Key Points
We report for the first time the biological features of MRD cells in MM and unravel that clonal selection is already present at the MRD stage.
MRD cells show a singular phenotypic signature that may result from persisting clones with different genetic and gene expression profiles.
Abstract
Persistence of chemoresistant minimal residual disease (MRD) plasma cells (PCs) is associated with inferior survival in multiple myeloma (MM). Thus, characterization of the minor MRD subclone may represent a unique model to understand chemoresistance, but to our knowledge, the phenotypic and genetic features of the MRD subclone have never been investigated. Here, we compared the antigenic profile of MRD vs diagnostic clonal PCs in 40 elderly MM patients enrolled in the GEM2010MAS65 study and showed that the MRD subclone is enriched in cells overexpressing integrins (CD11a/CD11c/CD29/CD49d/CD49e), chemokine receptors (CXCR4), and adhesion molecules (CD44/CD54). Genetic profiling of MRD vs diagnostic PCs was performed in 12 patients; 3 of them showed identical copy number alterations (CNAs), in another 3 cases, MRD clonal PCs displayed all genetic alterations detected at diagnosis plus additional CNAs that emerged at the MRD stage, whereas in the remaining 6 patients, there were CNAs present at diagnosis that were undetectable in MRD clonal PCs, but also a selected number of genetic alterations that became apparent only at the MRD stage. The MRD subclone showed significant downregulation of genes related to protein processing in endoplasmic reticulum, as well as novel deregulated genes such as ALCAM that is prognostically relevant in MM and may identify chemoresistant PCs in vitro. Altogether, our results suggest that therapy-induced clonal selection could be already present at the MRD stage, where chemoresistant PCs show a singular phenotypic signature that may result from the persistence of clones with different genetic and gene expression profiles. This trial was registered at www.clinicaltrials.gov as #NCT01237249.
Introduction
In multiple myeloma (MM) as in virtually all hematologic malignancies, there is a correlation between depth of response and prolonged survival.1 Over the last 20 years, significantly higher rates of complete response (CR) have been achieved contributing to improve MM patient outcome.2 However, long-term follow-up studies have shown that approximately two-thirds of these will not sustain their CR and will relapse before they can be considered as achieving “operational cure” (ie, >10-year progression-free survival).3 Relapses among MM patients in CR are now better understood and predicted with the advent of minimal residual disease (MRD) studies, which have shown an intrinsic correlation between the persistence of reduced numbers of clonal plasma cells (PCs) after therapy (ie, MRD) and inferior survival.4-7 Thus, MRD represents a very small fraction of all diagnostic tumor cells that are chemoresistant, potentially quiescent (not producing M-protein), and able to recapitulate the initial tumor burden at relapse.
Overall, there are 2 putative mechanisms by which chemoresistance may arise in cancer: (1) therapy-induced molecular alterations and (2) the presence of cellular heterogeneity within the tumor bulk, where several subclones coexist and compete with each other in such a way that treatment eradicates the major subclone (chemosensitive), but a minor, resistant, and initially dormant subclone subsequently expands and gives rise to disease relapse.8 In MM, the first mechanism has been suggested to explain chemoresistance to proteasome inhibition (eg, acquired mutations in genes encoding proteasome β subunits9 or decreased endoplasmic reticulum [ER] stress signaling10 ) and immunomodulatory agents (eg, decreased cereblon expression11,12 and overexpression hyaluronan binding proteins such as CD4413 ), or upregulation of the mitogen-activated protein kinase kinase/extracellular signal-regulated kinase pathway.14 In turn, the second mechanism would explain the shift in dominance of tumor subclones over time in paired samples from MM patients studied at diagnosis vs relapse.15-17 Of note, chemoresistance may also result from the combination of both mechanisms; for example, when less differentiated myeloma PC clones downregulate specific transcriptional factors to overcome treatment (eg, suppression of Xbp1s to diminish ER front-loading and cytotoxic susceptibility to proteasome inhibition).18 Despite all the above, both hypothetical mechanisms rely on data generated from the study of clonal PCs at the time of relapse, whereas no biological studies have been performed in primary chemoresistant clonal PCs that persist at MRD levels after frontline therapy before the patients’ relapse.
MRD monitoring has become one of the most relevant prognostic factors in MM, independently of patients’ serologic response and diagnostic cytogenetic profile.5 Thus, it has been shown that the outcome of patients with standard- or high-risk cytogenetics is modulated by the persistence vs eradication of MRD5 but also that, particularly among MRD-positive cases, survival is significantly inferior in patients with clonal PCs harboring high-risk cytogenetic abnormalities.5 Thus, both the depth of response and the genetic profile of chemoresistant subclones most likely play an important role in the outcome of MM patients. Thereby, a better understanding of the biological signature of MRD vs diagnostic cells could potentially contribute to gain insight in the mechanisms of chemoresistance at the MRD level and the potential discovery of novel therapeutic targets.
Here, we characterized chemoresistant MRD cells from patients included in the GEM2010MAS65 clinical trial by comparing their phenotypic and genetic profiles against paired diagnostic clonal PCs. Our results reveal that therapy-induced clonal selection is already present at the MRD stage, in which chemoresistant PCs show a singular phenotypic signature that may result from the persistence of clones with different cytogenetic and gene expression profiles. Furthermore, we show that such analyses also provide a novel approach to better understand chemoresistance, through the identification of novel candidate genes that could be potentially responsible for drug resistance and, consequently, impact on patients’ survival.
Methods
Patients and treatment
A total of 40 elderly, transplant-ineligible patients with newly diagnosed symptomatic MM staged according to the International Myeloma Working Group criteria19 were prospectively studied after inclusion in the GEM2010MAS65 trial (#NCT01237249). In all patients, bone marrow (BM) aspirates were collected at diagnosis and after 9 cycles of induction therapy. In brief, patient treatment consisted of either 9 identical induction cycles with bortezomib, melphalan, prednisone (VMP; n = 21) or alternating cycles of VMP and lenalidomide plus low-dose dexamethasone (Rd) for up to 9 courses (n = 19). Samples were collected after informed consent was given by each individual according to the local ethical committees and the Helsinki Declaration.
Multidimensional flow cytometry immunophenotyping
Approximately 400 μL EDTA-anticoagulated BM aspirated samples were immunophenotyped using 4 different 8-color combinations of monoclonal antibodies (mAbs) and direct immunofluorescence stain-and-then-lyse technique: Pacific Blue (PacB), Pacific Orange (PacO), fluorescein isothiocyanate (FITC), phycoerythrin (PE), peridinin chlorophyll protein-cyanin 5.5 (PerCP-Cy5.5), PE-cyanin 7 (PE-Cy7), allophycocyanin (APC), alexafluor 700 (AF700): (1) CD29, CD45, CD11a, β7, CD79b, CD49d, CD19, and CD38; (2) CD11c, CD45, CD41a, CD49e, CD33, CD117, CD19, and CD38; (3) CD20, CD45, CD81, CD54, CD138, CD56, CD19, and CD38, and (4) HLA-DR, CD45, CD44, CXCR4, CD27, CD28, CD19, and CD38. Markers were selected based on their role to identify the PC compartment (CD38 and CD138), discriminate between clonal and normal PCs within the PC compartment (CD19, CD20, CD27, CD28, CD45, CD56, CD81, and CD117), define B-cell/PC differentiation (CD19, CD20, CD27, CD45, CD79b, CD81, and HLADR), mediate cell-to-cell interactions (CD28 and CD117), and in cell adhesion-mediated drug resistance (CAM-DR: CD11a, CD11c, CD29, CD41a, CD44, CD49d, CD49e, CD54, CXCR4, and β7). Data acquisition was performed for ∼106 leukocytes per tube in a FACSCantoII flow cytometer (Becton Dickinson Biosciences [BDB], San Jose, CA) using the FACSDiva 6.1 software (BDB). Instrument performance was daily monitored using the Cytometer Setup Tracking (CST; BDB) and rainbow 8-peak beads (Spherotech, Lake Forest, IL) after laser stabilization, following the EuroFlow guidelines20 ; sample acquisition was performed only in case of longitudinal instrument stability. In supplemental Figure 1, available on the Blood Web site, the expression levels of CD19 in mature B cells from BM samples assessed at diagnosis and after induction are represented and confirm the longitudinal instrument stability. Per protocol, newly diagnosed MM patients included in the GEM2010MAS65 trial had BM samples studied after 9 induction cycles to monitor MRD levels; whenever persistent clonal PCs were detected (MRD positive), exactly the same immunophenotypic method performed at diagnosis was repeated for the characterization of the chemoresistant clone after therapy.
Generation of immunophenotypic protein expression profiles
To generate immunophenotypic protein expression profiles (iPEPs), we used first those 5 parameters measured in common for each aliquot (CD38, CD45, CD19, forward light scatter [FSC], and sideward light scatter [SSC]) to define the PC compartment. Then, the iPEP of diagnostic (baseline) and chemoresistant (MRD) cells for all 23 phenotypic markers analyzed plus FSC and SSC was generated for every single clonal PC, after merging of flow cytometry data files and calculation of data.21,22 First, the merge function of the Infinicyt software (Cytognos SL, Salamanca, Spain) was used to fuse the different data files corresponding to the 4 different 8-color mAb combinations studied per sample into a single data file containing all information measured for that sample. For any single cell in each 8-color mAb combination (aliquot), this included data about those antigens that were measured directly on it and antigens that were not evaluated directly (missing values) for that cell in the corresponding aliquot it was contained in. Then, the calculation function of the Infinicyt software was used to fill in the missing values, based on the nearest neighbor statistical principle,23,24 defined by the unique position (Euclidean distance) of individual PCs in the multidimensional space created by the 5 common (backbone) parameters (FSC, SSC, CD38, CD45, and CD19). The nearest neighbor for each individual clonal PC in a sample aliquot was calculated as that clonal PC in another aliquot showing the shortest distance to it in the 3-dimensional space generated by the 5 parameters measured in common in both aliquots. Then, for each individual single clonal PC in a sample aliquot, those values obtained for each of the closest single clonal PC in the other aliquot were assigned for each of those parameters not actually measured in the former single clonal PC.25,26 Ultimately, an iPEP was generated for every single PC, which included all 23 phenotypic markers analyzed plus FSC and SSC. Therapy-induced (phenotypic) clonal selection was further investigated by principal component analysis, based on the 25 parameters evaluated using the automated population separator (principal component 1 vs principal component 2), graphical representation of the Infinicyt software,20 and comparing merged patient-paired iPEPs of baseline (diagnostic) vs MRD myeloma PCs.
Copy number and gene expression profiling of baseline vs MRD patient-paired myeloma PCs
Per protocol, clonal PCs from diagnostic samples were isolated from BM aspirates using CD138+ immunomagnetic beads (Miltenyi Biotech, Bisley, UK). Following induction therapy, BM aspirates were obtained to determine patients’ MRD response at cycle 9, and clonal PCs from MRD-positive cases were fluorescence-activated cell sorted (FACS) (FACSAria II, BDB; purity ≥97%) according to patient-specific aberrant phenotypes. DNA from matched diagnostic and MRD clonal PCs was available in 8 of 40 patients, as well as in 4 additional cases treated with 6 induction cycles with VRD, making a total of 12 patients. Afterward, genome-wide detection of copy number abnormalities (CNAs) and loss of heterozygosity (LOH) were investigated using the standard Affymetrix Cytoscan 750K platform (Affymetrix, Santa Clara, CA). An unpaired analysis was performed using 240 Hapmap files as reference for normal DNA. The complete dataset was analyzed by visual inspection using the AGCC and ChAS software programs (Affymetrix). CNAs were reported when the 3 following criteria were met: ≥25 consecutive imbalanced markers per segment; ≥100-kb minimum genomic size; and <50% overlap with paired control DNA and/or genomic variants of Toronto DB (DGV).27 Only copy number neutral LOH (CNN-LOH) >5 Mb was considered.
Gene expression profiling (GEP) was performed in matched diagnostic and MRD clonal PCs from 7 of 40 cases with adequate RNA extracted from CD138+ and patient-specific phenotypically aberrant FACS-purified tumor cells, respectively. Briefly, the integrity of the extracted RNA was assessed using the Agilent 2100 Bioanalyzer. Afterward, RNA was amplified, labeled, and subsequently hybridized to the Human Gene 1.0 ST Array (Affymetrix).28 Normalization was carried out by using the expression console (Affymetrix) with the RMA algorithm, which includes background correction, normalization, and calculation of expression values (log2).29 The SIMFIT (http://www.simfit.org.uk/) statistical software was used to perform hierarchical clustering analyses based on Euclidean distances and the group average linkage method. Differentially expressed genes between classes were identified using the significant analysis of microarrays (SAM) algorithm (http://statweb.stanford.edu/∼tibs/SAM/), and significant genes were selected based on the lowest q-value (<0.05).28,29 We used the WebGestalt online suite30 to identify the most relevant functional pathways involved according to the differentially expressed genes. Full microarray data are available at the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo/; accession number GSE70399). The potential impact on disease survival of selected top-deregulated genes was investigated in a series of newly diagnosed MM patients who had been treated according to the Total Therapy (TT) programs 3A (#NCT00081939; n = 276) and 3B (#NCT00572169; n = 168) with GEP data on CD138+ purified PCs. Optimal gene expression cutoffs were first established in patients enrolled in TT3A (training set) and then tested in patients enrolled in TT3B (validation set). A complete description of the TT3 program has been reported elsewhere.31
In vitro evaluation of ALCAM expression in chemoresistant MM cells
Baseline levels of the ALCAM protein (CD166; BDB) were assessed by flow cytometry on the surface of 8 MM cell lines (NCI-H929, JJN3, KMS-11, MM1S, OPM2, RPMI-8226, U266, and U266-LR7). ALCAM expression was more extensively evaluated in RPMI-8226 MM cells after coculture with BM stromal cells (BMSCs) with or without simultaneous exposure to bortezomib (5 nM), lenalidomide (10 µM), dexamethasone (1 µM), and melphalan (1 µM) for 24 and 48 hours at 37°C. Stromal cells were obtained as previously described,32 and a confluent monolayer was generated by plating 10 × 105 BMSCs in a 12-well plate for 48 hours. ALCAM expression was measured in chemoresistant (Annexin-V−) and chemosensitive (Annexin-V+) RPMI-8226 cells after exclusion of debris and cell doublets based on their FSC and SSC characteristics and of CD38−CD138− BMSCs. Data acquisition was performed in a FACSCantoII flow cytometer (BDB) using the FACSDiva 6.1 software (BDB); for data analysis, the Infinicyt software (Cytognos) was used. All experiments were performed in triplicate.
Statistical analysis
The Wilcoxon signed rank test was used to evaluate the statistical significance of the phenotypic differences observed between baseline and MRD myeloma PCs, whereas the Mann-Whitney U and the Kruskal-Wallis tests were used to estimate the statistical significance of differences observed between 2 or more groups, respectively. The IBM SPSS software (version 20.0; IBM, New York, NY) was used for all statistical tests.
Results
MRD clonal PCs show a singular iPEP
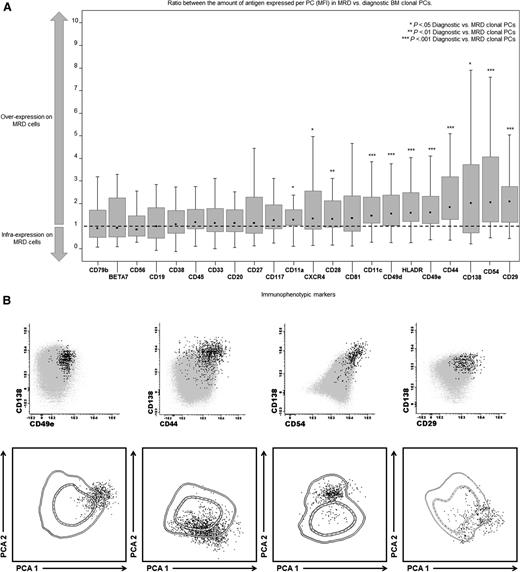
The iPEP of clonal PCs from patient-paired diagnostic and MRD samples were analyzed in 40 patients. Compared with the diagnostic tumor bulk, chemoresistant MRD clonal PCs showed significant upregulation (P < .05) of CD11a, CD11c, CD29, CD44, CD49d CD49e, CD54, CD138, CXCR4, and HLADR (Figure 1A). Of note, differences in the expression levels of proteins were not due to de novo expression on MRD cells of previously undetectable antigens on diagnostic clonal PCs, but rather because the MRD clone clustered among those cells that at diagnosis showed the highest levels of expression for such markers (Figure 1B); in fact, MRD cells typically clustered in a more restricted area of that occupied by the whole BM clonal PC compartment at diagnosis (Figure 1B). Interestingly, differences in the ratio between the amount of antigen expressed per cell in paired MRD vs diagnostic BM PCs for the top five deregulated markers were particularly evident in patients receiving only VMP vs those receiving alternating cycles of VMP and Rd (Table 1); in fact, differential expression of markers such as CD81 (P = .06) and CD117 (P = .03) was noted among patients exposed only to VMP and not Rd. Altogether, these results suggest that between diagnosis and the MRD stage, therapy-induced phenotypic selection of clonal PCs might occur within the initial tumor bulk, in which PCs showing stronger expression of integrin and adhesion molecules are more prone to survive (Figure 1B).
iPEP of MRD vs diagnostic MM clonal PCs (n = 40). (A) Detailed immunophenotypic features of paired diagnostic vs MRD clonal PCs from 40 MM patients. All markers were evaluated in all 40 cases at both time points (diagnosis vs MRD). Notched boxes represent the 25th and 75th percentile values of the ratio between the amount of antigen mean fluorescence intensity expression per paired MRD/diagnostic BM clonal PCs; the dot in the middle and vertical lines correspond to the median value and both the 10th and 90th percentiles, respectively. (B) Representative bivariate dot-plots for the top 5 overexpressed markers in MRD vs diagnostic BM clonal PCs are shown (CD29, CD54, CD138, CD44, CD49e), as well as the corresponding iPEPs for each of the 5 representative patients. The iPEP of diagnostic BM clonal PCs is represented by 1 and 2 standard deviation lines, whereas their paired individual MRD cells are represented by black dots. Each patient-specific iPEP is represented using the automated population separator (APS1) plot based on a graphical representation of principal component 1 (x-axis) vs principal component 2 (y-axis) for a total of 25 parameters.
iPEP of MRD vs diagnostic MM clonal PCs (n = 40). (A) Detailed immunophenotypic features of paired diagnostic vs MRD clonal PCs from 40 MM patients. All markers were evaluated in all 40 cases at both time points (diagnosis vs MRD). Notched boxes represent the 25th and 75th percentile values of the ratio between the amount of antigen mean fluorescence intensity expression per paired MRD/diagnostic BM clonal PCs; the dot in the middle and vertical lines correspond to the median value and both the 10th and 90th percentiles, respectively. (B) Representative bivariate dot-plots for the top 5 overexpressed markers in MRD vs diagnostic BM clonal PCs are shown (CD29, CD54, CD138, CD44, CD49e), as well as the corresponding iPEPs for each of the 5 representative patients. The iPEP of diagnostic BM clonal PCs is represented by 1 and 2 standard deviation lines, whereas their paired individual MRD cells are represented by black dots. Each patient-specific iPEP is represented using the automated population separator (APS1) plot based on a graphical representation of principal component 1 (x-axis) vs principal component 2 (y-axis) for a total of 25 parameters.
iPEP of diagnostic vs MRD BM clonal PCs in MM patients (n = 40) studied after induction therapy
| Markers . | Overall series (n = 40) . | Sequential chemotherapy (n = 21) . | Alternating chemotherapy (n = 19) . | |||
|---|---|---|---|---|---|---|
| Fold-change . | P value . | Fold-change . | P value . | Fold change . | P value . | |
| CD29 | 1.95 | <.001 | 1.95 | <.001 | 2.02 | .009 |
| CD54 | 1.92 | <.001 | 2.98 | <.001 | 1.31 | .04 |
| CD138 | 1.90 | .04 | 2.16 | .03 | 1.22 | .36 |
| CD44 | 1.79 | <.001 | 2.04 | <.001 | 1.50 | .008 |
| CD49e | 1.58 | <.001 | 1.97 | .002 | 1.49 | .084 |
| Markers . | Overall series (n = 40) . | Sequential chemotherapy (n = 21) . | Alternating chemotherapy (n = 19) . | |||
|---|---|---|---|---|---|---|
| Fold-change . | P value . | Fold-change . | P value . | Fold change . | P value . | |
| CD29 | 1.95 | <.001 | 1.95 | <.001 | 2.02 | .009 |
| CD54 | 1.92 | <.001 | 2.98 | <.001 | 1.31 | .04 |
| CD138 | 1.90 | .04 | 2.16 | .03 | 1.22 | .36 |
| CD44 | 1.79 | <.001 | 2.04 | <.001 | 1.50 | .008 |
| CD49e | 1.58 | <.001 | 1.97 | .002 | 1.49 | .084 |
Nine cycles of VMP vs 9 alternating cycles of VMP and RD. The fold-change in the ratio between the amount of protein per PC (MFI) for paired MRD/diagnostic BM clonal PCs is shown for the top 5 markers overexpressed on MRD cells in the overall patient series and the 2 different patient treatment groups. All markers were evaluated in all 40 patients at both time points (diagnosis vs MRD).
MFI, mean fluorescence intensity; PR, partial response; VGPR, very good partial response.
Copy number profile of patient-paired diagnostic vs MRD clonal PCs
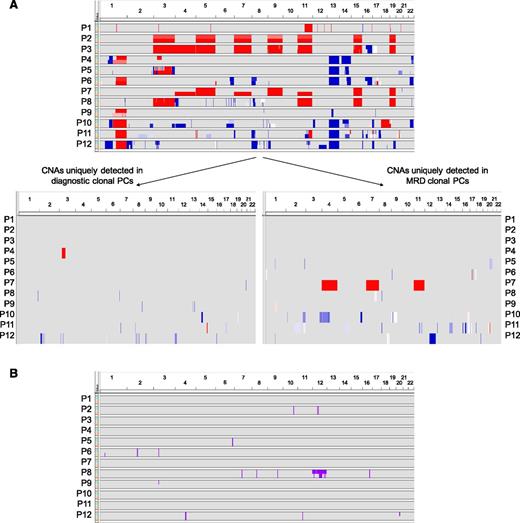
Comparison between iPEPs of diagnostic vs MRD clonal PCs suggested that the latter might represent a (phenotypic) subset of the whole diagnostic tumor population. To gain further insight into this novel hypothesis, we investigated CNA profiles of matched diagnostic vs MRD clonal PCs (Figure 2). The median number of CNAs in diagnostic and MRD clonal PCs were 11 and 14, respectively. Individual patient analyses revealed that in 3 of the 12 cases analyzed, both clones showed identical copy number profiles (1, 2, and 3), whereas in the remaining 9 patients, there were unique CNAs detected in ≥1 of the 2 PC populations. Namely, MRD clonal PCs from patients 4, 5, and 6 displayed all genetic alterations detected at diagnosis but 1 interstitial gain in chromosome 3 of patient 4, plus 4 and 5 additional CNAs that emerged at the MRD stage in cases 5 and 6, respectively (Figure 2). In the remaining six patients (7-12) there was further variability, with both CNAs present at diagnosis that were undetectable in MRD clonal PCs but also a selected number of genetic alterations that became apparent only at the MRD stage (total of 4, 4, 4, 10, 14, and 17 different CNAs between diagnostic vs MRD clonal PCs in patients 7, 8, 9, 10, 11, and 12, respectively). Overall, a total of 66 CNA (56 losses and 10 gains) were uniquely present in only 1 of the 2 PC populations (26 at diagnosis and 40 after therapy), in the absence of recurrent patterns of genetic evolution from baseline into MRD cells. We also investigated the profile of CNN-LOH in matched diagnostic and chemoresistant clones. As expected, these abnormalities were less frequently observed and were only noted in 6 of the 12 cases; in 3 of the latter 6 (8, 9, and 12), the MRD clonal PCs lacked CNN-LOH present at diagnosis, whereas patient 6 had a CNN-LOH in chromosome 1, which was not detected at diagnosis. All remaining cases had overlapping CNN-LOH profiles.
Clonal evolution from diagnosis to MRD clonal PCs. (A) Overview of CNAs and (B) CNN-LOH detected in clonal PCs from matched BM samples from 12 patients at diagnosis and during MRD monitoring. In A, copy number profile of patient-paired diagnostic and MRD clonal PCs for each patient are stacked, with the diagnostic sample shown on the top (green) and the MRD sample in the bottom (salmon). Sample pairs are indicated on the y-axis and chromosome location on the x-axis. Blue shading indicates the presence of copy number loss; red indicates copy number gain. In B, CNN-LOH detected in diagnostic (green) and MRD (salmon) clonal PCs are represented by pink bars.
Clonal evolution from diagnosis to MRD clonal PCs. (A) Overview of CNAs and (B) CNN-LOH detected in clonal PCs from matched BM samples from 12 patients at diagnosis and during MRD monitoring. In A, copy number profile of patient-paired diagnostic and MRD clonal PCs for each patient are stacked, with the diagnostic sample shown on the top (green) and the MRD sample in the bottom (salmon). Sample pairs are indicated on the y-axis and chromosome location on the x-axis. Blue shading indicates the presence of copy number loss; red indicates copy number gain. In B, CNN-LOH detected in diagnostic (green) and MRD (salmon) clonal PCs are represented by pink bars.
Gene expression profiling of chemoresistant MRD clonal PCs
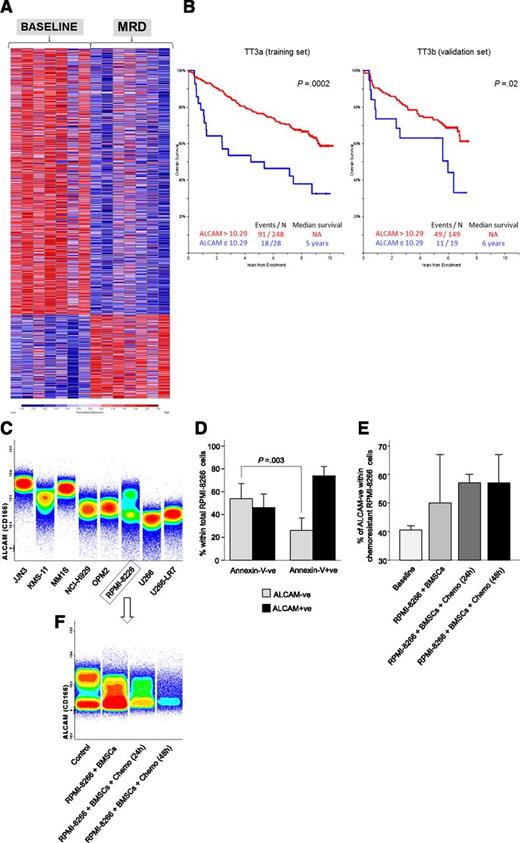
On demonstrating that chemoresistant MRD cells are characterized by a singular iPEP and they often carry CNAs that were undetectable at diagnosis or only became apparent after therapy, we sought to compare the GEP of diagnostic vs MRD clonal PCs in patient-paired (n = 7) samples of the GEM2010MAS65 study, to determine whether chemoresistant subclones have unique GEP signatures. As a result, MRD clonal PCs showed deregulated expression of 1336 genes (317 up- and 1019 downregulated; Figure 3A; supplemental Excel File 1). Functional analysis using all deregulated genes revealed that several of those being downregulated were related to cellular functions such as protein export, protein processing in the ER, and N-glycan biosynthesis (Table 2). In addition to downregulation of a large number of ER-related genes, MRD clonal PCs also showed lower expression of genes encoding for different proteasome subunits (ie, PAAF1, PSMA6, PSMB10, POMP, PSME1, PSMD10, PSMB6, PSME3, and PSMA2). Afterward, we focused on those genes that were maximally upregulated (fold-change ≥2) and downregulated (fold-change ≤0.5) in MRD vs diagnostic clonal PCs and that had been related to chemoresistance/aggressiveness in other neoplasms or related to proteasome inhibition and ER stress (n = 21 genes), and investigated their potential impact on the survival of newly diagnosed MM patients enrolled in the TT3 program (Table 3). Overall, 9 of the 21 genes tested showed a significant prognostic impact on overall survival (OS) in the training (TT3a) set, but only 3 of these 9 genes kept their prognostic significance in the validation (TT3b) MM patient set; thus, inferior OS rates were observed in both series for cases showing overexpression of both FLNA (median OS of 5 years vs not reached for patients with high vs low FLNA expression, respectively; P ≤ .002; supplemental Figure 2A) and FERMT3 (median OS not reached for patients with high vs low FERMT3 expression, respectively; P = .03; supplemental Figure 2B), as well as lower expression of ALCAM (median OS not reached vs 6 years for patients with high vs low ALCAM expression, respectively; P ≤ .02; Figure 3A). In line with the findings in newly diagnosed patients enrolled in the TT3 program, the MRD clone in the GEM2010MAS65 trial also showed overexpression of FERMT3 and FLNA plus downregulation of ALCAM vs diagnostic PCs.
Gene expression profiles of MRD vs diagnostic MM clonal PCs. (A) Heat map of genes with differential expression (false discovery rate q-value <0.05) between patient-paired diagnostic and MRD clonal PCs. (B) Overall survival according to low vs high (≤10.29 vs >10.29 normalized mRNA expression) ALCAM levels determined in CD138+ve purified PCs from newly diagnosed MM patients treated according to TT3A (n = 276; training set) and TT3B (n = 168; validation set). (C) ALCAM (CD166) protein surface expression in a panel of MM cell lines is shown using the total density graphic representation of the Infinicyt software. (D) Overall ALCAM (CD166) protein surface expression in chemoresistant (Annexin-V−ve) and apoptotic (Annexin-V+ve) RPMI-8226 cells; the percentage of ALCAM−ve and ALCAM+ve MM cells is represented by the black and gray bars, respectively. (E) Percentage of ALCAM−ve cells within total chemoresistant (Annexin-V−ve) RPMI-8226 cells at baseline, on coculture with BMSCs and in coculture with BMSCs after 24- and 48-hour exposure to bortezomib, lenalidomide, dexamethasone, and melphalan. All experiments were performed in triplicate. (F) Representative experiment (n = 1) of ALCAM (CD166) protein surface expression in chemoresistant (Annexin-V−ve) RPMI-8226 cells.
Gene expression profiles of MRD vs diagnostic MM clonal PCs. (A) Heat map of genes with differential expression (false discovery rate q-value <0.05) between patient-paired diagnostic and MRD clonal PCs. (B) Overall survival according to low vs high (≤10.29 vs >10.29 normalized mRNA expression) ALCAM levels determined in CD138+ve purified PCs from newly diagnosed MM patients treated according to TT3A (n = 276; training set) and TT3B (n = 168; validation set). (C) ALCAM (CD166) protein surface expression in a panel of MM cell lines is shown using the total density graphic representation of the Infinicyt software. (D) Overall ALCAM (CD166) protein surface expression in chemoresistant (Annexin-V−ve) and apoptotic (Annexin-V+ve) RPMI-8226 cells; the percentage of ALCAM−ve and ALCAM+ve MM cells is represented by the black and gray bars, respectively. (E) Percentage of ALCAM−ve cells within total chemoresistant (Annexin-V−ve) RPMI-8226 cells at baseline, on coculture with BMSCs and in coculture with BMSCs after 24- and 48-hour exposure to bortezomib, lenalidomide, dexamethasone, and melphalan. All experiments were performed in triplicate. (F) Representative experiment (n = 1) of ALCAM (CD166) protein surface expression in chemoresistant (Annexin-V−ve) RPMI-8226 cells.
Selected pathways and transcription targets predicted to be significantly inhibited according to the corresponding deregulated genes in patient-paired diagnostic vs MRD clonal PCs
| . | P value . | Genes down- or upregulated in MRD clonal PCs . |
|---|---|---|
| Pathways | ||
| Protein export | 1.50e−05 | SRPRB, SRP72, SEC61A1, SEC61G, SRP68, SPCS1, SEC11C, SPCS2, SPCS3, SEC63 |
| Protein processing in endoplasmic reticulum | 1.50e−05 | ERLEC1, SEC61G, MAN1A2, DNAJB11, SSR2, UGGT1, SEC63, EDEM2, CANX, SEC23B, OS9, SSR1, DERL1, SAR1B, RPN1, SEC61A1, STT3A, DERL2, DAD1, UBE2J1, SEL1L, EIF2S1, SSR3, LMAN2, UBE2G1, LMAN1, PDIA6 |
| N-glycan biosynthesis | .0004 | MAN2A2, RPN1, ALG5, DPM1, STT3A, ALG14, ALG13, MAN1A2, DAD1, ALG8, DOLK, ALG1 |
| Transcription targets | ||
| ELK1 | 1.91e−06 | ZNF35, TOMM20, YIPF5, PYROXD1, TRAF7, F11R, POMP, SND1, TMEM167A, HCCS, BECN1, MTMR2, UBLCP1, C14orf119, PHF5A, UBA2, DNAAF2, ATXN10, ACVR2A, RNF2, DNMT1, DDX1, RNF185, U2AF2, SLC39A9, RWDD2A, DPM1, UXT, TLK1, SDF2, TMEM59, GTF2A2, JAGN1, UBR4, NECAP2, AMZ2, TMCO1, WAS, DSCR3, TBCC, SEC61A1, C21orf59, ZBTB41, GMIP, E2F4, PEX2, POLR2H, MAD2L1BP, C11orf57, YEATS2, NXT2, CCDC25, UNC13D, MKI67IP, NDUFS1, PIGW, ZCCHC9, MUT, C17orf80, PHB2, ITFG1, UFC1, CBLL1, SDHAF2, UGGT1, HNRNPH1, SLC35A5, EIF1AD, GRPEL2, CEP164, ERH, LSM5, SHKBP1, VMP1, TMEM62, FTSJD1, COX6A1, BANF1, RPL27, FBXO38, MEA1, CKS1B, HARS2, LMAN2, MORN2, MRPL40, TOMM70A, ZNF184, MAP4K2, RHOA, PGS1, CALU, ZSCAN12, UFM1, TRMT112, TGDS, FANCF, RNF13, THUMPD3, COX17, ZFP3, RPL32, PALB2, MRPL33, LYL1, COX8A, EIF2S1, PITPNA, TXNDC12, PSME3, MRPL3 |
| . | P value . | Genes down- or upregulated in MRD clonal PCs . |
|---|---|---|
| Pathways | ||
| Protein export | 1.50e−05 | SRPRB, SRP72, SEC61A1, SEC61G, SRP68, SPCS1, SEC11C, SPCS2, SPCS3, SEC63 |
| Protein processing in endoplasmic reticulum | 1.50e−05 | ERLEC1, SEC61G, MAN1A2, DNAJB11, SSR2, UGGT1, SEC63, EDEM2, CANX, SEC23B, OS9, SSR1, DERL1, SAR1B, RPN1, SEC61A1, STT3A, DERL2, DAD1, UBE2J1, SEL1L, EIF2S1, SSR3, LMAN2, UBE2G1, LMAN1, PDIA6 |
| N-glycan biosynthesis | .0004 | MAN2A2, RPN1, ALG5, DPM1, STT3A, ALG14, ALG13, MAN1A2, DAD1, ALG8, DOLK, ALG1 |
| Transcription targets | ||
| ELK1 | 1.91e−06 | ZNF35, TOMM20, YIPF5, PYROXD1, TRAF7, F11R, POMP, SND1, TMEM167A, HCCS, BECN1, MTMR2, UBLCP1, C14orf119, PHF5A, UBA2, DNAAF2, ATXN10, ACVR2A, RNF2, DNMT1, DDX1, RNF185, U2AF2, SLC39A9, RWDD2A, DPM1, UXT, TLK1, SDF2, TMEM59, GTF2A2, JAGN1, UBR4, NECAP2, AMZ2, TMCO1, WAS, DSCR3, TBCC, SEC61A1, C21orf59, ZBTB41, GMIP, E2F4, PEX2, POLR2H, MAD2L1BP, C11orf57, YEATS2, NXT2, CCDC25, UNC13D, MKI67IP, NDUFS1, PIGW, ZCCHC9, MUT, C17orf80, PHB2, ITFG1, UFC1, CBLL1, SDHAF2, UGGT1, HNRNPH1, SLC35A5, EIF1AD, GRPEL2, CEP164, ERH, LSM5, SHKBP1, VMP1, TMEM62, FTSJD1, COX6A1, BANF1, RPL27, FBXO38, MEA1, CKS1B, HARS2, LMAN2, MORN2, MRPL40, TOMM70A, ZNF184, MAP4K2, RHOA, PGS1, CALU, ZSCAN12, UFM1, TRMT112, TGDS, FANCF, RNF13, THUMPD3, COX17, ZFP3, RPL32, PALB2, MRPL33, LYL1, COX8A, EIF2S1, PITPNA, TXNDC12, PSME3, MRPL3 |
Terms that are bold and underlined represent upregulated genes.
Deregulated genes in patient-paired MRD vs diagnostic clonal PCs and their prognostic value according to mRNA expression levels in PCs from newly diagnosed patients treated according to TT3A (training set) and TT3B (validation set)
| Genes . | Expression in MRD vs diagnostic clonal PCs . | Expression in TT3 patients . | TT3A (training set) . | TT3B (validation set) . | ||
|---|---|---|---|---|---|---|
| Median (years) . | P value . | Median (years) . | P value . | |||
| Cancer related | ||||||
| FERMT3 | (↑) | >9.8 | NR | .03 | NR | .03 |
| ≤9.8 | NR | NR | ||||
| TSPO | (↑) | >11.55 | 7 | .06 | 3 | .0002 |
| ≤11.55 | NR | NR | ||||
| ALOX5 | (↑) | >12.39 | NR | .07 | NR | .11 |
| ≤12.39 | NR | NR | ||||
| NCF4 | (↑) | >10.58 | NR | .09 | 7 | .2 |
| ≤10.58 | NR | NR | ||||
| FLNA | (↑) | >10.56 | 7 | .0002 | 6 | .003 |
| ≤10.56 | NR | NR | ||||
| PREX1 | (↑) | >5.33 | NR | .02 | NR | .79 |
| ≤5.33 | 7 | NR | ||||
| PYCARD | (↑) | >9.37 | 9 | .001 | NR | .35 |
| ≤9.37 | NR | NR | ||||
| MYO1G | (↑) | >4.66 | NR | .01 | NR | .09 |
| ≤4.66 | 7 | NR | ||||
| SERPINI1 | (↓) | >9.85 | NR | .11 | NR | .47 |
| ≤9.85 | NR | NR | ||||
| DUSP11 | (↓) | >11.04 | NR | .06 | NR | .86 |
| ≤11.04 | NR | NR | ||||
| ALCAM | (↓) | >10.29 | NR | .0002 | NR | .02 |
| ≤10.29 | 5 | 6 | ||||
| CCNC | (↓) | >11.71 | NR | .07 | NR | .67 |
| ≤11.71 | 9 | NR | ||||
| COPZ1 | (↓) | >11.29 | NR | .38 | NR | .71 |
| ≤11.29 | NR | NR | ||||
| FZD3 | (↓) | >9.20 | NR | .01 | NR | .18 |
| ≤9.20 | 9 | NR | ||||
| E2F5 | (↓) | >9.92 | 9 | .10 | NR | .88 |
| ≤9.92 | NR | NR | ||||
| ER related | ||||||
| PDIA5 | (↓) | >11.76 | NR | .17 | NR | .10 |
| ≤11.76 | 9 | NR | ||||
| EMC7 | (↓) | >13.72 | 9 | .11 | 7 | .77 |
| ≤13.72 | NR | NR | ||||
| PI related | ||||||
| PSMD10 | (↓) | >10.68 | 9 | .003 | NR | .25 |
| ≤10.68 | NR | NR | ||||
| SMILE/TMTC3 | (↓) | >4.0 | NR | .08 | NR | .06 |
| ≤4.0 | 9 | 6 | ||||
| PSME3 | (↓) | >8.69 | NR | .03 | NR | .33 |
| ≤8.69 | NR | NR | ||||
| PSMB6 | >12.42 | 9 | .11 | NR | .07 | |
| (↓) | ≤12.42 | NR | NR | |||
| CAV1 | (↓) | >12.46 | 7 | .11 | NR | .43 |
| ≤ 12.46 | NR | NR | ||||
| Genes . | Expression in MRD vs diagnostic clonal PCs . | Expression in TT3 patients . | TT3A (training set) . | TT3B (validation set) . | ||
|---|---|---|---|---|---|---|
| Median (years) . | P value . | Median (years) . | P value . | |||
| Cancer related | ||||||
| FERMT3 | (↑) | >9.8 | NR | .03 | NR | .03 |
| ≤9.8 | NR | NR | ||||
| TSPO | (↑) | >11.55 | 7 | .06 | 3 | .0002 |
| ≤11.55 | NR | NR | ||||
| ALOX5 | (↑) | >12.39 | NR | .07 | NR | .11 |
| ≤12.39 | NR | NR | ||||
| NCF4 | (↑) | >10.58 | NR | .09 | 7 | .2 |
| ≤10.58 | NR | NR | ||||
| FLNA | (↑) | >10.56 | 7 | .0002 | 6 | .003 |
| ≤10.56 | NR | NR | ||||
| PREX1 | (↑) | >5.33 | NR | .02 | NR | .79 |
| ≤5.33 | 7 | NR | ||||
| PYCARD | (↑) | >9.37 | 9 | .001 | NR | .35 |
| ≤9.37 | NR | NR | ||||
| MYO1G | (↑) | >4.66 | NR | .01 | NR | .09 |
| ≤4.66 | 7 | NR | ||||
| SERPINI1 | (↓) | >9.85 | NR | .11 | NR | .47 |
| ≤9.85 | NR | NR | ||||
| DUSP11 | (↓) | >11.04 | NR | .06 | NR | .86 |
| ≤11.04 | NR | NR | ||||
| ALCAM | (↓) | >10.29 | NR | .0002 | NR | .02 |
| ≤10.29 | 5 | 6 | ||||
| CCNC | (↓) | >11.71 | NR | .07 | NR | .67 |
| ≤11.71 | 9 | NR | ||||
| COPZ1 | (↓) | >11.29 | NR | .38 | NR | .71 |
| ≤11.29 | NR | NR | ||||
| FZD3 | (↓) | >9.20 | NR | .01 | NR | .18 |
| ≤9.20 | 9 | NR | ||||
| E2F5 | (↓) | >9.92 | 9 | .10 | NR | .88 |
| ≤9.92 | NR | NR | ||||
| ER related | ||||||
| PDIA5 | (↓) | >11.76 | NR | .17 | NR | .10 |
| ≤11.76 | 9 | NR | ||||
| EMC7 | (↓) | >13.72 | 9 | .11 | 7 | .77 |
| ≤13.72 | NR | NR | ||||
| PI related | ||||||
| PSMD10 | (↓) | >10.68 | 9 | .003 | NR | .25 |
| ≤10.68 | NR | NR | ||||
| SMILE/TMTC3 | (↓) | >4.0 | NR | .08 | NR | .06 |
| ≤4.0 | 9 | 6 | ||||
| PSME3 | (↓) | >8.69 | NR | .03 | NR | .33 |
| ≤8.69 | NR | NR | ||||
| PSMB6 | >12.42 | 9 | .11 | NR | .07 | |
| (↓) | ≤12.42 | NR | NR | |||
| CAV1 | (↓) | >12.46 | 7 | .11 | NR | .43 |
| ≤ 12.46 | NR | NR | ||||
NR, not reached.
Because loss of ALCAM has been linked to more aggressive disease in different solid tumors, we further investigated its potential relationship with MM chemoresistance in vitro. ALCAM expression was found to be strong positive in the JJN3, KMS-11, and MM1S cell lines, dim positive in NCI-H929 and OPM2, and almost absent in the U266 and U266-LR7 cell lines (Figure 3B). RPMI-8266 MM cells showed heterogeneous ALCAM expression with both a negative and a positive subset; therefore, we used this latter cell line to investigate whether after drug exposure, chemoresistant (Annexin-V−ve) MM cells would become enriched with the ALCAM−ve subset compared with chemosensitive (Annexin-V+ve) PCs. Interestingly, the relative percentage of ALCAM− cells within total RPMI-8226 MM cells was significantly increased (due to selective decrease of the ALCAM+ve subset) among chemoresistant vs chemosensitive PCs cocultured with BMSCs and exposed in vitro to the combination of bortezomib, lenalidomide, dexamethasone, and melphalan for 24 or 48 hours (median of 57% vs 27%, respectively; P = .003; Figure 3D). In fact, the increment in ALCAM−ve cells was more pronounced when RPMI-8266 were exposed to all drugs administered simultaneously compared with each drug individually (supplemental Figure 3). Moreover, there was a trend toward progressively increased numbers of ALCAM−ve MM cells within chemoresistant (Annexin-V−ve) RPMI-8266 cells at baseline, after coculture with BMSCs, and after exposure to chemotherapy (medians of 41%, 50%, and 57%, respectively; P = .09; Figure 3E-F). A similar pattern was noted on comparing ALCAM expression in the chemoresistant KMS11/BTZ, U266-LR7, and MM1R cell lines vs their chemosensitive counterparts (KMS11, U266, and MM1S) at baseline and on coculture with BMSCs (supplemental Figure 4).
Discussion
Understanding MM genomic and molecular heterogeneity is required to develop treatment strategies that could target dominant but also subclonal reservoirs of chemoresistant cells. Although clonal heterogeneity at diagnosis and clonal tiding from diagnosis to patients’ relapse has been recently described,15-17,21,33 no attention has been paid to primary chemoresistant PCs that persist even among patients in serologic response (ie, MRD). Because persistence of MRD is strongly linked to an inferior survival,4-7 a better understanding of MRD clonal PCs is warranted to potentially overcome their chemoresistant phenotype. Here, we report for the first time the biological features of MRD cells in MM. Overall, our results revealed that chemoresistant clones are enriched in cells with a distinct phenotypic signature, but also that there is frequent tiding of genetically different subclones. Ultimately, MRD PCs have a deregulated GEP that allows them to survive after multidrug chemotherapy.
In MM, disease recurrence after initial (complete) response to treatment has been typically hypothesized to be associated with more immature progenitor cells that escape conventional antimyeloma therapy.18,34-36 Interestingly, although early studies suggested that persistent CD19+CD138− clonotypic B cells represented the MM cancer stem cell compartment and were responsible for disease relapse,34-36 more recent data indicate that CD138+ PCs have a complex architecture linked to interconvertible phenotypic and functional compartments with different chemoresistant potential.18,21,37 In particular, it has been suggested that mature CD138+ PCs give rise to more immature CD138lo pre-PCs/plasmablasts that secrete lower immunoglobulin levels (and are thereby less susceptible to ER stress)18 and could be specifically involved in disease dissemination.37 In line with this hypothesis, we recently showed that MM circulating tumor cells represent a singular phenotypic and functional subset of MM tumor cells characterized by lower CD138 expression22,38 Here, we used new analytic tools developed by the EuroFlow Consortium20 to simultaneously evaluate the pattern of expression of 23 markers at the single-cell level and longitudinally compare the iPEPs from diagnostic vs MRD clonal PCs. In contrast to the abovementioned hypothesis, our results demonstrate that MM cells with stronger capacity to survive frontline chemotherapy were those with the highest expression of CD138; because MRD clonal PCs are responsible for the disease relapse, these results point out the clonogenic potential of mature CD138+ PCs. Furthermore, MRD subclones showed enhanced expression of several integrins (eg, CD11a, CD11c, CD29, CD49d, and CD49e), chemokine receptors (eg, CXCR4), adhesion molecules (eg, CD44 and CD54), CD28 (a prosurvival mediator through PC-dendritic cell interaction39 ), and HLADR. The expression of these class of markers has been shown to be intrinsically related to different PC chemoresistant potential in vitro and in vivo,40-45 as well as to patients’ survival,46-48 which would be potentially due to a stronger attachment of MRD clonal PCs to the BM stroma, which provides higher protection from chemotherapy-induced apoptosis.49,50 In fact, it could be hypothesized that a subset of chemoresistant MRD cells strongly attached to the stroma might be potentially underestimated in BM aspirates. By contrast, a different role has been assigned to CD44 in cell adhesion-mediated drug resistance in several cancers including MM, where CD44 may partially contribute to dexamethasone resistance51 and becomes upregulated in lenalidomide-resistant MM cells to stabilize the expression of P-glycoprotein, a multidrug resistance efflux pump responsible for the cellular uptake of lenalidomide.13,52 Interestingly, differences between diagnostic vs MRD cells were more pronounced among patients treated only with VMP vs cases exposed to VMP and Rd. Altogether, these findings suggest that, from a phenotypic standpoint, chemoresistant PCs represent a uniquely defined subclone of all diagnostic tumor cells, but also that the more drugs being simultaneously used the higher the chances to debulk all different phenotypic tumor subclones. Further studies are warranted to confirm if the enhanced expression of integrins in MRD clonal PCs is biologically meaningful; in vitro, we noted that chemoresistant myeloma cell lines slightly increase their expression of CD11c, CD29, CD44, CD49d, and CD49e on short-term (24 hour) coculture with BMSCs (supplemental Figure 5).
Longitudinal comparison between the genomic profiles of PCs studied at diagnosis and during patient relapse has revealed 3 models of clonal dynamics: genetic stability over time, acquired CNAs at relapse, and heterogeneous fluctuation of different genetic clones from earlier to later time points.16,53 Here, we show that such genetic instability is already detectable at the MRD level, with 3 of 12 cases analyzed showing identical copy number profiles, whereas in the remaining patients there was a median of 4 different CNAs between diagnostic and MRD clonal PCs. However, it is unlikely that all the differences observed were acquired on treatment exposure; instead, it is most likely that a significant number of such CNAs are present at subclonal levels, but because the methodology used in the present study can only detect abnormalities that are present in 20% or more of clonal PCs cells,17 they may have gone undetected (supplemental Figures 6 and 7). Altogether, these findings are of high relevance because they illustrate that small subclones carrying unique cytogenetic abnormalities may become the predominant population after chemotherapy, and according to the specific type of genetic alteration acquired or lost, influence patients’ prognosis. Longitudinal analyses between diagnostic vs MRD vs relapse patient-paired samples are now warranted to understand if the clonal distribution of PCs at MRD mimics that of relapse, or if additional changes are observed between both time points. Accordingly, our observations suggest that monitoring of MM patients should become more comprehensive, not only gathering information on MRD levels to determine the depth of response, but also combining it with the cytogenetic characterization of MRD cells to better understand their malignant potential and identify potential therapeutic targets.21 In this regard, we also hypothesized that understanding the molecular signature of MRD cells could contribute to the discovery of novel genes/pathways that are responsible for drug resistance in individual patients. Thus, comparison of the GEP of patient-paired diagnostic vs MRD cells revealed >1000 deregulated genes, several of which are related to protein export, protein processing, and N-glycan biosynthesis. These findings are consistent with those reported by Leung-Hagesteijn et al,18 who showed that PCs with lower protein production have lower ER protein load, rendering them less vulnerable to lethal ER stress induced by proteasome inhibitors. In vitro studies have also reported similar observations in which attenuation of the eukaryotic initiation factor (eIF)-2α phosphorylation and ER stress were required for MM cells to survive proteasome inhibition10,54-56 ; accordingly, MRD clonal PCs showed significant downregulation of different eIF-related genes (supplemental Excel File 1). One potential caveat of the present study is the sole focus on clonal and mature PCs, because the MRD compartment may also reside among CD138− cells that are expected to have lower expression of ER genes.18 However, our experience on the comparison of MRD levels determined by multidimensional flow cytometry (that focus on the CD138+ PC compartment) vs molecular methods (that measure clonotypic VDJ rearrangements in whole BM samples) shows a highly significant correlation between the number of MRD cells detected by both techniques.4,6 Accordingly, if CD138− MRD “stem cells” exist, they would be present at levels <10−6 and therefore virtually impossible to identify and study using current technology.
The transcriptomic characterization of chemoresistant tumor cells might represent a different strategy to identify novel molecular pathways potentially related to patients’ outcome. Accordingly, mRNA expression levels in newly diagnosed MM patients of 3 of the top-deregulated genes in MRD clonal PCs (FERMT3, FLAN, and ALCAM) were of significant prognostic value, suggesting that patients with a PC compartment that at diagnosis was potentially enriched in chemoresistant cells have inferior overall survival. Noteworthy, such genes have been previously linked to more aggressive phenotypes in solid tumors but not in MM. In line with the singular iPEP of MRD clonal PCs potentially favoring their attachment to the BM stroma, FERMT3 and FLNA have been assigned a role in integrin activation and cell migration, respectively. Similarly, ALCAM has been implicated in cell migration and invasion in melanoma, as well as several other tumors including those of the prostate, esophagus, colon, bladder, pancreas, and lung.57 Interestingly, in an in vitro model where MM cells cocultured with BMSCs were exposed to the same drugs as the MRD clone from patients enrolled in the GEM2010MAS65 trial, a progressive accumulation of ALCAM−ve cells among chemoresistant cells was observed. Thus, the understanding of the molecular signature of MRD cells along with the investigation of whether these cells were detectable before treatment (intrinsic resistant cells) or not (acquired resistant cells) will hopefully provide novel insight into the field of chemoresistance in MM; that notwithstanding, such knowledge is most likely treatment dependent, because different molecular pathways will potentially emerge from the study of MRD cells after different treatment strategies. Thus, the results of the GEP comparison between patient-paired diagnostic and chemoresistant MRD clonal PCs, herein performed in a limited number of cases, should be interpreted as hypothesis generating rather than conclusive. This also applies for ALCAM, and further studies in larger series of patients are warranted to confirm this hypothesis.
In summary, by characterizing patient-paired diagnostic vs MRD clonal PCs, we unravel that therapy-induced clonal selection is already present at the MRD stage, in which chemoresistant myeloma PCs show a specific phenotypic signature that may result from the persistence of subclones with different cytogenetic and gene expression profiles. Characterization of the MRD clone may represent a unique model for a better understanding of chemoresistance and the characteristics of the cellular source of relapses after response to frontline therapy, as well as to ultimately design therapeutic strategies to overcome resistance already at the MRD stage.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all the investigators of the Spanish Myeloma Group participating in the PETHEMA/GEM2010MAS65 clinical trial.
This study was supported by Cooperative Research Thematic Network grants RD12/0036/0048, RD12/0036/0058, RD12/0036/0046, and RD12/0036/0069 of the Red de Cancer (Cancer Network of Excellence); Instituto de Salud Carlos III, Spain; Instituto de Salud Carlos III/Subdirección General de Investigación Sanitaria (FIS: PI060339; 06/1354; 02/0905; 01/0089/01-02; PS09/01897/01370; G03/136; Sara Borrell: CD13/00340); and Asociación Española Contra el Cáncer (GCB120981SAN), Spain. The study was also supported internationally by the International Myeloma Foundation (IMF) junior grant, the Black Swan Research Initiative of the IMF, the Multiple Myeloma Research Foundation research fellow award, the Qatar National Research Fund award 7-916-3-237, the American Association of Cancer Research–Millennium Fellowship in Multiple Myeloma Research (15-40-38-PAIV), and the European Research Council 2015 starting grant.
Authorship
Contribution: B.P., J.J.L., and J.F.S.M. conceived and designed the study; B.P., L.A.C., P.M., I.R., D.A., L.B., M.-L.S., P.B., A. Orfao, S.K.J., and J.E. developed the methodology; B.P., L.A.C., M.-B.V., P.M., I.R., D.A., M.-L.S., P.B., M.-A.E., M.T.H., R.G.-S., E.M.O., N.P., A. Oriol, M.G., L.P., F.D.A., Y.G., J.J.L., J.B., A. Orfao, S.K.J., J.E., B.B., M.-V.M., and J.F.S.M. acquired the data; B.P., L.A.C., P.M., A. Orfao, J.E., B.B., M.-V.M., and J.F.S.M. analyzed and interpreted the data; B.P. and J.F.S.M. wrote the manuscript; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the members of the Grupo Español de Mieloma/Programa para el Estudio de la Terapéutica en Hemopatías Malignas (GEM/PETHEMA) Cooperative Study Groups appears in “Appendix.”
Correspondence: Jesus F. San Miguel, Clinica Universidad de Navarra, Centro de Investigacion Médica Aplicada, Av. Pio XII 36, 31008 Pamplona, Spain; e-mail: sanmiguel@unav.es.
Appendix: study group members
The members of the Grupo Español de Mieloma/Programa para el Estudio de la Terapéutica en Hemopatías Malignas (GEM/PETHEMA) Cooperative Study Groups are: Ernesto Pérez Persona, Antonia Sampol Mayol, Joan Bargay Lleonart, Eugenia Abella Monreal, Joan Bladé Creixentí, Miguel Granell Gorrochategui, Albert Oriol Rocafiguera, Albert Altes Hernández, Elena Rámila Herrero, Mercedes Gironella Mesa, Anna Sureda Balari, Carmen Cabrera Silva, Francisco Javier Capote Huelva, José Luis Guzmán Zamudio, María Mas Esteve, Carmen Calle Primo, José Luis Bello López, Carlos Cerveró Santiago, Yolanda González Montes, Dunia de Miguel Llorente, María Asunción Echeveste Gutiérrez, Fernando Escalante Barrigón, Raquel de Paz Arias, María Jesús Blanchard Rodríguez, Juan José Lahuerta Palacios, Isabel Krsnik Castello, Rafael Martínez Martínez, Adrián Alegre Amor, Cristina Encinas Rodríguez, Juan José Gil Fernández, Carolina Bombín Canal, Pilar Bravo Barahona, Francisco Javier Peñalver Párraga, Rebeca Iglesias del Barrio, Jaime Pérez de Oteyza, Eugenio Giménez Mesa, José Ángel Hernández Rivas, Ana Paz Lafuente Guijosa, María Casanova Espinosa, José María Moraleda Jiménez, Felipe de Arriba de la Fuente, María Ángeles Goñi Herranz, Felipe Prósper Cardoso, Jesús F. San Miguel Izquierdo, Alexia Suárez Cabrera, Marivi Mateos Mateos, Miguel Teodoro Hernández García, Eulogio Conde García, José Mariano Hernández Martín, Eduardo Ríos Herranz, Jesús Martín Sánchez, José Luis Bueno Cabrera, Felipe Casado Montero, Javier de la Rubia Comos, Paz Ribas García, Aurelio López Martínez, Ana Isabel Teruel Casasus, María Ángeles Ruíz Guinaldo, Elena Amutio Díez, Monserrat Pérez Sánchez, Luis Palomera Bernal, and Pilar Delgado Beltrán.