Key Points
Cord blood content of ALDHbr cells correlates well with CFUs and may act as a surrogate potency assay for cord blood units.
ALDHbr cells in segments are assayed rapidly, allowing potency results to be used for release of the unit from a public cord blood bank.
Abstract
Banked, unrelated umbilical cord blood provides access to hematopoietic stem cell transplantation for patients lacking matched bone marrow donors, yet 10% to 15% of patients experience graft failure or delayed engraftment. This may be due, at least in part, to inadequate potency of the selected cord blood unit (CBU). CBU potency is typically assessed before cryopreservation, neglecting changes in potency occurring during freezing and thawing. Colony-forming units (CFUs) have been previously shown to predict CBU potency, defined as the ability to engraft in patients by day 42 posttransplant. However, the CFU assay is difficult to standardize and requires 2 weeks to perform. Consequently, we developed a rapid multiparameter flow cytometric CBU potency assay that enumerates cells expressing high levels of the enzyme aldehyde dehydrogenase (ALDH bright [ALDHbr]), along with viable CD45+ or CD34+ cell content. These measurements are made on a segment that was attached to a cryopreserved CBU. We validated the assay with prespecified criteria testing accuracy, specificity, repeatability, intermediate precision, and linearity. We then prospectively examined the correlations among ALDHbr, CD34+, and CFU content of 3908 segments over a 5-year period. ALDHbr (r = 0.78; 95% confidence interval [CI], 0.76-0.79), but not CD34+ (r = 0.25; 95% CI, 0.22-0.28), was strongly correlated with CFU content as well as ALDHbr content of the CBU. These results suggest that the ALDHbr segment assay (based on unit characteristics measured before release) is a reliable assessment of potency that allows rapid selection and release of CBUs from the cord blood bank to the transplant center for transplantation.
Introduction
Banked unrelated donor cord blood is an important source of allogeneic cells for hematopoietic stem cell transplantation (HSCT). Benefits of cord blood include rapid procurement, minimal risk to the donor, lower risk of graft-versus-host disease in the recipient, and more permissive HLA matching.1-5 Despite these benefits, delays in engraftment or primary graft failure are observed in 10% to 15% of patients after transplant.6-8 Many graft failures may be due to low potency, the ability of the cord blood unit (CBU) to restore hematopoiesis after HSCT.
CBUs are selected by transplant centers (TCs) using HLA matching along with CBU characteristics of potency measured before cryopreservation, including the total nucleated cell count (TNCC), myeloerythroid and mixed hematopoietic progenitor colony-forming units (CFUs), and CD34+ cell content, often adjusted for cell dose. Freezing, storage, and thawing of CBUs reduce the overall potency. Measurements taken before cryopreservation would not reflect potential insults that could affect the potency of the transplanted unit.9-11 Potency markers measured on a representative sample of the thawed CBU closer to the time of transplant would more accurately predict the capacity of the unit to engraft in the transplant setting. Previous work in our laboratory showed that CFUs measured on thawed CBUs are predictive of neutrophil engraftment and survival posttransplant.12 However, CFU assays are typically scored 2 weeks after the unit has been transplanted. Therefore, an assay is needed to assess the potency of the CBU before final selection and release to the TC.
Aldehyde dehydrogenase (ALDH) is highly expressed (ie, ALDH bright [ALDHbr]) in hematopoietic stem and progenitor cells.13-17 Flow-sorted ALDHbr CBU cells coexpress CD34 and CD133 antigens that are associated with primitive repopulating and progenitor cell functions.15 Furthermore, ALDHbr populations isolated from CBUs are highly enriched for CFUs, whereas ALDHdim cells possess little colony-forming ability.18 Transplant of purified ALDHbr human cord blood progenitors into immunodeficient, interleukin-2 receptor-γ null mice results in lymphoid and myeloid reconstitution.19 Accordingly, we developed a potency assay based on ALDHbr expression in cord blood cells.
For the past 10 years, it has been common practice at cord blood banks to heat-seal the tubing through which a cryobag is filled so that a series of blood-containing segments remain attached to the unit. These segments can be removed individually without compromising the integrity of the CBU and are used for confirmatory typing (CT) of the HLA match prior to final selection of a CBU for transplantation. Because HLA-typing requires only a small amount of blood, the remainder of blood in the segment is available for potency testing.
We now describe the development and validation of an in vitro flow cytometric assay measuring ALDHbr cell content of a CBU segment for evaluating the potency of a CBU during CT. We consider the CFU assay the best available laboratory surrogate for clinical engraftment; therefore, we prospectively compared the correlation of CFU content of 3908 segments from a public cord blood bank with several flow parameters. Our results suggest that the ALDHbr cell content of segments is the parameter that is most highly correlated with CFU. Therefore, ALDHbr content is likely to be the best parameter measured to predict the ability of a CBU to engraft.
Materials and methods
Control segment preparation
Fresh CBUs excluded from banking in the Carolinas Cord Blood Bank (CCBB) were processed to generate segments that served as assay controls. Hetastarch (6%; Hospira, Lake Forest, IL) was sterilely added to the cord blood collection bag at a 1:5 ratio. The cord blood bag was docked to a Sepax cell separation kit (CS-530.1; Biosafe Systems, Houston, TX) and processed on a Sepax-1 system using the program for cord blood volume reduction and red blood cell (RBC) depletion. Mononuclear cells were collected in a volume of 20 mL. The output bag was inserted into a Coolmix device (Biosafe), and 5 mL of dimethyl sulfoxide (DMSO; Akron Biotech, Boca Raton, FL) (55% DMSO in 5% dextran 40) was added (1 mL/min) using a Fusion One Syringe Infusion Pump (Chemyx, Stafford, TX).
Segments with ALDHbr cells (purified by magnetic-activated cell sorting [MACS] according to the manufacturer’s protocol) spiked into stocks of CBU cells were prepared for validation experiments. Fresh CBUs were processed using Ficoll-Paque Premium (GE Healthcare Life Sciences, Piscataway, NJ). Cells from the mononuclear layer were incubated with Fc receptor blocking reagent, CD34, and CD133 MicroBeads (100 μL each; Miltenyi Biotec, Bergisch Gladbach, Germany), and passed over a LS column (Miltenyi Biotec). MACS buffer (75 mL of MACS BSA Stock Solution added to 1450 mL of autoMACS Rinsing Solution (both Miltenyi Biotec) was used for washing and elution. The column pass-through, containing unbound cells, and the column elute, containing purified cells, were collected. The percentage of ALDHbr cells in each fraction was determined by flow cytometry using Aldecount and cell counts obtained with an XE-1000i Automated Hematology System (Sysmex America, Lincolnshire, IL). The eluate was spiked into the pass-through to create segments with specific ALDHbr values and cryopreserved for validation studies.
Segments were prepared using ethylene vinyl acetate tubing (provided by Medsep, Pall, Port Washington, NY), the same tubing used in CBU cryopreservation bags. Four-inch lengths of tubing were heat-sealed at one end. Processed cord blood was dispensed into the tubing using a syringe and a 16-gauge blunt needle (BD Biosciences, San Jose, CA), and segments were created by heat-sealing the tubing in thirds. The segments were inserted in cryotubes, placed into a Cryo 1°C Freezing Container (Nalgene, Rochester, NY), and placed in a −80°C freezer. The resulting freezing rate of 1°C/min simulated control-rate freezers. The following day, the segments were transferred to long-term storage in the vapor phase of liquid nitrogen (−196°C).
Thawing, washing, and staining of cord blood removed from CBU segments
After removal from liquid nitrogen storage, the segments were kept on dry ice until ready to be thawed. A segment was thawed by holding it between the fingers, and then it was wiped with alcohol and placed on a sterile gauze pad. The blood was aspirated into a syringe using a 25-gauge needle and dispensed into dextran-albumin (DA, 100 μL). DA was prepared by adding human serum albumin (25%; Octapharma, Hoboken, NJ) to dextran 40 (10% in 0.9% sodium chloride, Hospira) for a final concentration of 5% human serum albumin. All segments were processed before proceeding to the next steps. The blood-and-DA mixture was shaken to mix and then incubated at room temperature for 10 minutes, followed by step-wise DA additions, mixing, and incubations: 200 μL of DA for 3 minutes, 400 μL of DA for 3 minutes, and 200 μL of DA for 3 minutes.
An aliquot (120 μL) was removed and placed into a separate tube (CFU assay tube). The remaining sample was centrifuged at 2000g for 30 minutes at 4°C. The cell pellet was washed in 2 mL of Aldecount Assay Buffer (STEMCELL Technologies, Vancouver, BC, Canada) and recentrifuged at 250g for 5 minutes at 4°C. The cell pellet was resuspended in 0.5 mL of Aldecount buffer for ALDH staining (Figure 1), which was done according to the manufacturer’s instructions. The sample was added to an activated Aldecount Reagent tube (STEMCELL Technologies), and then 0.5 mL of sample was transferred to the diethylaminobenzaldehyde (DEAB) control tube. All tubes were incubated in a water bath at 37°C for 30 minutes.
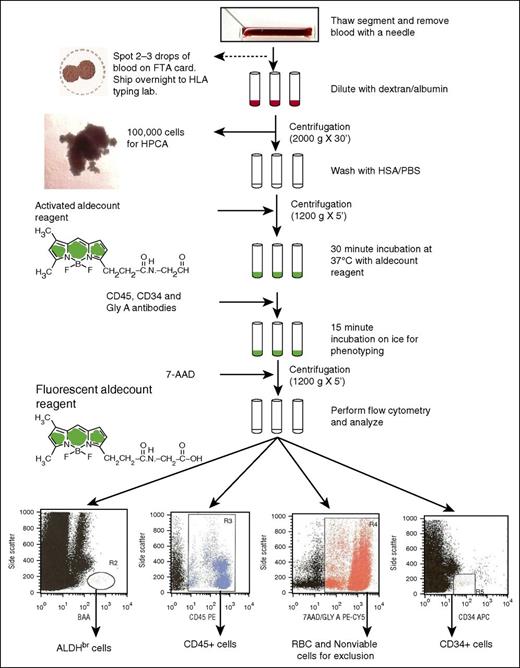
Flowchart of the ALDH potency assay performed on attached segments of CBUs requested for CT for donor selection. 7-AAD, 7-aminoactinomycin D; FTA, Fast Technology Analysis; GlyA, glycophorin A; HSA/PBS, human serum albumin/phosphate-buffered saline; HPCA, hematopoietic progenitor cell assay.
Flowchart of the ALDH potency assay performed on attached segments of CBUs requested for CT for donor selection. 7-AAD, 7-aminoactinomycin D; FTA, Fast Technology Analysis; GlyA, glycophorin A; HSA/PBS, human serum albumin/phosphate-buffered saline; HPCA, hematopoietic progenitor cell assay.
Multiparameter flow antibody staining
After the 37°C incubation, all of the tubes were placed in an ice bath. The Aldecount Reagent tubes were stained with the phenotyping antibodies anti-CD45-phycoerythrin (PE) (mouse, clone 5B1; Miltenyi Biotec), anti-GlyA-PE-Cy5 (diluted 100-fold from antibody stock, mouse, clone GA-R2; BD Biosciences), and anti-CD34-allophycocyanin (APC) (mouse, clone 8G12, BD Biosciences). The tubes were incubated in an ice bath for 15 minutes, followed by centrifugation (250g for 5 minutes at 4°C). The cell pellet in each tube was resuspended in Aldecount Assay buffer. Via-Probe viability solution (7-AAD, 10 μL; BD Bioscience) was added to each phenotyping tube prior to flow cytometry analysis.
Flow cytometry
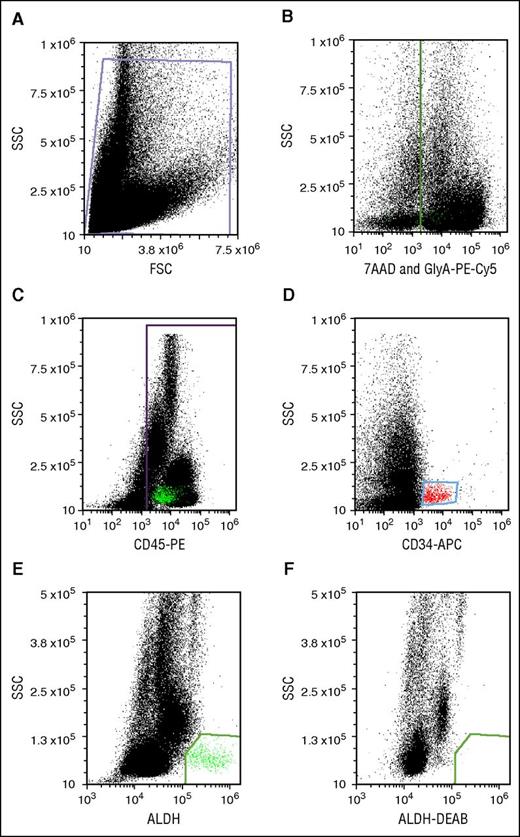
Flow cytometry was performed on an Accuri C6 digital flow cytometer (BD Biosciences) equipped with 488- and 635-nm lasers with appropriate color detectors/filters. Gates for antigen-expressing cells were set using appropriate isotype-specific antibody control tubes. For each sample, 100 000 viable CD45+ events were collected. Data were analyzed using FCS Express (De Novo Software, Glendale, CA). All dot plots were gated off R1 in a side-scatter vs forward-scatter light-scatter cytogram; Figure 2A). Lysis of RBCs in thawed segments caused high cell death and cell debris, which interfered with flow cytometric analysis. We circumvented the problem of RBC interference by staining RBCs with GlyA-PE-Cy5 and creating a dump gate. Detection of GlyA and the viability stain in the same channel (Figure 2B) allowed simultaneous exclusion of RBCs and nonviable cells. CD45+ (Figure 2C) and CD34+ populations (Figure 2D) were identified after excluding events in the dump gate. CD34+ cells were low in SSC and medium-bright for APC. CD45+ cells were spread across the SSC spectrum. ALDHbr cells were detected as a well-separated low-SSC and BODIPY aminoacetate–bright population (Figure 2E). The cells in this group were mostly CD34+ and CD45+, though characteristically dim in CD45-PE fluorescence. This cell population was eliminated with a specific ALDH inhibitor, DEAB (Figure 2F).
Potency assay gating. (A) Debris-free gate. (B) Gating of the nonviable (7-AAD+) and RBC (PE-Cy5-GlyA+) cells. (C) Gating of the PE-CD45+ cells. (D) Gating of APC-CD34+ cells. (E) Gating of ALDHbr cells. (F) Gating of cells stained with the Aldecount reagent in the presence of the inhibitor DEAB. BAA, BODIPY aminoacetate; FSC, forward scatter; SSC, side scatter.
Potency assay gating. (A) Debris-free gate. (B) Gating of the nonviable (7-AAD+) and RBC (PE-Cy5-GlyA+) cells. (C) Gating of the PE-CD45+ cells. (D) Gating of APC-CD34+ cells. (E) Gating of ALDHbr cells. (F) Gating of cells stained with the Aldecount reagent in the presence of the inhibitor DEAB. BAA, BODIPY aminoacetate; FSC, forward scatter; SSC, side scatter.
Plating and enumeration of postthaw CFU assay
An aliquot of cells from the CFU assay tube was diluted 1:10 in Cellpack solution (Sysmex America) and counted on an XE-1000i. The volume containing 1.0 × 105 cells was calculated and added to 2 mL of MethoCult (H4434; STEMCELL Technologies). The sample was mixed on a vortex mixer, allowed to sit for 2 minutes, and plated into 4 wells (0.4 mL each) in the middle of a 24-well plate. Sterile water was added to the outer wells for humidification. The plates were incubated at 37°C in a humidified incubator with 5% carbon dioxide for 14 days. CFUs, including erythroid burst-forming units, granulocyte-macrophage CFUs, and granulocyte-erythrocyte-macrophage-megakaryocyte CFUs were counted using a CKX41 inverted, phase-contrast microscope (Olympus America, Center Valley, PA) and a 40× objective, and reported as the mean colony count per 1.0 × 105 white blood cells. Total CFUs were calculated by adding the totals of the granulocyte-macrophage CFUs, granulocyte-erythrocyte-macrophage-megakaryocyte CFUs, and erythroid burst-forming units.
Validation of the potency assay
A validation plan for the ALDH potency assay was written according to the Harmonized Tripartite Guidelines Q2(R1)20 set by the International Conference on Harmonization, in consultation with regulatory authorities. Acceptance criteria were set for accuracy (ALDHbr within 20% of the expected value), repeatability (≤20% of the coefficients of variation [CVs] within a single operator on 3 replicates at 5 different concentrations), linearity (between the samples’ target and tested values of R2 ≥0.75), intermediate precision (CVs between operators, samples, and instruments needed to be ≤20%), and specificity (the number of ALDHbr cells detected within a contaminating background of cells was required to be within 20% of the expected value).
Assessment of accuracy, repeatability, and linearity
Commercially available control standards for measuring ALDHbr cells are not available; therefore, reference standards were created in our laboratory. For validating accuracy, repeatability, and linearity, standardized control segments reflecting a specified range of concentrations of ALDHbr cells (0.05%, 0.1%, 0.5%, 1.0%, and 3.0%) were prepared from sorted CD34+/CD133+ cells to mimic the range of values obtained on segments from banked CBUs. Three segments were assayed at each concentration for the percentage of ALDHbr cells in the viable CD45+ gate. Accuracy was determined by comparing the predicted ALDHbr percentage, calculated based on the number of ALDHbr cells added to each control sample, with the actual ALDHbr percentages obtained by analysis for each sample. These data were used to assess repeatability by a single operator at each of the concentrations, using the CV. Linearity was assessed by linear regression using the R2 statistic, comparing the target ALDHbr and actual values.
Assessment of intermediate precision
Intermediate precision was measured by 2 operators, each testing segments prepared from 5 CBUs on 2 Accuri C6 flow cytometers on 6 different days. All test samples consisted of segments prepared for the validation from randomly chosen CBUs. Segments were stored in liquid nitrogen to mirror storage of the actual segments on a CBU. Variability was assessed by comparing the ALDHbr (as a percentage of viable CD45) from each sample. The CVs were reported for segments from each CBU as a whole for both operators and instruments, and by assessing operators and instruments separately.
Assessment of specificity
ALDHbr cells make up <1% of total nucleated cells in a typical CBU. The primary impurities in the CBU segment are other cell types that are present in CBUs (eg, RBCs and monocytes that express low levels of ALDHbr). To evaluate the impact of these impurities, the ability to measure 5000 ALDHbr cells in increasing numbers of contaminating cells was evaluated. ALDHbr cells (20 000 within a population of CD34+/CD133+-selected cells) were spiked into 1 × 106, 3 × 106, 5 × 106, and 10 × 106 nucleated cord blood cells taken from the CD34/CD133-negative fraction. Triplicate samples were run at each concentration. Results were reported as number of ALDHbr cells detected.
Assaying thawed or fresh units
Before cryopreservation, 100 μL of fresh blood taken from a CBU was diluted in 4 mL of Aldecount Lysis Buffer (Aldagen, Durham, NC). After a 15-minute incubation at 37°C, the sample was centrifuged at 250g for 5 minutes at 4°C, washed with 2 mL of Aldecount Assay Buffer, and recentrifuged. The sample was then stained for ALDHbr, CD45, 7-AAD, and CD34.
Cryopreserved CBUs selected for assaying were thawed with the modified procedure used for preparation of units for transplant.12 The washed cells were resuspended in dextran 40/5 albumin in a final volume of 50 mL. Aliquots (100 μL) were diluted in 2 mL of Aldecount Assay Buffer prior to centrifugation (250g for 5 minutes at 4°C), with staining as described earlier for segments.
Correlation of segment potency with engraftment
A search of our clinical database identified 78 single-cord transplants performed at Duke University Medical Center using single-CBU grafts that represented either the initial or the only transplant for that patient. Characteristics measured on segments from the transplanted units were examined to determine whether they were predictive of neutrophil engraftment (3 consecutive days of absolute neutrophil count >500/μL), using plots of the cumulative incidence function of each segment biomarker and differences determined by Gray’s test. Death before engraftment (1 patient) and death before graft failure (4 patients) were treated as competing risks.
Results
We developed and validated a segment-based rapid assay to determine the potency of cryopreserved CBUs selected for consideration for HSCT before release from a US Food and Drug Administration–licensed public cord blood bank. The flow cytometric–based assay enumerates ALDHbr, CD34+, CD45+, and viable cells measured on a thawed segment attached to a CBU. A validation plan addressing accuracy, repeatability, linearity, intermediate precision, and specificity was created and executed.
Accuracy, repeatability, and linearity
Segments containing cord blood with 5 known percentages of ALDHbr (3 segments per percentage) were assayed and analyzed for accuracy, repeatability, and linearity. At each test concentration, ALDHbr measured by the assay fell within 20% of the expected value as required by our predefined validation specification for accuracy (Table 1). The CVs for repeatability, indicating a single operator’s ability to achieve similar results on multiple replicates at each concentration, were well within specifications for acceptability, ranging from 1.2% to 7.5%, and there was no clear relationship between CV and concentration of ALDHbr. Results of the linear regression analysis showed excellent linearity between ALDHbr measured by the assay and the true concentration (R2 = 0.9878).
Statistics for segments used for accuracy, linearity, and repeatability
| Test ALDHbr concentration, % . | Actual ALDHbr concentration, % . | Mean . | Standard deviation . | CV . | Meets specifications? . | ||
|---|---|---|---|---|---|---|---|
| Segment 1 . | Segment 2 . | Segment 3 . | |||||
| 0.05 | 0.052 | 0.054 | 0.049 | 0.051 | 0.002 | 3.3 | Yes |
| 0.10 | 0.108 | 0.108 | 0.105 | 0.106 | 0.001 | 1.2 | Yes |
| 0.50 | 0.524 | 0.535 | 0.481 | 0.513 | 0.023 | 4.5 | Yes |
| 1.00 | 0.974 | 1.101 | 1.133 | 1.069 | 0.069 | 6.4 | Yes |
| 3.00 | 2.692 | 3.084 | 2.596 | 2.791 | 0.211 | 7.5 | Yes |
| Test ALDHbr concentration, % . | Actual ALDHbr concentration, % . | Mean . | Standard deviation . | CV . | Meets specifications? . | ||
|---|---|---|---|---|---|---|---|
| Segment 1 . | Segment 2 . | Segment 3 . | |||||
| 0.05 | 0.052 | 0.054 | 0.049 | 0.051 | 0.002 | 3.3 | Yes |
| 0.10 | 0.108 | 0.108 | 0.105 | 0.106 | 0.001 | 1.2 | Yes |
| 0.50 | 0.524 | 0.535 | 0.481 | 0.513 | 0.023 | 4.5 | Yes |
| 1.00 | 0.974 | 1.101 | 1.133 | 1.069 | 0.069 | 6.4 | Yes |
| 3.00 | 2.692 | 3.084 | 2.596 | 2.791 | 0.211 | 7.5 | Yes |
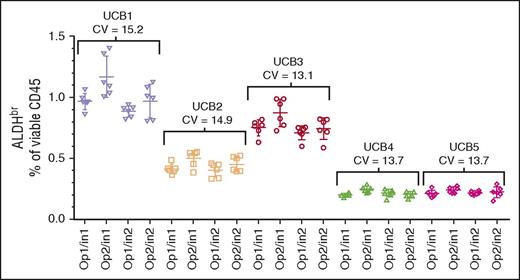
Intermediate precision
The segments from 5 CBUs were assayed by 2 operators on 2 cytometers on 6 different days, analyzed separately, and collated. The data were reanalyzed with respect to overall sample precision across both operators and instruments (Figure 3; CVs, 13.1%-15.2%). In addition, intraoperator precision (data not shown; CVs, 10.2%-15.2%), and intrainstrument precision (data not shown; CVs, 7.5%-16.7%) were analyzed. All analyses met the ≤20% CV acceptance criteria. Because the individual CBUs used in this experiment were chosen randomly, comparisons between the CBUs were not performed.
Intermediate precision assessment. CV of segments from 5 CBUs (UCB1-UCB5) analyzed over 6 days using 2 operators (Op1 and Op2) and 2 Instruments (In1 and In2). The target specification was a CV ≤20%.
Intermediate precision assessment. CV of segments from 5 CBUs (UCB1-UCB5) analyzed over 6 days using 2 operators (Op1 and Op2) and 2 Instruments (In1 and In2). The target specification was a CV ≤20%.
Specificity
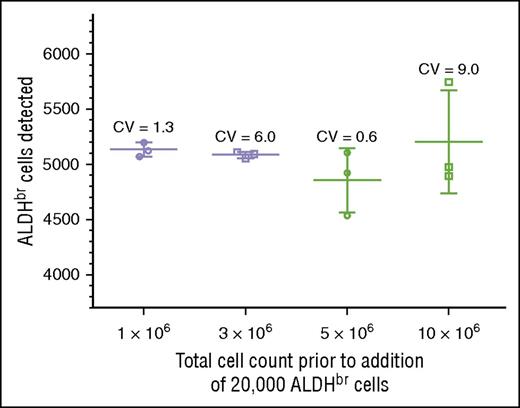
We tested specificity by determining whether we could detect 5000 ALDHbr cells spiked into varying concentrations of cord blood mononuclear cells. Acceptance was scored by detecting between 4000 and 6000 ALDHbr cells in a background of 1 million to 10 million contaminating cells. This criterion was met at all levels of cell background, with CVs ranging from 0.6% to 9.0% (Figure 4). The largest variation (9.0%) was seen in the samples with the highest background contamination, in which the ALDHbr concentration was 100-fold less than that seen in routine clinical samples. In the range of typical CBU samples, the CVs were much tighter, ranging from 0.6% to 1.3%. RBC contamination was low in these samples due to the use of Ficoll.
Validation of specificity. Purified ALDHbr cells (20 000) were spiked into varying amounts of CBU cells (1 × 106, 3 × 106, 5 × 106, and 10 × 106). Segments were created and cryopreserved. The segments were subsequently thawed and analyzed with the potency assay, with a target of collecting 5000 ALDHbr cells. The target of 4000 to 6000 cells was met at all concentrations.
Validation of specificity. Purified ALDHbr cells (20 000) were spiked into varying amounts of CBU cells (1 × 106, 3 × 106, 5 × 106, and 10 × 106). Segments were created and cryopreserved. The segments were subsequently thawed and analyzed with the potency assay, with a target of collecting 5000 ALDHbr cells. The target of 4000 to 6000 cells was met at all concentrations.
Experience with the ALDHbr potency assay in a public cord blood bank: can the ALDHbr assay substitute for the CFU assay?
Since February 2010, we performed the potency assay on segments from CBUs requested for CT in the CCBB. Segments were detached from the CBU in vapor phase, thawed, and a small aliquot spotted onto an FTA card and sent to the National Marrow Donor Program reference laboratory for CT. The remaining blood was used for the potency assay. Over a 5-year period, 3908 segments were tested for ALDHbr, CD45+, and CD34+ cells; viability; and CFUs. We detected a range of ALDHbr cells within the CD45+ viable population (0.00%-2.30%; median, 0.37%). CD45+ cells had a median cell viability of 66.10% (range, 8.72%-99.02%). Of the viable CD45+ cells, 0.66% (range, 0.00%-3.49%) coexpressed CD34 (Table 2).
Data obtained of cell subsets as determined by flow cytometric analysis of the potency assay and CFUs
| . | Median . | Minimum . | Maximum . |
|---|---|---|---|
| ALDHbr as a percentage of viable CD45+, % | 0.37 | 0.00 | 2.30 |
| CD34+ as a percentage of viable CD45+, % | 0.66 | 0.00 | 3.49 |
| CD45+ viability, % | 66.10 | 8.72 | 99.02 |
| CD34+ viability, % | 82.90 | 21.37 | 100 |
| Mean total CFUs (per 1 × 105 white blood cells) | 46.25 | 0 | 530 |
| . | Median . | Minimum . | Maximum . |
|---|---|---|---|
| ALDHbr as a percentage of viable CD45+, % | 0.37 | 0.00 | 2.30 |
| CD34+ as a percentage of viable CD45+, % | 0.66 | 0.00 | 3.49 |
| CD45+ viability, % | 66.10 | 8.72 | 99.02 |
| CD34+ viability, % | 82.90 | 21.37 | 100 |
| Mean total CFUs (per 1 × 105 white blood cells) | 46.25 | 0 | 530 |
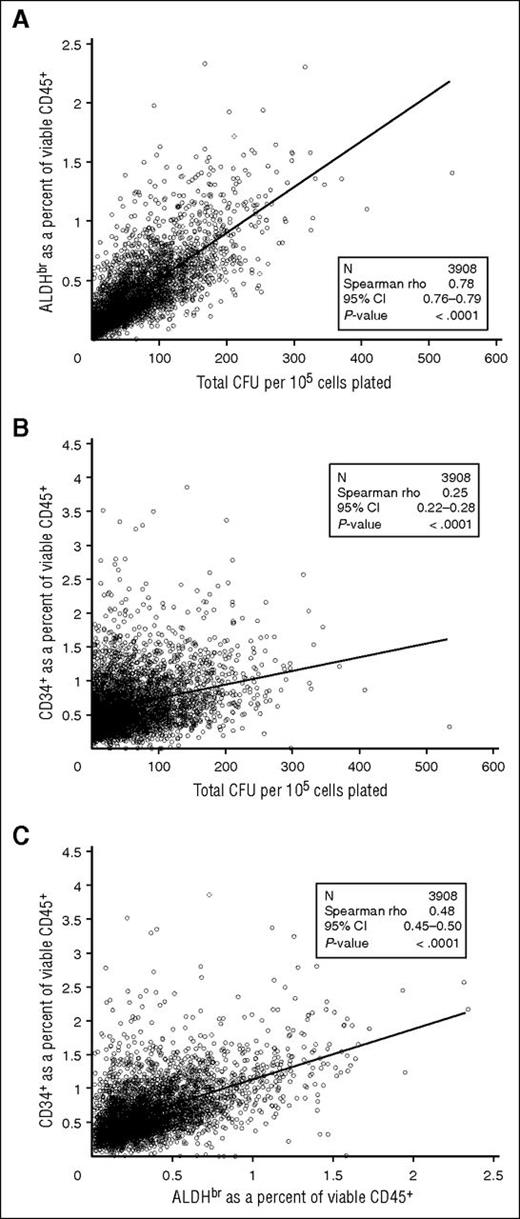
We examined the correlation of ALDHbr cells to CFUs in the segments tested. ALDHbr (as a percentage of viable CD45+ cells) correlated strongly with total CFU (per 105 cells) (Spearman r = 0.78; 95% confidence interval [CI], 0.76-0.79; Figure 5A). However, the correlation between CD34+ (as a percentage of viable CD45+ cells) and CFU was substantially weaker (r = 0.25; 95% CI, 0.22-0.28; Figure 5B). ALDHbr correlated more modestly with CD34+ (r = 0.48; 95% CI, 0.45-0.50; Figure 5C). The differences in correlation likely result from ALDHbr and CFU needing metabolically active cells, whereas CD34+ is an antibody-based test. These correlations with CFUs were consistent regardless of how long the cryopreserved CBUs were stored, suggesting that the potency of properly stored CBUs is stable over at least 15 years (Table 3).
Comparison of cellular components of CT segments. (A) ALDHbr in viable CD45+ vs CFUs. (B) CD34+ in viable CD45+ vs CFUs. (C) ALDHbr in viable CD45+ vs CD34+ in viable CD45+. N = 3908.
Comparison of cellular components of CT segments. (A) ALDHbr in viable CD45+ vs CFUs. (B) CD34+ in viable CD45+ vs CFUs. (C) ALDHbr in viable CD45+ vs CD34+ in viable CD45+. N = 3908.
Relationship of ALDHbr and viable CD34+ to CFUs with time in storage
| . | Spearman r (P) . | |||
|---|---|---|---|---|
| All samples, N = 3908 . | Storage duration, y . | |||
| <5, n = 3188 . | 5-10, n = 579 . | >10, n = 141 . | ||
| ALDHbr as a percentage of viable CD45+ | 0.78 (<.0001) | 0.78 (<.0001) | 0.78 (<.0001) | 0.77 (<.0001) |
| CD34+ as a percentage of viable CD45+ | 0.25 (<.0001) | 0.25 (<.0001) | 0.25 (<.0001) | 0.29 (.0004) |
| . | Spearman r (P) . | |||
|---|---|---|---|---|
| All samples, N = 3908 . | Storage duration, y . | |||
| <5, n = 3188 . | 5-10, n = 579 . | >10, n = 141 . | ||
| ALDHbr as a percentage of viable CD45+ | 0.78 (<.0001) | 0.78 (<.0001) | 0.78 (<.0001) | 0.77 (<.0001) |
| CD34+ as a percentage of viable CD45+ | 0.25 (<.0001) | 0.25 (<.0001) | 0.25 (<.0001) | 0.29 (.0004) |
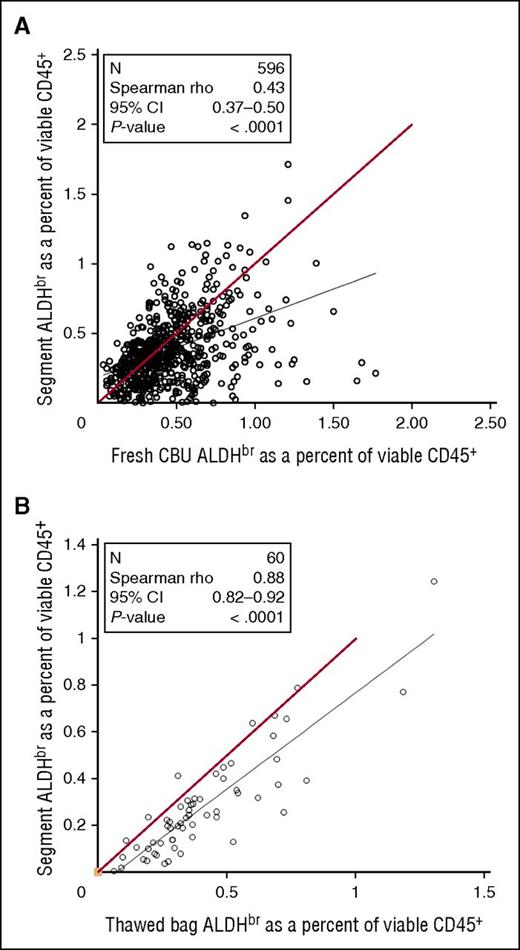
Between July 2007 and August 2009, samples from fresh CBUs before cryopreservation were assayed for ALDHbr. As the CT was done, segments frozen with those CBUs were tested for ALDHbr. Comparison of the data from fresh CBUs and frozen segments (Figure 6A) revealed a weak correlation (Spearman r = 0.43), with segments tending to have a lower ALDHbr content. To determine the correlation of the potency assay between cryopreserved segments and bags, research units from the CCBB were randomly selected and the entire unit was thawed and then assayed along with the distal segment that would normally be assayed in for CT (Figure 6B). Although there was a strong overall correlation between segments and units (Spearman r = 0.88), segments still tended to underestimate the ALDHbr content of the unit.
Comparison of CT segments in fresh or thawed CBUs. (A) Samples taken from fresh CBUs before cryopreservation between July 2007 and August 2009 were assayed for ALDHbr and compared with the results of the CT assay of segments from those units. N = 596. Red line indicates theoretical equality. (B) Research CBUs stored at the CCBB were selected. The bags were thawed with the segments and tested for ALDHbr. N = 60. Red line indicates theoretical equality.
Comparison of CT segments in fresh or thawed CBUs. (A) Samples taken from fresh CBUs before cryopreservation between July 2007 and August 2009 were assayed for ALDHbr and compared with the results of the CT assay of segments from those units. N = 596. Red line indicates theoretical equality. (B) Research CBUs stored at the CCBB were selected. The bags were thawed with the segments and tested for ALDHbr. N = 60. Red line indicates theoretical equality.
ALDHbr impact on engraftment
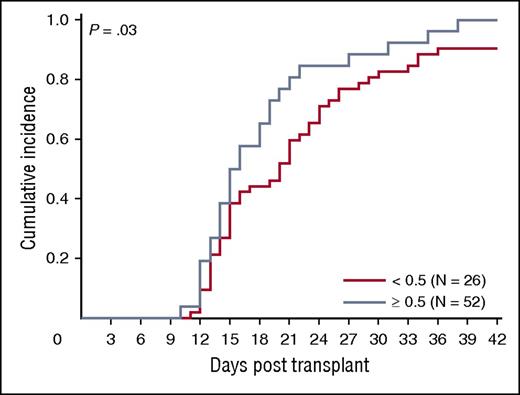
We asked whether ALDHbr, CD34, and CFUs from the segment potency assay correlated with engraftment in a cohort of 78 patients transplanted at Duke University Medical Center with a single CBU. Ninety-four percent (73/78) of patients engrafted (18 malignant and 55 nonmalignant diagnoses). The median time to engraftment was 16 days (range, 10-38 days). The association between segment-based ALDHbr and CFUs in this subset of segments was similar to that observed in the entire sample of segments included in our study (r = 0.70; P < .001). We observed a trend toward faster engraftment with increased expression of ALDHbr (P = .03; Figure 7), although caution is warranted in interpretation of this result due to small subgroup sizes. CFUs and CD34 did not predict neutrophil engraftment in the entire cohort (P > .05 for all; data not shown).
ALDHbr impact on engraftment. Impact of ALDHbr measured on segments during CT on engraftment of the corresponding unit. Probability plots are shown for the units with an ALDHbr >0.5% of viable CD45+ or the units with an ALDHbr <0.5% of viable CD45+ in reaching an absolute neutrophil count of 500/μL. N = 78.
ALDHbr impact on engraftment. Impact of ALDHbr measured on segments during CT on engraftment of the corresponding unit. Probability plots are shown for the units with an ALDHbr >0.5% of viable CD45+ or the units with an ALDHbr <0.5% of viable CD45+ in reaching an absolute neutrophil count of 500/μL. N = 78.
Discussion
Our results demonstrate the successful development of a segment-based potency assay that meets the International Conference on Harmonization guidelines for accuracy, specificity, precision, repeatability, and linearity. Furthermore, we have demonstrated through a prospective evaluation of almost 4000 segments from cryopreserved CBUs that the assay (1) is highly correlated with CFUs, which have been previously shown to best predict neutrophil engraftment after umbilical cord blood transplant; and (2) can be efficiently integrated into routine cord blood banking release procedures during the CT process before final selection and release of the CBU to the TC.
Currently, CBUs are selected3,21-23 by a TC after review of the precryopreservation TNCC, HLA matching, and/or CD34+ cell content. These data fail to reflect insults to the CBU caused by freezing and thawing of the unit. Therefore, we focused on postthaw CBU characteristics or surrogates for these measures. We previously reported that postthaw CFU content best correlated with engraftment for both neutrophils and platelets.12,24 Although postthaw CFU content showed the strongest correlation with engraftment, it is not feasible to use this parameter for CBU selection because it is generally scored 14 days after plating the assay.
The established correlation of CFU and ALDHbr cells in fresh CBU and the availability of segments at the time of CBU selection led us to develop and optimize the ALDHbr assay on thawed CBU segments. This approach posed some technical challenges that had to be resolved for successful measurement of potency. To maintain cell viability and protect the cells from DMSO cytotoxicity during thawing, we followed a DA wash protocol based on a method by Rubinstein et al,25 which emulates CBU washing for transplantation. We demonstrated that the temperature during this step was critical and that cells and DA needed to be at room temperature to avoid loss of detectable ALDHbr cells. This methodology is in contrast to protocols applied to CBUs that require washing the cells with ice-cold DA.
In these studies, we also observed that the ALDHbr content of the segments correlates well with the CFU content. This finding strongly suggests that measurement of ALDHbr can serve as a surrogate for the CFU assay. The stronger correlation between segments and frozen units, as opposed to fresh units, likely means that cryopreservation impacts both segments and units, with a possible greater impact on the segments. This means that the segment assay may disqualify some units with adequate potency from being used. However, the segment assay is highly unlikely to identify low-potency units as high-potency units. Because the potency assay data are available at the time of final donor selection, it can be used as a prospective potency assay for selecting CBUs. Although confirmatory studies are needed, our data suggest that CBUs rich in ALDHbr cells are predictive of engraftment, which would be expected because this enzyme is characteristically highly expressed in the hematopoietic stem and progenitor cells from human umbilical cord blood,15,26 bone marrow,27,28 and mobilized peripheral blood.29,30
Other methods to assess engraftment potential of CBUs have been reported in the literature. Higher postthaw CD34+ viability has been correlated with the dominant unit after double-CBU transplantation, and low CD34+ viability is associated with an increased risk of graft failure.31,32 Dominant-unit CD34+-infused cell dose has been found to predict engraftment after double-unit transplantation.33 In addition, the use of the HALO adenosine triphosphate bioluminescence assay on segments and units indicates that potency assessment on the mononuclear cell fraction may be more robust and less variable than on the TNCC fraction.34 Further refining of the CFU assay to decrease the length of the assay is also under investigation.35,36 The creation of the Cord Blood Apgar scoring system, which utilizes precryopreservation and postthaw graft variables (TNCC, mononuclear cell, CFU, CD34+, and volume), showed that a composite scoring system could be used to optimize selection of CBUs for transplantation.37
The present studies demonstrated rapid, reliable measurement of ALDHbr cell content in thawed CBU segments by flow cytometry. The assay requires a small quantity of blood from a segment, and the integrity of the entire unit is unaffected. The assay is performed in less than 4 hours, providing an important advantage over the traditional CFU assay. Implementation of this complex assay in other cord blood banks will require focused technology transfer with targeted training and validation to ensure optimal performance of the assay. Additional studies to examine correlations with engraftment in clinical transplantation are under way to further validate this potency assay for use in selecting CBUs for transplantation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the staff of the Carolinas Cord Blood Bank and the Duke Stem Cell Laboratory for providing and testing CBUs used in the experiments described in this manuscript.
This project was partially supported by Health Resources and Services Administration, National Cord Blood Inventory grant HHSH-234007002C 07-N234-5754 and by National Heart, Lung, and Blood Institute, National Institutes of Health grant K23HL104575. Additional support was provided by the Robertson Foundation and the National Marrow Donor Program.
Authorship
Contribution: K.S. took the lead role in adapting the ALDHbr assay for use with segments during CT, performed much of the assays and data analysis, and wrote the first draft of the manuscript; P.N. performed many of the assays and helped with data analysis and editing of the manuscript; J.D.T. provided statistical analysis of the data and advice for the validation; L.C. and A.P. provided support in the design, analysis, and interpretation of the data for the validation studies; K.P. contributed to the conception of the assay method, interpretation of the data for clinical use, and editing of the manuscript; T.G. and A.E.B. worked on developing the original ALDHbr assay, contributed to the design of the CT assay and the validation, and assisted in drafting and editing the manuscript; and J.K. was instrumental in the conception, design, and interpretation of the studies presented here and in provided editing and critical revisions of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kevin Shoulars, Duke University Medical Center, 1400 N. Pavilion Bldg, Durham, NC 27710; e-mail: kevin.shoulars@dm.duke.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal