was introduced into MM1.S cells. Subsequent IP/IB assays identified a multiply ubiquitinated species of c-Maf (Figure 2B). To view the ubiquitination of c-Maf in vitro, purified c-Maf and HERC4 were added to the ubiquitination reaction mixture containing Ub, E1, E2, and ATP. As shown in Figure 2C, polyubiquitinated c-Maf was generated in the complete reaction system, but not when the E1/E2 components were omitted.14,15 This result further suggested that c-Maf was polyubiquitinated under the direction of HERC4.
HERC4 is required for c-Maf ubiquitination. (A) HEK293 cells were transfected with an HA-c-Maf with or without a Flag-HERC4 plasmid. Forty-eight hours later, cells were treated with MG132 for 2 hours, followed by lysate preparation and co-IP with an anti-Ub–specific antibody and IB with indicated antibodies. (B) MM1.S cells were infected with HERC4 lentivirus. Ninety-six hours later, cell lysates were prepared for IP with an anti–c-Maf antibody followed by IB against indicated antibodies. (C) HA-c-Maf and Flag-HERC4 proteins were separately isolated and purified from overexpressing HEK293 cells with an anti-HA and Anti-Flag antibody, respectively. The purified proteins were then incubated in a 1.5-mL tube containing ATP and HA-Ub with or without E1-E2 mixture. Two hours later, the reaction was terminated and subjected to IP with an anti-Flag antibody followed by an anti–c-Maf antibody. (D) HEK293 cells were transfected with c-Maf, HERC4, wild-type (WT) or K48R or K63R mutated ubiquitin (Ub) plasmids. Twenty-four hours later, cells lysates were subjected to IP with an anti-HA antibody followed by IB with an anti-Flag antibody. (E) HEK293 cells were infected with individual HERC4 shRNAs; 48 hours later, cell lysates were prepared for evaluation of HERC4 protein levels by IB. (F) c-Maf, HERC4, and or HERC4 shRNA (#1) were introduced into HEK293 cells; 48 hours later, cell lysates were prepared for IP (with a Ub antibody) and IB against c-Maf. Cells were treated with MG132 before cell lysis.
HERC4 is required for c-Maf ubiquitination. (A) HEK293 cells were transfected with an HA-c-Maf with or without a Flag-HERC4 plasmid. Forty-eight hours later, cells were treated with MG132 for 2 hours, followed by lysate preparation and co-IP with an anti-Ub–specific antibody and IB with indicated antibodies. (B) MM1.S cells were infected with HERC4 lentivirus. Ninety-six hours later, cell lysates were prepared for IP with an anti–c-Maf antibody followed by IB against indicated antibodies. (C) HA-c-Maf and Flag-HERC4 proteins were separately isolated and purified from overexpressing HEK293 cells with an anti-HA and Anti-Flag antibody, respectively. The purified proteins were then incubated in a 1.5-mL tube containing ATP and HA-Ub with or without E1-E2 mixture. Two hours later, the reaction was terminated and subjected to IP with an anti-Flag antibody followed by an anti–c-Maf antibody. (D) HEK293 cells were transfected with c-Maf, HERC4, wild-type (WT) or K48R or K63R mutated ubiquitin (Ub) plasmids. Twenty-four hours later, cells lysates were subjected to IP with an anti-HA antibody followed by IB with an anti-Flag antibody. (E) HEK293 cells were infected with individual HERC4 shRNAs; 48 hours later, cell lysates were prepared for evaluation of HERC4 protein levels by IB. (F) c-Maf, HERC4, and or HERC4 shRNA (#1) were introduced into HEK293 cells; 48 hours later, cell lysates were prepared for IP (with a Ub antibody) and IB against c-Maf. Cells were treated with MG132 before cell lysis.
There are 2 common types of polyubiquitination, which are distinguished by the lysine residue through which branching occurs: K48-chain and K63-chain. To determine which type(s) of polyubiquitination in c-Maf is mediated by HERC4, plasmids containing c-Maf, HERC4, and wild-type (WT) or K48R, K63R ubiquitin constructs were transfected into HEK293 cells. Immunoblotting revealed that both WT and K63R led to polyubiquitinated c-Maf, but polyubiquitination was impaired in the presence of K48R ubiquitin (Figure 2D), suggesting that HERC4-mediated c-Maf polyubiquitination is mainly branched through K48.
The aforementioned studies demonstrated that HERC4 was able to mediate c-Maf polyubiquitination. To find whether c-Maf could be ubiquitinated when HERC4 protein expression was knocked down, HERC4 was targeted by a specific shRNA that was introduced into cells after lentivirus infection. As shown in Figure 2E, 3 of 4 HERC4 shRNAs effectively abolished HERC4 expression, one of which (#1) was used to study HERC4-mediated c-Maf ubiquitination. As shown in Figure 2F, HERC4 led to polyubiquitinated c-Maf, which was abolished by shHERC4. These results verified HERC4 as a responsible E3 ligase for cellular c-Maf polyubiquitination.
HERC4 mediates c-Maf degradation
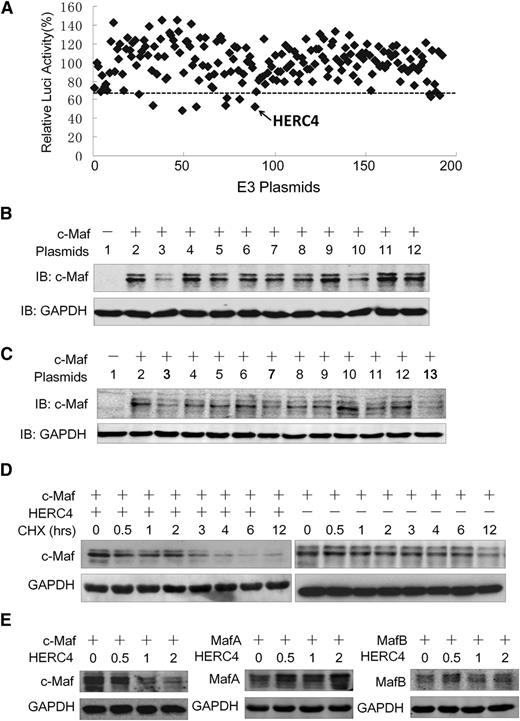
The aforementioned studies demonstrated that HERC4 is an E3 ligase for c-Maf ubiquitination. To determine whether there are other E3s that mediate c-Maf degradation, we established a functional screen in which an E3 array was transfected into NIH3T3 cells expressing c-Maf and a luciferase reporter system driven by a cyclin D2 promoter containing a c-Maf recognition element. Because c-Maf modulates cyclin D2 transactivation, thus increasing the expression of cyclin D2 promoter-driving luciferase, an E3 ligase that decreased c-Maf stability would decrease luciferase activity driven by the cyclin D2 promoter. With an arbitrary cutoff line (65%) (Figure 3A), E3 ligases of 180 were selected at the first screen. These 9 E3 ligases were further evaluated in transfected NIH3T3 cells expressing c-Maf. As shown in Figure 3B, HERC4 was the only E3 that decreased c-Maf at the protein level. Because HERC4 is a HECT domain–containing E3 ligase, we wondered whether other HECT-type E3 ligases were capable of mediating c-Maf degradation. Representative HECT-type E3s tested included HACE1, UREB1, FLJ21156, HERC4, ITCH, and WWP2. Three RING-family E3s including TRIM8, RNF111, ZNF313 were also tested. As shown in Figure 3C, c-Maf protein was markedly decreased by HERC4 (including 3 independently sourced expression plasmid/cDNAs). The RING family E3s and the HECT family E3s other than HERC4 had little or no effect on c-Maf proteins (Figure 3C). Therefore, these results further demonstrated that HERC4 specifically mediates c-Maf degradation. To bolster this conclusion, c-Maf was transfected with or without HERC4, followed by a CHX pulse-chase assay during which new protein synthesis was inhibited by CHX. As shown in Figure 3D, the presence of HERC4 significantly accelerated c-Maf degradation and shortened the half-life of c-Maf. In the presence of CHX, c-Maf protein appeared to decrease by roughly half after approximately 12 hours compared with ∼3 hours in the presence of HERC4 (Figure 3D).
HERC4 regulates c-Maf degradation. (A) NIH3T3 cells expressing c-Maf and the cyclin D2 promoter–driving luciferase were transfected with a selected subset of E3 ligase plasmids. Twenty-four hours later, cyclin D2 transactivation was assessed by the luciferase assay. (B) NIH3T3 cells were cotransfected with c-Maf and E3 candidates for 48 hours, and cell lysates were then prepared for IB to measure c-Maf protein level. 1, pcDNA3.1; 2, pcDNA3.1; 3, HERC4; 4, Itch; 5, CDKN1B; 6, COPS8; 7, CXCR4; 8, EDN1; 9, FBXL14; 10, WWP2; 11, HIF1A; 12, pcDNA 3.1. (C) NIH3T3 cells were transfected with HERC4 and several HECT-containing E3 plasmids for 48 hours before cell lysate preparation for c-Maf protein determination with IB. 1, pcDNA3.1; 2, pcDNA3.1; 3, HERC4; 4, HACE1; 5, FLJ21156; 6, UREB1; 7, DKFZP564G092 (HERC4); 8, ITCH; 9, WWP2; 10, ZNF313; 11.TRIM8; 12. RNF111; 13. HERC4-myc. (D) HEK293 cells were transfected with c-Maf, HERC4, or vector control. Forty-eight hours later, cells were treated with CHX (100 μg/mL) for 0, 0.5, 1, 2, 3, 4, 6, and 12 hours to inhibit de novo protein synthesis and were then harvested for IB. (E) HEK293 cells were transfected with MafA, MafB, or c-Maf along with increased HERC4. Forty-eight hours later, cells were harvested for IB to determine the Maf protein levels.
HERC4 regulates c-Maf degradation. (A) NIH3T3 cells expressing c-Maf and the cyclin D2 promoter–driving luciferase were transfected with a selected subset of E3 ligase plasmids. Twenty-four hours later, cyclin D2 transactivation was assessed by the luciferase assay. (B) NIH3T3 cells were cotransfected with c-Maf and E3 candidates for 48 hours, and cell lysates were then prepared for IB to measure c-Maf protein level. 1, pcDNA3.1; 2, pcDNA3.1; 3, HERC4; 4, Itch; 5, CDKN1B; 6, COPS8; 7, CXCR4; 8, EDN1; 9, FBXL14; 10, WWP2; 11, HIF1A; 12, pcDNA 3.1. (C) NIH3T3 cells were transfected with HERC4 and several HECT-containing E3 plasmids for 48 hours before cell lysate preparation for c-Maf protein determination with IB. 1, pcDNA3.1; 2, pcDNA3.1; 3, HERC4; 4, HACE1; 5, FLJ21156; 6, UREB1; 7, DKFZP564G092 (HERC4); 8, ITCH; 9, WWP2; 10, ZNF313; 11.TRIM8; 12. RNF111; 13. HERC4-myc. (D) HEK293 cells were transfected with c-Maf, HERC4, or vector control. Forty-eight hours later, cells were treated with CHX (100 μg/mL) for 0, 0.5, 1, 2, 3, 4, 6, and 12 hours to inhibit de novo protein synthesis and were then harvested for IB. (E) HEK293 cells were transfected with MafA, MafB, or c-Maf along with increased HERC4. Forty-eight hours later, cells were harvested for IB to determine the Maf protein levels.
c-Maf protein shares a high similarity in amino acid sequences with MafA and MafB, to find out whether HERC4 also modulates the stability of MafB and MafA, these 2 proteins were evaluated after HERC4 was overexpressed. As shown in Figure 3E, HERC4 decreased the level c-Maf protein, but not MafA or MafB. In contrast with its specific effect on c-Maf, HERC4 slightly increased the level MafA protein (Figure 3E). Therefore, HERC4 specifically decreased c-Maf but not MafA or MafB proteins.
HERC4 mediates c-Maf ubiquitination at K85 and K297
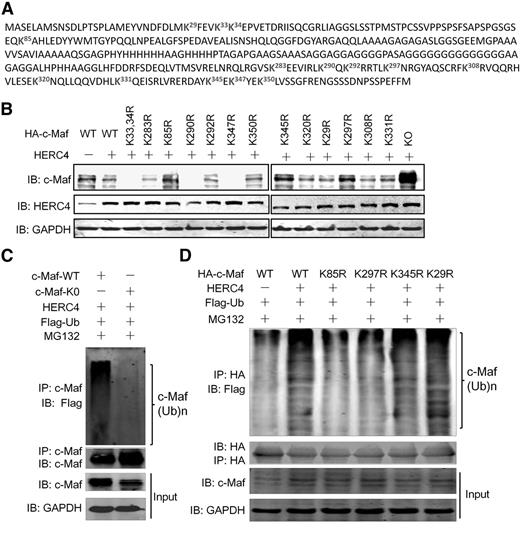
There are 14 lysine residues in the c-Maf protein. Our previous study found that c-Maf polyubiquitination is probably mediated by more than a single specific lysine residue, such as K85 and K350.8 To find out which lysine residue(s) mediated HERC4-induced c-Maf ubiquitination, we constructed 14 c-Maf mutants encoding specific K to R substitutions (Figure 4A). Wild-type and mutant (mt) c-Maf plasmids were cotransfected into HEK293 cells along with HERC4. As shown in Figure 4B, the protein level of WT c-Maf was decreased by HERC4, while the K0 c-Maf variant (with all K residues changed to R) was not affected by HERC4 because c-Maf was not ubiquitinated (Figure 4C). Compared with other K to R mutants, the protein levels of K85R, K297R, and K345R, c-Maf remained unchanged (Figure 4B) in the presence of HERC4, implying that these mutants did not undergo polyubiquitination mediated by HERC4. To confirm that the degradation of the K85R, K297R, and K345R mutants were caused by ubiquitination, individual K85R, K297R, K345R, and WT c-Maf plasmids were coexpressed with HERC4, followed by evaluation of c-Maf ubiquitination. As shown in Figure 4D, HERC4 mediated the polyubiquitination of WT and K29R c-Maf (a negative control), whereas polyubiquitination of K85R and K297R were markedly decreased to levels comparable with that seen with WT c-Maf in the absence of HERC4. However, the K345R displayed a level of HERC4-dependent polyubiquitination similar to WT c-Maf. Therefore, these results suggest HERC4 mediates c-Maf ubiquitination at K85 and K297.
K85 and K297 were required for HERC4-mediated c-Maf ubiquitination. (A) Lysine residues in c-Maf protein. Each K was changed to arginine (R) by site-direct mutagenesis. (B) Wild-type (WT) and mutant c-Maf plasmids were transfected into HEK293 cells with or without a HERC4 plasmid. Forty-eight hours later, cell lysates were subjected to an IB assay for c-Maf. (C) WT or all-lysine-substituted (K0) c-Maf plasmids were transfected with HERC4 and Ub-Flag. Forty-eight hours later, cells were treated with MG132 for 2 hours before cell lysate preparation. Cell lysates were immunoprecipitated with an anti–c-Maf antibody, followed by IB against Flag (Ub) or c-Maf. (D) HEK293 cells were transfected with HERC4, Flag-Ub, and WT or the indicated c-Maf variant. Forty-eight hours later, cells were treated with or without MG132 before cell lysate preparation. Cell lysates were then subjected to IP with an anti-HA antibody (for c-Maf), and a subsequent IP assay against Flag (for Ub).
K85 and K297 were required for HERC4-mediated c-Maf ubiquitination. (A) Lysine residues in c-Maf protein. Each K was changed to arginine (R) by site-direct mutagenesis. (B) Wild-type (WT) and mutant c-Maf plasmids were transfected into HEK293 cells with or without a HERC4 plasmid. Forty-eight hours later, cell lysates were subjected to an IB assay for c-Maf. (C) WT or all-lysine-substituted (K0) c-Maf plasmids were transfected with HERC4 and Ub-Flag. Forty-eight hours later, cells were treated with MG132 for 2 hours before cell lysate preparation. Cell lysates were immunoprecipitated with an anti–c-Maf antibody, followed by IB against Flag (Ub) or c-Maf. (D) HEK293 cells were transfected with HERC4, Flag-Ub, and WT or the indicated c-Maf variant. Forty-eight hours later, cells were treated with or without MG132 before cell lysate preparation. Cell lysates were then subjected to IP with an anti-HA antibody (for c-Maf), and a subsequent IP assay against Flag (for Ub).
The deubiquitinase USP5 abolishes HERC4-mediated c-Maf ubiquitination
Polyubiquitin-modified proteins may be subjected to deubiquitination by specific deubiquitinating enzymes. The Dub USP5 was identified in the c-Maf IPs, suggesting it may interact with and possibly deubiquitinates c-Maf (Figure 1A). USP5 is a 96-Kd protein, of which >57% amino acid sequence coverage was identified by AP-MS analysis of c-Maf IPs (Figure 5A). To test the hypothesis that USP5 modulates c-Maf polyubiquitination, c-Maf was cotransfected with USP5 into HEK293 cells. As shown in Figure 5B, c-Maf polyubiquitination was decreased by USP5. Consistently, total c-Maf protein was increased by the introduction of USP5 (Figure 5B, middle panel). To test whether USP5 attenuates HERC4-mediated ubiquitination of c-Maf, these 3 plasmids were coexpressed in HEK293 cells. The results confirmed that HERC4 decreased c-Maf protein, but it was abolished by USP5 (Figure 5C). Notably, USP5 mediated c-Maf degradation in a concentration- and time-dependent manner. As shown in supplemental Figure 1 (available on the Blood Web site), the ratio of USP5 to c-Maf plasmid was important for USP5 to accumulate c-Maf protein. When the ratio was >0.75, USP5 could significantly prevent c-Maf degradation. This finding was further supported by the CHX chase assay. As shown in Figure 5D, when cells were treated with CHX, USP5 slowed down c-Maf degradation by HERC4. Therefore, USP5 antagonizes HERC4-induced c-Maf degradation. This is consistent with a model wherein the USP5 catalytic activity removes ubiquitin and/or prevents the accumulation of polyubiquitin chains on c-Maf, resulting in its stabilization.
USP5 abolishes c-Maf degradation induced by HERC4. (A) USP5 was identified in the c-Maf immunoprecipitates by liquid chromatography-MS/MS. Unique peptides in USP5 identified by MS were highlighted. (B) HEK293 cells were transfected with c-Maf, USP5, and Ub plasmids. Forty-eight hours later, cell lysates were prepared for IP with an anti-HA antibody, followed by IB assay against ubiquitin (anti-Flag). (C) HEK293 cells were cotransfected with c-Maf, HERC4, and USP5 alone or together. Forty-eight hours later, cell lysates were prepared for IB. (D) HEK293 cells were transfected with c-Maf, HERC4, with or without USP5 for 24 hours followed by CHX treatment for indicated duration. Cell lysates were then prepared for IB assay.
USP5 abolishes c-Maf degradation induced by HERC4. (A) USP5 was identified in the c-Maf immunoprecipitates by liquid chromatography-MS/MS. Unique peptides in USP5 identified by MS were highlighted. (B) HEK293 cells were transfected with c-Maf, USP5, and Ub plasmids. Forty-eight hours later, cell lysates were prepared for IP with an anti-HA antibody, followed by IB assay against ubiquitin (anti-Flag). (C) HEK293 cells were cotransfected with c-Maf, HERC4, and USP5 alone or together. Forty-eight hours later, cell lysates were prepared for IB. (D) HEK293 cells were transfected with c-Maf, HERC4, with or without USP5 for 24 hours followed by CHX treatment for indicated duration. Cell lysates were then prepared for IB assay.
HERC4 is steadily decreased during the development of MM
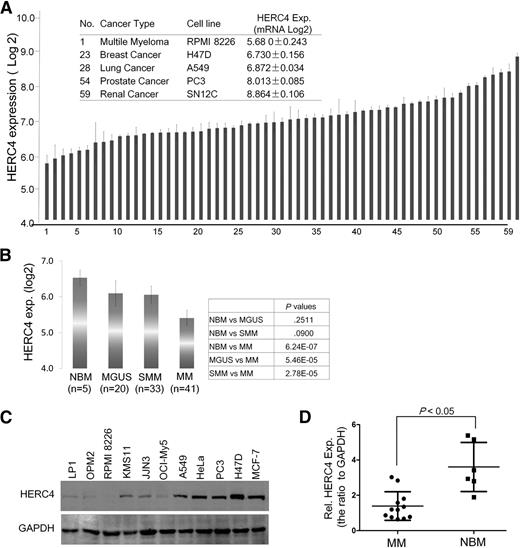
c-Maf is an oncoprotein associated with MM and mantle cell lymphoma, but the HERC4 expression pattern in MM cells is unknown. To evaluate HERC4 in cancer cells, we retrieved and analyzed HERC4 expression in 60 NCI selected cell lines by DNA microarray, in which RPMI 8226 was the representative MM cell line. As shown in Figure 6A, HERC4 was expressed at the lowest level in RPMI 8226 among the selected 60 cancer cell lines. To further characterize HERC4 expression in MM, we analyzed HERC4 mRNA in 99 DNA microarray datasets including healthy donor (n = 5), MGUS (n = 20), SMM (n = 34), and MM (n = 40) patients. Analysis of the DNA microarray data revealed that HERC4 was expressed at a high level in the normal bone marrow, but it was steadily downregulated along the development of MM and reached the lowest level in the bone marrow species of MM patients (Figure 6B). The HERC4 mRNA level was significantly decreased in MM patients compared with the healthy donors and MGUS patients (Figure 6B). To verify this finding, a panel of MM cell lines and selected solid tumors were subjected to IB using HERC4-specific antibody. As shown in Figure 6C, only very weak expression of HERC4 was detected in MM cell lines compared with a generally higher level of expression in non-MM cell lines, such as those derived from lung cancer (A549), breast cancer (H47D), cervical cancer (HeLa), and prostate cancer (PC-3). To further verify HERC4 expression in primary cells, bone marrow cells from both healthy donors and MM patients were collected for RNA evaluation. As shown in Figure 6D, the RNA levels of HERC4 in human MM patients were significantly lower than those in healthy humans. This result was consistent with the DNA microarray assay (Figure 6B). Therefore, HERC4 is downregulated during the development of MM and a pattern of diminished HERC4 expression was cancer- and tissue-specific.
HERC4 is downregulated in MM cells. (A) The HERC4 expression profile in 60 NCI cell lines. #1 was RPMI 8226, a typical MM cell line. These data were retrieved from GEO dataset (http://www.ncbi.nlm.nih.gov/gds/). (B) HERC4 expression in the process of MM development. HERC4 expression levels were retrieved from a DNA microarray with 99 primary bone marrow samples including normal donors, MGUS, SMM, and MM patients. (C) Selected solid-tumor cell lines and MM cell lines were subjected to IB against HERC4. (D) Primary bone marrow cells from MM patients and healthy donors were applied for the evaluation of HERC4 RNA expression by reverse-transcription PCR followed by density analysis with ImageJ software.
HERC4 is downregulated in MM cells. (A) The HERC4 expression profile in 60 NCI cell lines. #1 was RPMI 8226, a typical MM cell line. These data were retrieved from GEO dataset (http://www.ncbi.nlm.nih.gov/gds/). (B) HERC4 expression in the process of MM development. HERC4 expression levels were retrieved from a DNA microarray with 99 primary bone marrow samples including normal donors, MGUS, SMM, and MM patients. (C) Selected solid-tumor cell lines and MM cell lines were subjected to IB against HERC4. (D) Primary bone marrow cells from MM patients and healthy donors were applied for the evaluation of HERC4 RNA expression by reverse-transcription PCR followed by density analysis with ImageJ software.
HERC4 inhibits MM cell proliferation and delays MM tumor growth
The aforementioned analyses showed that HERC4 was downregulated in MM cells, but its action on MM was unclear. To determine whether HERC4 affected MM cell growth, MM cell lines RPMI 8226 and MM1.S, both of which express endogenous c-Maf, were infected with HERC4 lentivirus. c-Maf was decreased in a multiplicity of infection–dependent manner; after an increase of the multiplicity of infection of HERC4 virus, less c-Maf protein was detected (Figure 7A). The effects of HERC4 in 5 MM cell lines including RPMI 8226, MM1.S, JJN3, OPM2, and H929 were also evaluated. Ectopic expression of HERC4 markedly decreased MM cell viability as measured by the MTT assay in RPMI 8226, MM1.S, and JJN3 cells, but not marked in OPM2 and H929 cells (Figure 7B and supplemental Figure 2). This discrepancy was probably a result of the diverse genetic backgrounds of different MM cell lines. To find out whether anti-MM drugs modulated HERC4 expression, RPMI 8226 was treated by dexamethasone (Dex), a mainstay in MM treatment, followed by evaluation of HERC4 expression with both reverse-transcription PCR and IB assays. The results showed that Dex upregulated HERC4 expression at both the RNA and protein levels (supplemental Figure 3). Because USP5 antagonized HERC4 in modulating c-Maf stability, we wondered whether USP5 knockdown could enhance HERC4 activity. To this end, RPMI 8226 cells were treated with siUSP5 and/or HERC4 lentiviruses, followed by MTT assay. As shown in supplemental Figure 4, USP5 knockdown decreased RPMI 8226 cell proliferation, and it significantly enhanced HERC4-suppressing MM cell proliferation.
HERC4 decreases MM cell proliferation and delays MM tumor growth in nude mice. (A) RPMI 8226 and MM1.S cells were infected with increased pLVX-HERC4 lentivirus. Cells were subjected to IB for the expression of HERC4 and c-Maf. (B) RPMI 8226 and MM1.S cells were infected with HERC4 lentivirus followed by MTT assay for cell viability. (C) RPMI 8226 cells infected with HERC4 or mock lentivirus were injected into SCID mice subcutaneously. Tumor weights were then measured at the end of the experiment. (D) Tumor samples from the myeloma xenografts were subjected to IB analysis for c-Maf, HERC4, and cyclin D2 with specific antibodies.
HERC4 decreases MM cell proliferation and delays MM tumor growth in nude mice. (A) RPMI 8226 and MM1.S cells were infected with increased pLVX-HERC4 lentivirus. Cells were subjected to IB for the expression of HERC4 and c-Maf. (B) RPMI 8226 and MM1.S cells were infected with HERC4 lentivirus followed by MTT assay for cell viability. (C) RPMI 8226 cells infected with HERC4 or mock lentivirus were injected into SCID mice subcutaneously. Tumor weights were then measured at the end of the experiment. (D) Tumor samples from the myeloma xenografts were subjected to IB analysis for c-Maf, HERC4, and cyclin D2 with specific antibodies.
To further evaluate the effect of HERC4 on MM tumor growth in vivo, RPMI 8226 cells expressing HERC4 were inoculated into SCID mice. During the study course of 14 days, MM tumor growth rate was markedly decreased by HERC4. At the end of the experiment, the mean volumes of the HERC4-expressing tumors were >50% smaller than those of the mock-infected MM cells with lentivirus (Figure 7C). Notably, subsequent analysis of the tumor tissues from the xenografts showed that in the HERC4 lentivirus–infected cells, a high level of HERC4, along with decreased c-Maf and cyclin D2 proteins, was detected (Figure 7D). Therefore, HERC4 inhibits MM cell viability and delays MM tumor growth in association with c-Maf downregulation.
Discussion
HERC4 belongs to the HECT family of ubiquitin ligases featured by the presence of a HECT domain and one or more RCC1-like domains.18 It was reported that HERC4 is associated with spermatogenesis because disruption of HERC4 causes immature sperm with less motility.19 Recently, HERC4 is found in advanced lung cancers20 and breast cancers.21 However, the biological functions of HERC4 are not well known in terms of its ubiquitin ligation activity. Our present studies demonstrated that HERC4 interacts with and mediates c-Maf polyubiquitination and subsequent degradation in a serial set of in vitro and in vivo studies.
The ubiquitination process of a specific protein is strictly regulated by a serial enzyme cascade in which the ubiquitin ligase recognizes its specific substrate protein and catalyzes the attachment of the ubiquitin molecule to the specific lysine residue, thus modifying the specific function of the substrate protein. For example, the androgen receptor (AR) could be ubiquitinated by both Mdm2 and RNF6; however, the effects on AR function are quite different. Mdm2 leads to AR polyubiquitination and subsequent degradation,22 whereas RNF6 mediates an atypical ubiquitination chain on AR, thus enhancing its transcriptional activity.23 In the present study, we identified that HERC4 specifically mediates c-Maf ubiquitination but has no effect on MafA and MafB protein degradation, suggesting that HERC4 can differentiate its substrate although c-Maf shares a very high similarity with MafA and MafB. Moreover, in a panel of HECT-E3 ligases, HERC4 is the only one to mediate c-Maf degradation, further demonstrating that HERC4 specifically identifies c-Maf and mediates its ubiquitination and degradation, although other E3 ligases not in the examined panel will also mediate c-Maf ubiquitination (such as HUWE1 which was found in the c-Maf IP complexes), the current study at least suggests c-Maf stability and function is strictly regulated. This finding is consistent with c-Maf expression profile. Previous studies have demonstrated that c-Maf is specific in developmental stages and cell types.1 In the fetus, c-Maf is mainly involved in organogenesis such as the lens of the eyes, whereas it is mainly involved in Th cell differentiation in adults. This expression indicates that c-Maf function is well regulated. This specific expression pattern and the selectivity of E3 ligase–mediating c-Maf ubiquitination indicates that c-Maf stability and function is critically important, in which ubiquitination is a major modulating manner.
The specificity of c-Maf ubiquitination is also found in its lysine residues mediating ubiquitination. There are 13 lysine residues in c-Maf protein, and our previous studies found that all lysine residues in c-Maf protein could mediate c-Maf ubiquitination, including K85 and K350.8 However, HERC4 only mediates c-Maf ubiquitination at K85 and K297. This finding implies that c-Maf ubiquitination could be mediated by other E3 ligases, which warrants further investigation. In our AP-MS studies, there were other E3s existing in the c-Maf IPs, including RNF114, HUWE1, and UBR5 (Figure 1A and data not shown). Whether these E3s also mediate c-Maf ubiquitination remains unknown. These findings and unsolved questions suggest that c-Maf ubiquitination on individual lysine residues is probably determined by a specific E3 ligase, further demonstrating c-Maf stability and function is strictly regulated.
Post-translational modifications such as SUMOylation, phosphorylation, and ubiquitination are key methods to regulating the biological activity of c-Maf—for example, SUMOylation on c-Maf can negatively modulate its effects on IL-4 transcription,24,25 whereas phosphorylation increases c-Maf–transforming activity in embryonic mouse cells26 and enhances IL-4.27 In MM cells, both c-Maf and MafB can be phosphorylated by the Ser/Thr kinase GSK3, thus increasing MM cell proliferation and colony formation.28 The ubiquitination of c-Maf could lead to its degradation which could possibly decrease its oncogenicity in MM cells.9 HERC4 is a ubiquitin ligase for c-Maf and it modulates c-Maf degradation via the ubiquitin-proteasomal pathway. As demonstrated from various aspects, HERC4 binds to c-Maf and mediates its polyubiquitination. However, HERC4 is usually localized to the cytoplasm and is recruited to the nuclei upon c-Maf expression, which suggests that HERC4 is well responsive to overexpressed c-Maf in the nuclei in which c-Maf is probably quickly degraded upon ubiquitination in the presence of HERC4. This finding is confirmed by the CHX chase study in which HERC4 markedly increases c-Maf degradation.
Current studies show that HERC4 is probably an oncoprotein because it is expressed at a high level in solid cancers and associated with the advanced stage of these cancers, including lung cancer20 and breast cancer.21 This finding was confirmed by IB assays that showed HERC4 is upregulated in solid cancers. However, HERC4 is weakly expressed in MM. DNA microarray indicates HERC4 is expressed in normal bone marrow cells, but it is steadily decreased with the development from MGUS, SMM to MM. More importantly, enforced HERC4 in MM cell lines decreases MM cell proliferation and delays MM tumor growth in nude mice. These outcomes probably come from HERC4-induced c-Maf degradation. It is known that c-Maf as a transcription factor modulates the transcription of several genes including cyclin D2, integrin β7, and CCR1, which are critical for MM cell cycle progress, cell proliferation, and stromal cell adhesion.7 Because c-Maf has been well documented as an oncoprotein for MM,7 HERC4 probably acts as a tumor suppressor in MM cells, although it is highly associated with oncogenesis in some solid cancers.
In summary, to the best of our knowledge, this is the first report on HERC4 as an E3 ubiquitin ligase and the first report that identifies a ubiquitin ligase and Dub for c-Maf ubiquitin modification. This study suggests that modulation of c-Maf stability may be a therapeutic modality worthy of investigation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study supported in part by the National Natural Science Foundation of China (81320108023 and 81272632), the Natural Science Foundation of Jiangsu Province (BE2014630), the National Basic Research Program of China (2011CB933501), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and the Jiangsu Key Laboratory for Translational Research and Therapeutics of Neuro-Psycho-Diseases (BK2013003).
Authorship
Contribution: X.M. designed the study; Z.Z., J.T., X.T., J.J., B.C., R.H., G.C., P.T., X.X. J.D., and C.-x.S. conducted experiments; G.W. provided essential ubiquitin plasmids and analyzed the data; J.H., D.W., A.K.S., A.D.S., M.F.M,. and X.M. analyzed data; and X.M. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Xinliang Mao, Department of Pharmacology, Soochow University, 199 Ren Ai Rd, Suzhou 215123, China; e-mail: xinliangmao@suda.edu.cn.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal