Key Points
RHOA mutations are common in ATLL and show a unique distribution compared with other T-cell lymphomas.
Depending on patients, functionally distinct RHOA mutations are clonally selected and involved in the pathogenesis of ATLL.
Abstract
Adult T-cell leukemia/lymphoma (ATLL) is a distinct form of peripheral T-cell lymphoma with poor prognosis, which is caused by the human T-lymphotropic virus type 1 (HTLV-1). In contrast to the unequivocal importance of HTLV-1 infection in the pathogenesis of ATLL, the role of acquired mutations in HTLV-1 infected T cells has not been fully elucidated, with a handful of genes known to be recurrently mutated. In this study, we identified unique RHOA mutations in ATLL through whole genome sequencing of an index case, followed by deep sequencing of 203 ATLL samples. RHOA mutations showed distinct distribution and function from those found in other cancers. Involving 15% (30/203) of ATLL cases, RHOA mutations were widely distributed across the entire coding sequence but almost invariably located at the guanosine triphosphate (GTP)-binding pocket, with Cys16Arg being most frequently observed. Unexpectedly, depending on mutation types and positions, these RHOA mutants showed different or even opposite functional consequences in terms of GTP/guanosine diphosphate (GDP)-binding kinetics, regulation of actin fibers, and transcriptional activation. The Gly17Val mutant did not bind GTP/GDP and act as a dominant negative molecule, whereas other mutants (Cys16Arg and Ala161Pro) showed fast GTP/GDP cycling with enhanced transcriptional activation. These findings suggest that both loss- and gain-of-RHOA functions could be involved in ATLL leukemogenesis. In summary, our study not only provides a novel insight into the molecular pathogenesis of ATLL but also highlights a unique role of variegation of heterologous RHOA mutations in human cancers.
Introduction
Adult T-cell leukemia/lymphoma (ATLL) is a distinct form of peripheral T-cell lymphomas (PTCLs) caused by human T-lymphotropic virus type 1 (HTLV-1). It originates from T cells immortalized by HTLV-1 infection during early infancy. Although HTLV-1 can effectively immortalize T cells, there is a long latency period of 30 to ∼50 years prior to the onset of ATLL, suggesting that HTLV-1 infection alone may not be sufficient for the development of ATLL, but that additional acquired genetic hits are thought to be essential for its pathogenesis.1,2 In this regard, mutations in several genes, such as TP53,3 FAS,4 ZEB1,5 and CCR4,6 have been implicated in the latter process. However, the entire sequence of genetic events that shape an ATLL genome is still unclear. In this study, we first identified a RHOA mutation (Gly17Val) combined with a TET2 mutation in a case of ATLL using whole genome sequencing (WGS), which has been closely implicated in angioimmunoblastic T-cell lymphoma (AITL) and other PTCL not otherwise specified (PTCL-NOS). However, a subsequent large-scale analysis disclosed a very different nature of RHOA and TET2 mutations in ATLL cases, in terms of their distributions and functional consequences. We describe unique features of RHOA mutations in ATLL in comparison with other cancers, including AITL and PTCL-NOS together with their functional implications.
Methods
Tumor specimens
A total of 203 patients with different ATLL subtypes, including 74 acute, 54 lymphoma, 52 chronic, 6 smoldering, and 17 unknown types, were included in this study after written informed consent was obtained. Survival data were available for 113 of the 203 patients. This study was approved by the Institutional Review Board at Kyoto University and conducted in accordance with the Declaration of Helsinki. Tumor DNA was extracted from bone marrow, peripheral blood, and lymphoid organs. Genomic DNA samples from buccal mucosa were also obtained for 38 patients and used as germline controls.
WGS
DNA samples were prepared for paired-end DNA Sample Prep kit (Illumina) according to the manufacturer’s protocol with some modifications. Sequence data were generated using the Illumina Hisequation 2000 platform in 2×100 paired-end reads. Data processing, variant calling, copy number estimation, and structural alteration detection were performed as described previously.7 All somatic mutations in protein coding regions were validated by deep sequencing (see supplemental Table 1 on the Blood Web site). The results of the analysis were integrated and visualized using Circos.8
Targeted deep sequencing
Entire coding exons of RHOA, TET2, and DNMT3A, and mutational hotspots of IDH1/2 were polymerase chain reaction-amplified using NotI-tagged primers (supplemental Table 2) and subjected to high-throughput random sequencing using Hisequation 2000. Mutation calling was performed as previously described,2 where the data from 95 normal individuals were used to eliminate single nucleotide polymorphisms (SNPs). All candidate variants were validated by Sanger sequencing or independent deep sequencing using non-amplified DNA.
SNP array-karyotyping
Mutagenesis and vector construction
Plasmids bearing human wild-type (WT), Gly14Val, Gly17Val, and Ala161Glu mutant RHOA complementary DNA were described previously.2 Mutagenesis to create constructs encoding the Cys16Arg, Cys16Phe, Lys118Glu, Ala161Val, and Arg161Pro mutants was carried out with the PrimeSTAR Mutagenesis Basal kit (Takara) according to the manufacturer’s protocol. All constructs were verified by Sanger sequencing. These constructs were subcloned into the tetracycline-inducible lentivirus-based expression vector CS-TRE-PRE-Ubc-tTA-I2G7,11 the pEF-neo expression vector,2 and the pGEX-6P-1 vector (GE Healthcare). In addition, the last four amino acids corresponding to the CAAX motif were deleted for the pGEX-6P-1 vector.
Biochemical analyses of recombinant RHOA proteins
Purification and biochemical characterization of RHOA proteins were performed as described previously with minor modifications.12 Glutathione S-transferase-fusion proteins of WT and mutant RHOA were induced with 0.1 mM isopropyl-1-thio-β-d-galactopyranoside at 20°C for 16 hours in Escherichia coli BL21-CodonPlus DE3 (Stratagene) and purified with glutathione-Sepharose 4B beads (GE Healthcare Life Sciences). For measuring the guanosine triphosphate (GTP)-binding properties of Gly17Val and Ala161Glu RHOA mutants, the protein concentrations were estimated based on the total amount of proteins, because most of them were present in a nucleotide-free form. In other guanine nucleotide exchange assays, the active protein concentrations were estimated based on the amounts of proteins bound to the guanine nucleotides. For [35S]GTPγS-binding assays, purified glutathione S-transferase-fusion proteins (5 pmol) were incubated with 5 µM of the radiolabeled nucleotide (4000 cpm/pmol). After incubation for the indicated periods, samples were diluted, and filtered through a nitrocellulose membrane (0.45-µm pore size; Advantech MFS, Dublin, CA). The membrane was washed 4 times and the radioactivity retained on the membrane was determined by a liquid scintillation counter.
For [35S]GTPγS- and [3H] guanosine diphosphate (GDP)-dissociation assays, purified proteins (1.5 pmol) were pre-incubated with 5 µM of the radiolabeled nucleotides (4000 cpm/pmol for [35S]GTPγS and 7000 dpm/pmol for [3H]GDP, respectively) for 2.5 hours. Dissociation of the radiolabeled nucleotides from RHOA proteins was then initiated by the addition of unlabeled GTPγS (final concentration of 50 µM) to the solution. After incubation for the indicated periods, the amounts of the radioactivity associated with the proteins were determined as described above.
Filamentous-actin staining
NIH3T3 cells were transfected with plasmids expressing green fluorescent protein alone, green fluorescent protein-fused WT, or mutant RHOA. After pre-cultured for 24 hours, cells were induced for gene expression with doxycycline, serum-starved for 24 hours, fixed, permeabilized, and stained by Rhodamine Phalloidin (Cytoskeleton). Nuclei were stained with 4′,6-diamidino-2-phenylindole, dihydrochloride (Dojindo). Images were obtained by confocal-laser scanning microscopy (Olympus).
Reporter assay
For luciferase reporter assay, 1.2 × 105/well 293T cells were seeded in 12-well plates and co-transfected with WT or mutant RHOA in pEF-neo together with pSRα/β-galactosidase and serum response factor-response element (SRF-RE)/pGL4.34 [luc2P/SRF-RE/Hygro] Vector (Promega), by using X-tremeGENE 9 DNA Transfection Reagents (Roche) according to the manufacturer’s protocol. Activity of firefly luciferase in cell lysates was measured as previously described.2
Flow cytometry
Peripheral blood- and bone marrow-derived cells were stained with fluorescent-labeled antibodies and analyzed by flow cytometry using an LSRFortressa (BD Biosciences) with standard filter sets. Antibodies used were fluorescein isothiocyanate-labeled anti-CD4 (RPA-T4; BD Biosciences), phycoerythrin Cy7-labeled anti-CD25 (2A3; BD Biosciences), and phycoerythrin-labeled anti–PD-1 (MIH4; BD Biosciences). For intracellular staining, Alexa Fluor 647 mouse anti-human FOXP3 and human Foxp3 Buffer Set (259D/C7BD; Biosciences) was used after surface staining. All flow cytometry data were analyzed with FloJo software (Tree Star).
Immunohistochemistry (IHC)
For immunostaining, 3 µm sections were cut, deparaffinized, subjected to a heat-induced antigen retrieval with Target Retrieval Solution (Dako), and pretreated with 0.3% hydrogen peroxide for blocking. They incubated with primary antibodies against CD25 (clone 4C9, dilution 1:1; Nichirei Bioscience) and CD45RO (clone UCLH-1, dilution 1:100; DAKO).13,14 The bound antibodies were detected using the Histofine Simple Stain MAX PO reagent with diaminobenzidine as a chromogen, and the sections were counterstained with Mayer’s hematoxylin. IHC analysis with anti-CD3 (clone 2GV6, dilution 1:1; Roche) and anti-CD4 (clone SP35, dilution 1:1; Roche) antibodies was performed with the Ventana Benchmark automated staining system (Ventana Medical Systems) according to the manufacturer’s instruction.13 For double staining of FOXP3 and PD-1, the sections were first incubated with anti-FOXP3 antibody (clone 236A/E7, dilution 1:100; Abcam) and visualized using ImmPRESS HRP-conjugated anti-mouse polymeric system (Vector Laboratories) together with diaminobenzidine. Then, the sections were stained with anti–PD-1 antibody (clone NAT105, dilution 1:50; Abcam), detected with anti-rabbit ImmPRESS-AP polymer reagent and Vector Blue (Vector Laboratories), and followed by the Nuclear Fast Red counterstaining.15 The tissue section images were captured using the NanoZoomer 2.0-HT (Hamamatsu Photonics).
Statistical analysis
Overall survivals were assessed using Kaplan–Meier curves and two-sided log-rank tests. Hazard ratios (given in numbers) and their 95% confidential intervals were calculated by Cox regression. Analyses were performed using SPSS version 19.0.0 (IBM Corporation, Armonk, NY).
Results
Identification of RHOA mutations in ATLL
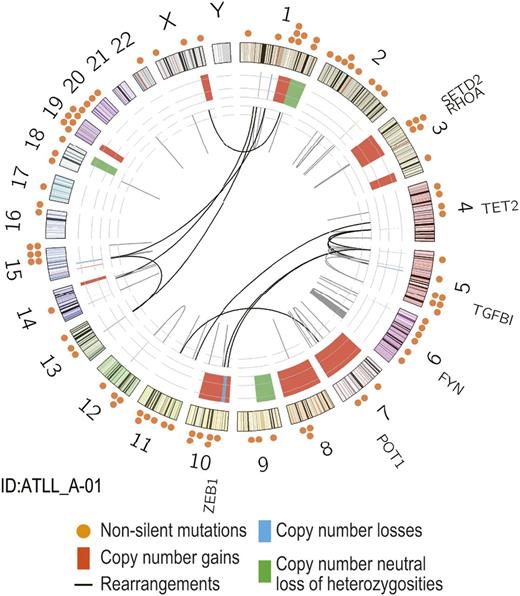
To obtain an insight into the ATLL genome, we performed WGS of paired tumor/normal DNA from a single case with ATLL. The mean coverage of WGS for paired tumor/normal DNA was 60.4× and 33.1×, with which 96% and 88% of the entire genomes were analyzed at more than 20 independent reads on average, respectively. WGS revealed a high complexity of this particular ATLL genome, in which 15 848 single nucleotide variants, 1012 insertions/deletions, 91 structural variations, and numerous copy number variations (CNVs), and allelic imbalances were detected (Figure 1). The single nucleotide variants included a number of potential drivers that has been previously implicated in ATLL and other T-cell malignancies, as well as solid cancers such as ZEB1, POT1, TET2, SETD2, TGFB1, FYN, and RHOA genes (supplemental Table 1). Among these, especially intriguing were mutations of RHOA (p.Gly17Val) and TET2 (p.Gln232*), because this combination of mutations were characteristically found in other distinct types of PTCLs, ie, AITL and PTCL-NOS with follicular helper T-cell (TFH-cell) phenotype2,16,17 ; being mutated in 83% (TET2) and 70% (RHOA) of the cases, both were among the most frequently mutated genes in the latter tumors, together with DNMT3A and IDH2.
Identification of RHOA mutations in ATLL. Circos plot of genomic alterations obtained from WGS for a single ATLL case. Locations of somatic non-silent mutations (n = 96) are indicated (orange circles). Abnormalities in genomic copy number are shown in red for gains, blue for losses, and green for copy number neutral loss of heterozygosities. Chromosomal rearrangements are indicated in the inner circle (black lines).
Identification of RHOA mutations in ATLL. Circos plot of genomic alterations obtained from WGS for a single ATLL case. Locations of somatic non-silent mutations (n = 96) are indicated (orange circles). Abnormalities in genomic copy number are shown in red for gains, blue for losses, and green for copy number neutral loss of heterozygosities. Chromosomal rearrangements are indicated in the inner circle (black lines).
Frequent mutations of RHOA in ATLL
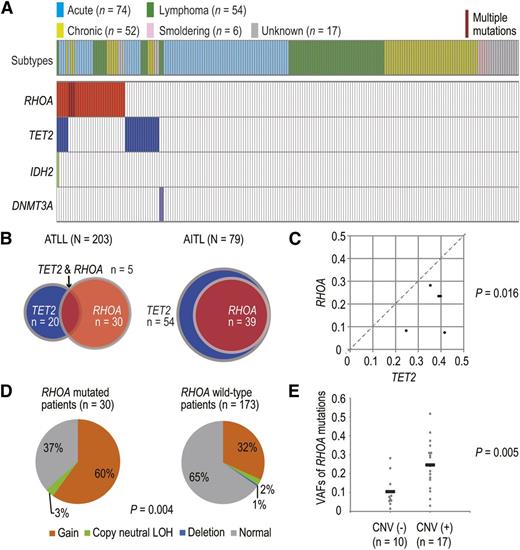
Prompted by this finding, we first interrogated mutations of RHOA and TET2, as well as DNMT3A and IDH2, in a cohort of 203 ATLL cases using targeted deep sequencing, in which their entire coding exons were individually amplified and subjected to deep sequencing (mean coverage, 18 346×) (supplemental Table 2). All patients had documented HTLV-1 infection, where HTLV-1 integration had been confirmed in 201 cases (data not shown). RHOA and TET2 mutations were detected and validated in 30 (15%) and 20 (10%) cases of our cohort, respectively, whereas DNMT3A and IDH2 were rarely mutated and found only in a few different cases (Figure 2A; supplemental Tables 3 and 4). Somatic origin of mutations was confirmed for nine RHOA and three TET2 variants, and all mutated cases had viral integration (Figure 2A and 3A; supplemental Figure 1). None of the RHOA variants detected in our ATLL series were observed in 95 normal Japanese samples (data not shown) or in other SNP databases, including dbSNP Build 131 and the National Heart, Lung, and Blood Institute GO Exome Sequencing Project (February 2012 release), as well as our in-house database from 127 germline controls from cancer-borne patients, except for two variants recorded in Catalogue of Somatic Mutations in Cancer version 71 (November 2014 release). In AITL and related lymphomas, RHOA mutations were always accompanied by TET2 mutations,2 whereas the latter mutations were less common in ATLL, accounting for only 17% of RHOA-mutated cases (Figure 2B). Nevertheless, TET2 mutations, if present, had a higher variant allele frequency (VAF) than RHOA mutations, suggesting that TET2 mutations predated the RHOA mutations (Figure 2C). There were no significant correlations of RHOA and TET2 mutations between disease subtypes or clinical outcomes (supplemental Figure 2). To examine the association of RHOA mutations and CNVs, SNP-array karyotyping was performed for the entire sample set (N = 203). Overall, a high frequency of copy number gains (36%) and copy neutral loss of heterozygosities (2%) was observed at the RHOA locus, although most CNVs affected large regions of the 3p chromosome. CNVs at the RHOA locus were more frequently detected in RHOA-mutated patients (63%) than in RHOA WT patients (35%) (Figure 2D). In addition, patients with CNVs at the RHOA locus had significantly higher VAFs of RHOA mutations than those without CNVs (Figure 2E), suggesting the preferential amplification of the mutant allele. These results suggest that RHOA is one of the potential targets of copy number change involving 3p, thus emphasizing the relevance of RHOA mutations in the pathogenesis of ATLL.
Spectrum and correlation of RHOA mutations in ATLL. (A) Mutational status of RHOA, TET2, IDH2, and DNMT3A genes in 203 ATLL cases. Subtype of each case is shown by indicated colors. (B) Comparison of distributions of RHOA and TET2 mutations between AITL and ATLL. Co-existence of RHOA and TET2 mutations are shown in AITL and ATLL. Red and blue circles indicated cases with RHOA and TET2 mutations, respectively. (C) Comparison of VAFs between RHOA and TET2 mutations. ATLL cases with both TET2 and RHOA mutations (n = 5) were analyzed. Each axis shows the VAFs of mutations. VAFs with genomic copy number abnormalities were corrected according to the usual methods. P value was calculated by Mann–Whitney U test. (D) Frequency of CNVs at the RHOA locus in ATLL cases with or without RHOA mutations. P value was calculated by Fisher’s exact test. (E) VAFs of RHOA mutations in cases with or without CNVs at the RHOA locus. Only patients with a single RHOA mutation were considered. P value was determined using Student t test.
Spectrum and correlation of RHOA mutations in ATLL. (A) Mutational status of RHOA, TET2, IDH2, and DNMT3A genes in 203 ATLL cases. Subtype of each case is shown by indicated colors. (B) Comparison of distributions of RHOA and TET2 mutations between AITL and ATLL. Co-existence of RHOA and TET2 mutations are shown in AITL and ATLL. Red and blue circles indicated cases with RHOA and TET2 mutations, respectively. (C) Comparison of VAFs between RHOA and TET2 mutations. ATLL cases with both TET2 and RHOA mutations (n = 5) were analyzed. Each axis shows the VAFs of mutations. VAFs with genomic copy number abnormalities were corrected according to the usual methods. P value was calculated by Mann–Whitney U test. (D) Frequency of CNVs at the RHOA locus in ATLL cases with or without RHOA mutations. P value was calculated by Fisher’s exact test. (E) VAFs of RHOA mutations in cases with or without CNVs at the RHOA locus. Only patients with a single RHOA mutation were considered. P value was determined using Student t test.
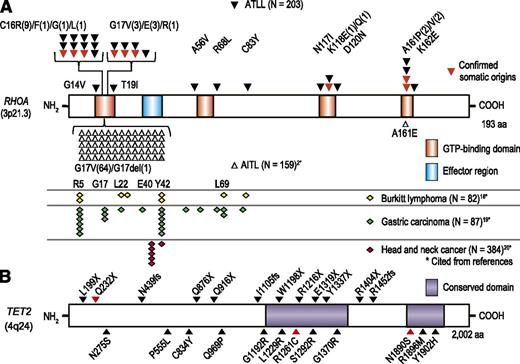
Distribution of RHOA and TET2 mutations in ATLL. (A) Distribution of RHOA mutations in ATLL and other cancers such as AITL,2 Burkitt lymphoma,18 gastric carcinoma,19 and head and neck cancers.20 (B) Distribution of TET2 mutations identified in ATLL is shown with triangle. Confirmed somatic mutations are marked by red triangles. Asterisk shows citations of references.
Distribution of RHOA and TET2 mutations in ATLL. (A) Distribution of RHOA mutations in ATLL and other cancers such as AITL,2 Burkitt lymphoma,18 gastric carcinoma,19 and head and neck cancers.20 (B) Distribution of TET2 mutations identified in ATLL is shown with triangle. Confirmed somatic mutations are marked by red triangles. Asterisk shows citations of references.
Unique distribution of RHOA mutations in ATLL
Unexpectedly and conspicuously, RHOA mutations in ATLL exhibited a very different pattern of distribution from those reported in AITL, even though both mature T-cell neoplasms are very similar and often indistinguishable from each other on histopathology except for HTLV-1 infection (Figure 3A).2,18-20 RHOA mutations in AITL almost invariably involved the Gly17 residue that participates in the GTP-binding pocket, causing an identical amino acid change (Gly17Val). In contrast, mutations in ATLL were widely distributed but clearly targeted to the GTP-binding pocket, showing discrete mutational hotspots at the Cys16, Gly17, and Ala161 residues with Cys16Arg mutations being the most prevalent (n = 9). The Gly17Val mutation was also found in 3 ATLL cases but other Gly17-involving mutations had never been reported in AITL, including Gly17Glu (n = 3) and Gly17Arg (n = 1). Because the same amino acid is commonly involved within the GTP binding site, Ala161Pro and Ala161Glu mutations were exclusively found in ATLL and AITL, respectively. Two independent RHOA mutations were detected in 3 cases. In ATLL-C-05, Gly17Arg, and Gly14Val, mutations were on the same allele in the majority of tumor cells (Figure 4A), whereas a minority carried a Gly14Val mutation alone, indicating that a Gly14Val-carrying cell that acquired the next Gly17Arg mutation had outgrown and dominated the tumor population (Figure 4A). In the remaining cases, harboring Cys16Arg/Thr19Ile and Cys16Leu/Lys118Gln mutations, a low allelic burden of the second RHOA mutation precluded accurate determination of the clonal structure (supplemental Table 3). Almost all mutations found in ATLL were distributed across all the highly conserved amino acids within or in the vicinity of the GTP-binding pocket (Figures 3A and 4B), most likely affecting GTP-binding capacity (Figure 4C).
Feature and structural modeling of RHOA mutations in ATLL. (A) Representative two independent co-occurring RHOA mutations in ATLL cases. ATLL_C-05 (top) and ATLL_A-07 (bottom) cases have Gly17Arg/Gly14Val and Thr19Ile/Cys16Arg mutations, respectively. (B) Amino acid alignment of RHOA proteins from different species. Evolutionally conserved amino acids among species are shown in blue and the amino acid positions of mutation in ATLL are highlighted by arrowhead. Conserved functional domains are also indicated. (C) Mutated amino acid positions identified in ATLL are mapped to the 3D structure of the GTP-biding pocket of human RHOA protein (Protein Data Bank identification, 1A2B). GTPγS and the magnesium ion are shown as white sticks and a green sphere.
Feature and structural modeling of RHOA mutations in ATLL. (A) Representative two independent co-occurring RHOA mutations in ATLL cases. ATLL_C-05 (top) and ATLL_A-07 (bottom) cases have Gly17Arg/Gly14Val and Thr19Ile/Cys16Arg mutations, respectively. (B) Amino acid alignment of RHOA proteins from different species. Evolutionally conserved amino acids among species are shown in blue and the amino acid positions of mutation in ATLL are highlighted by arrowhead. Conserved functional domains are also indicated. (C) Mutated amino acid positions identified in ATLL are mapped to the 3D structure of the GTP-biding pocket of human RHOA protein (Protein Data Bank identification, 1A2B). GTPγS and the magnesium ion are shown as white sticks and a green sphere.
Kinetics of different RHOA mutants for GDP/GTP-binding
RHOA encodes a ras-related GTP-binding protein that functions as a molecular switch in a wide variety of biological processes through cycling between active (GTP-bound) and inactive (GDP-bound) states.21 It has been demonstrated that the AITL-associated Gly17Val mutant does not bind GTP and inhibit WT RHOA function in a dominant-negative manner.2,16,17 To evaluate the functional impact of different RHOA mutations, we first examined the affinity of WT and mutant RHOA proteins to GTP and GDP by measuring kinetics of their binding to a radio-labeled nonhydrolyzable GTP analog such as [35S]GTPγS and [3H]GDP in vitro. The Gly17Val mutant hardly bound [35S]GTPγS and another AITL-associated RHOA mutant (Ala161Glu) also showed little binding to [35S]GTPγS (Figure 5A). On the other hand, two mutants newly identified in ATLL (Cys16Arg and Ala161Pro) bound to [35S]GTPγS more rapidly than WT RHOA (Figure 5B). Moreover, dissociation of [35S]GTPγS and [3H]GDP were prominently accelerated for Ala161Pro and to a lesser extent for Cys16Arg mutants (Figure 5C-D). Given a much higher intracellular concentration of GTP compared with GDP, these data indicated that Cys16Arg and Ala161Pro RHOA mutants function as a fast-cycling mutant having an increased intrinsic GDP/GTP exchange rate, favoring the active GTP-bound form.
Functional impact of RHOA mutants. (A-D) Kinetics of GDP/GTP-binding for different RHOA mutants in the presence of 0.8 mM Mg2+. (A-B) 5 µM of radio-labeled GTP analog ([35S]GTPγS) was incubated with indicated RHOA mutants, and the amounts of RHOA-bound [35S]GTPγS at indicated time points are plotted. (C-D) RHOA-bound [35S]GTPγS and [3H]GDP were measured at indicated time points after their dissociation was initiated by the addition of unlabeled GTPγS. Data represent mean ± standard deviation (n = 3 for each group). The significance of difference was determined by two-tailed unpaired Student t test. *P < .05; **P < .005; ***P < .0005; 3 independent experiments. (E) Activity of the SRF-RE reporter in 293T cells transduced with indicated RHOA mutants in luciferase assay. The differences between mutants and the WT RHOA were all statistically significant (*P < .05; **P < .005). Data represent mean ± standard deviation (n = 4). P value was determined using Student t test. Three independent experiments were performed.
Functional impact of RHOA mutants. (A-D) Kinetics of GDP/GTP-binding for different RHOA mutants in the presence of 0.8 mM Mg2+. (A-B) 5 µM of radio-labeled GTP analog ([35S]GTPγS) was incubated with indicated RHOA mutants, and the amounts of RHOA-bound [35S]GTPγS at indicated time points are plotted. (C-D) RHOA-bound [35S]GTPγS and [3H]GDP were measured at indicated time points after their dissociation was initiated by the addition of unlabeled GTPγS. Data represent mean ± standard deviation (n = 3 for each group). The significance of difference was determined by two-tailed unpaired Student t test. *P < .05; **P < .005; ***P < .0005; 3 independent experiments. (E) Activity of the SRF-RE reporter in 293T cells transduced with indicated RHOA mutants in luciferase assay. The differences between mutants and the WT RHOA were all statistically significant (*P < .05; **P < .005). Data represent mean ± standard deviation (n = 4). P value was determined using Student t test. Three independent experiments were performed.
Biological activity of different RHOA mutants
Because RHOA is one of the master regulators of actin cytoskeleton dynamics in various cell types and regulates gene transcription through activation of the SRF signaling pathway,2 we next assessed the impact of RHOA mutations on filamentous-actin formation and transcriptional activation of SRF. When examined by immunostaining, Gly17Val- and Ala161Glu-transduced NIH3T3 cells showed attenuated actin stress fiber formation, whereas similar to WT RHOA, the Cys16Arg and Ala161Pro mutants exhibited enhanced actin fiber formation (supplemental Figure 3). We further examined transcriptional activity of these mutations from an SRF-RE by luciferase assay using mutant-transduced 293T cells. The Gly17Val and Ala161Glu RHOA mutants downregulated transcription from the SRF-RE, supporting that both mutants can act as a dominant-negative molecule (Figure 5E).2 In contrast, the Cys16Arg and Ala161Pro mutants significantly enhanced transcriptional activity, similar to a known constitutively active mutant (Gly14Val),22,23 suggesting that these fast-cycling Cys16Arg and Ala161Pro mutants represent gain-of-function molecules that augment intrinsic RHOA functions and modulate the downstream transcriptional activity.24 In addition, Cys16Phe, Lys118Glu, and Ala161Val mutants, which were specifically observed in ATLL cases, also significantly enhanced SRF-RE luciferase activity, suggesting that these mutations possess a gain-of-function effect similar to Cys16Arg and Ala161Pro mutations.
Surface and intracellular markers of primary ATLL cells with unique RHOA mutations
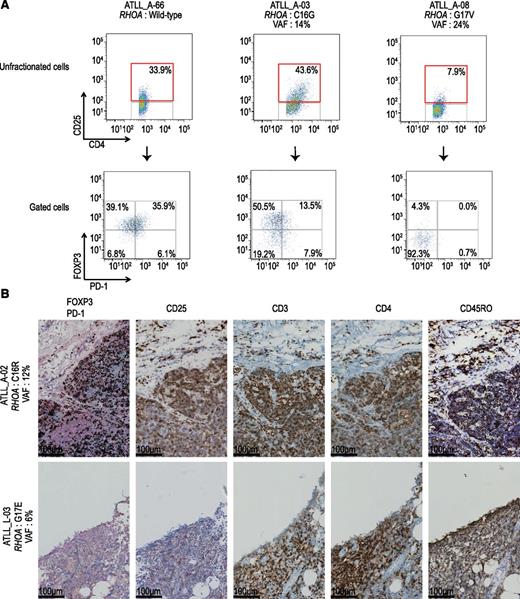
As for the cell-of-origin of ATLL, it has been suggested that ATLL may arise from different subsets of CD4+ T cells, ie, naïve helper, activated, or regulatory T-cells (Tregs),1 whereas TFH cells are assumed to be the cell-of-origin of AITL and a subset of PTCL-NOS.2 Thus, a possible explanation of unique variegation of RHOA mutations in ATLL would be that different mutations reflect different origins of ATLL cells. To address this hypothesis, we performed flow cytometric analysis of 12 patient-derived ATLL cells carrying different RHOA mutations (Table 1; Figure 6A). Most of the ATLL samples carrying WT RHOA, Cys16Arg, Cys16Gly, and Ala161Pro mutations showed CD4+CD25+FOXP3+PD-1− or CD4+CD25+FOXP3−PD-1+ phenotypes, suggesting their origin as being Tregs or effector T cells.25,26 In contrast, Gly17Val-positive ATLL cells had a CD4+CD25− phenotype, supporting that memory T cells are likely to be their cell-of-origin.25-29 Moreover, formalin-fixed paraffin-embedded specimens obtained from the lymph nodes from 2 RHOA-mutated ATLL cases were examined by IHC. The neoplastic cells of the patient with a Cys16Arg mutation (ATLL_A-02) showed the CD4+CD25+FOXP3+PD-1−CD3+CD45RO+ phenotype, which was consistent with the flow cytometry results (Figure 6B). On the other hand, tumor cells with a Gly17Glu mutation (ATLL_L-03) exhibited the CD4+CD25−FOXP3−PD-1−CD3+CD45RO+ phenotype, suggesting that they are derived from memory T cells.27-29 These results support the hypothesis that differential RHOA mutations are linked to distinct cells-of-origin.
Flow cytometry data of patient-derived primary ATLL cells
| ID . | Subtype . | RHOA mutations . | VAFs . | CD25 (% of positive) . | FOXP3 (% of positive) . | PD1 (% of positive) . |
|---|---|---|---|---|---|---|
| ATLL_A-02 | Acute | C16R | 0.12 | 43 | 26 | 0 |
| ATLL_A-04 | Acute | C16R | 0.31 | 15 | 26 | 1 |
| ATLL_A-03 | Acute | C16G | 0.14 | 44 | 64 | 21 |
| ATLL_C-08 | Chronic | A161V | 0.42 | 42 | 36 | 16 |
| ATLL_A-06 | Acute | C16F | 0.18 | 80 | 3 | 41 |
| ATLL_C-06 | Chronic | K118E | 0.29 | 60 | 27 | 47 |
| ATLL_A-13 | Acute | A161P | 0.04 | 63 | 8 | 73 |
| ATLL_A-08 | Acute | G17V | 0.24 | 8 | 4 | 1 |
| ATLL_A-01* | Acute | G17V | 0.31 | 25 | 11 | 6 |
| ATLL_A-20 | Acute | WT | — | 81 | 3 | 77 |
| ATLL_A-66 | Acute | WT | — | 34 | 75 | 42 |
| ATLL_C-42 | Chrnoic | WT | — | 23 | 21 | 6 |
| Control | Carrier | WT | — | 9 | 9 | 5 |
| ID . | Subtype . | RHOA mutations . | VAFs . | CD25 (% of positive) . | FOXP3 (% of positive) . | PD1 (% of positive) . |
|---|---|---|---|---|---|---|
| ATLL_A-02 | Acute | C16R | 0.12 | 43 | 26 | 0 |
| ATLL_A-04 | Acute | C16R | 0.31 | 15 | 26 | 1 |
| ATLL_A-03 | Acute | C16G | 0.14 | 44 | 64 | 21 |
| ATLL_C-08 | Chronic | A161V | 0.42 | 42 | 36 | 16 |
| ATLL_A-06 | Acute | C16F | 0.18 | 80 | 3 | 41 |
| ATLL_C-06 | Chronic | K118E | 0.29 | 60 | 27 | 47 |
| ATLL_A-13 | Acute | A161P | 0.04 | 63 | 8 | 73 |
| ATLL_A-08 | Acute | G17V | 0.24 | 8 | 4 | 1 |
| ATLL_A-01* | Acute | G17V | 0.31 | 25 | 11 | 6 |
| ATLL_A-20 | Acute | WT | — | 81 | 3 | 77 |
| ATLL_A-66 | Acute | WT | — | 34 | 75 | 42 |
| ATLL_C-42 | Chrnoic | WT | — | 23 | 21 | 6 |
| Control | Carrier | WT | — | 9 | 9 | 5 |
A case was analyzed by WGS.
Analysis of cell surface and intracellular markers in RHOA-mutated cases. (A) Analysis of cell surface marker in RHOA mutated cases. Expression of PD-1 and FOXP3 (bottom) in the CD4+CD25+ tumor cell fraction (top) in primary ATLL samples. Results of 3 representative cases are shown. Case ID, type of RHOA mutations, and their VAFs are also indicated. Numbers in boxes show percentage of cells in the indicated fractions. (B) Immunohistochemical analysis of formalin-fixed paraffin-embedded specimens obtained from the lymph nodes of two RHOA-mutated ATLL cases. Representative images of double staining with FOXP3 (intra-nuclei, brown) and PD-1 (cytosol, blue), and single staining with CD25, CD3, CD4, and CD45RO (brown) are shown.
Analysis of cell surface and intracellular markers in RHOA-mutated cases. (A) Analysis of cell surface marker in RHOA mutated cases. Expression of PD-1 and FOXP3 (bottom) in the CD4+CD25+ tumor cell fraction (top) in primary ATLL samples. Results of 3 representative cases are shown. Case ID, type of RHOA mutations, and their VAFs are also indicated. Numbers in boxes show percentage of cells in the indicated fractions. (B) Immunohistochemical analysis of formalin-fixed paraffin-embedded specimens obtained from the lymph nodes of two RHOA-mutated ATLL cases. Representative images of double staining with FOXP3 (intra-nuclei, brown) and PD-1 (cytosol, blue), and single staining with CD25, CD3, CD4, and CD45RO (brown) are shown.
Discussion
We have described novel recurrent RHOA mutations in ATLL, accounting for 15% of the current cohort. RHOA mutations have been most frequently reported in AITL and PTCL-NOS with the TFH phenotype, where the mutations almost invariably affect the Gly17 residue and is accompanied by co-existing TET2 mutations.2,16,17 In contrast, RHOA mutations in ATLL are more widely distributed near or within the GTP-binding pocket, suggesting the distinct pathogenesis of ATLL compared to other PTCLs. Also, the RHOA gene is a plausible target of 3p gain, which is one of the most frequent CNVs in ATLL.30
In this context, a notable finding in the current study is the variegated nature of RHOA mutations in ATLL. We identified two types of RHOA mutations in ATLL; loss-of-function (Gly17Val and Ala161Glu) and gain-of-function (Cys16Arg, Cys16Phe, Lys118Glu, Ala161Val, and Ala161Pro) mutations. Currently, there is no clear explanation to the underlying molecular mechanism for this variegation of RHOA mutations within the same tumor type that shows a unique distribution compared with other PTCLs histologically closely related to ATLL. A correlation between cellular phenotypes and mutation types might suggest the different cells-of-origin of ATLL depending on mutation types. ATLL cells with gain-of-function mutations had cellular phenotypes similar to effector or Tregs, whereas those cells with loss-of-function mutations were consistent with memory T cells. These data suggest that some HTLV-1–infected T-cell subsets could favor gain-of-function RHOA mutations, whereas others favor loss-of-function type mutations, both being selected for neoplastic proliferation to develop ATLL. Alternatively, it is also possible that altered downstream signaling by RHOA mutations may result in a change of immunophenotype in ATLL cells. Indeed, RHOA is a key regulator in T-cell–receptor signaling, which has been known to affect the expression of several T-cell subset markers, such as PD-1 or FOXP3.31
Another interesting aspect of the variegated nature of the RHOA mutation in ATLL are the two functionally different mutations detected in the same tumors. Particularly, in one case (ATLL_C-05), Gly14Val and Gly17Arg mutations were found to be present on the same allele. The Gly14Val is a previously known gain-of-function mutant,32 as it prevents intrinsic and GTPase-activating protein-induced GTP hydrolysis, thus keeping the protein in its active state,21 whereas Gly17 is located in the P-loop, which is a highly conserved consensus sequence essential for GDP/GTP-binding and GTPase activities.33 Because several kinds of mutations at Gly17 (ie, Gly17Val and Gly17Ala) have been shown to act in a dominant-negative manner by disrupting the binding of GTP/GDP,2,16,17,34 the Gly17Arg mutant is predicted to represent a dominant-negative RHOA molecule, although the exact functional consequence of this mutant has not been elucidated. The mechanism for the clonal selection of these double mutants having apparently opposite functional consequences is totally unknown also and requires further studies.
Recently, different RHOA mutations have been reported in a wide variety of human cancers other than PTCLs. These mutations exhibit different distributions depending on tumor types; in Burkitt lymphoma18 and gastric adenocarcinoma,19,35,36 Arg5 and Tyr42 are predominantly mutated, whereas head and neck cancers have a different mutational hotspot at Glu40.20 The strong correlation between phenotype (tumor type) and mutational distribution indicates that the functional consequence of a RHOA mutation during tumor development could be conditional on their derived tissues and cells. Several genes are affected by mutations having apparently opposite functional consequences, such as both gain-of-function mutations and loss-of-function mutations. For example, NOTCH1 mutations found in acute T-lymphoblastic leukemia exclusively reside in the carboxyl-terminal proline, glutamic acid, serine, and threonine (PEST) and heterodimerization domains, leading to constitutive activation of NOTCH1 signaling,37 whereas those found in many solid cancers show mutational hotspots within the epidermal growth factor-like domain and are thought to result in loss-of-function. Another example of tumor-type–specific mutations of the same gene is gain-of-function mutations of EZH2 at Tyr641 associated with diffuse large B-cell lymphoma and loss-of-function mutations found in myeloid neoplasms.38 In contrast to the functionally dichotomous nature of these mutations, a unique and more complex feature of RHOA mutation is a higher order of functional heterogeneity among different mutations that are differentially selected in specific tumor types. More comprehensive and detailed functional characterization of different RHOA mutants in proper cell contexts should be warranted in order to understand the oncogenic role of different RHOA mutations in ATLL and other RHOA-mutated human cancers, and to better target this molecule for therapy.
The genome sequence data reported in this article have been deposited in the European Genome-phenome Archive (accession number EGAS00001001210).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank K. Ishiyama and S. Miyawaki for providing specimens, and Maki Nakamura, Hitomi Higashi, and Miki Sago for their technical assistance.
This study was supported by grants-in-aid from the Ministry of Health, Labor and Welfare of Japan, grants-in-aid for Scientific Research on Innovative Areas, the Japan Society for the Promotion of Science through the Funding Program for World-Leading Innovative Research and Development on Science and Technology (ie, the FIRST Program), and by the Medical Research Support Center, Graduate School of Medicine, Kyoto University.
Authorship
Contribution: Y.N., Y. Shiozawa, T.Y., H.S., A. Kon, K.Y., Y. Sato, A.S.-O., and M.S. performed experiments and data analysis; Y. Shiraishi, K.C., H.T., Y.T., W.M., H.N., N.H., S.M., and T.S. were committed to bioinformatics analysis of the sequencing data; K. Kontani, T.E., T.R.K., M.H., M.S.-Y., S.E., K.A., O.N., H.H., T.K., and S.C. performed the functional analyses of RHOA mutants; A. Kitanaka, K.N., K.S., and T.W. collected specimens and were involved in planning the project; Y.N., K. Kataoka., R.I., and S.O. generated figures and tables, and wrote the manuscript; S.O. lead the entire project; and all authors participated in discussions and interpretation of the data and results.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Seishi Ogawa, Department of Pathology and Tumor Biology, Graduate School of Medicine, Kyoto University, Yoshida-Konoe-cho, Sakyo-ku, Kyoto 606-8501, Japan; e-mail: sogawa-tky@umin.ac.jp.


![Figure 5. Functional impact of RHOA mutants. (A-D) Kinetics of GDP/GTP-binding for different RHOA mutants in the presence of 0.8 mM Mg2+. (A-B) 5 µM of radio-labeled GTP analog ([35S]GTPγS) was incubated with indicated RHOA mutants, and the amounts of RHOA-bound [35S]GTPγS at indicated time points are plotted. (C-D) RHOA-bound [35S]GTPγS and [3H]GDP were measured at indicated time points after their dissociation was initiated by the addition of unlabeled GTPγS. Data represent mean ± standard deviation (n = 3 for each group). The significance of difference was determined by two-tailed unpaired Student t test. *P < .05; **P < .005; ***P < .0005; 3 independent experiments. (E) Activity of the SRF-RE reporter in 293T cells transduced with indicated RHOA mutants in luciferase assay. The differences between mutants and the WT RHOA were all statistically significant (*P < .05; **P < .005). Data represent mean ± standard deviation (n = 4). P value was determined using Student t test. Three independent experiments were performed.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/5/10.1182_blood-2015-06-644948/4/m_596f5.jpeg?Expires=1765905693&Signature=uyep5Rb3KzmbrFPRKj2TF345o0jX0Nj7C7Ya13fZUNM4DyPWuwNqPHpsqJMal9wMSh5EqtK3Nfqrqs~udlVJbOlE4lQuCw0pzrmRVxETN7FcFUW8Xj~vCBOzzAd5HlpOyaxz8tD7FNypGj7Os3TRbFgMUus2NmL8uNvdgUa1nwi0hecdTtm5iaWXCKm-Prn5nMUkIUa-JwAjytRcM4S-9977jMuchvCmOl6tef2lvv8AJzYrx2tg3TL0wpxNAql7tWkUPhmDZnUwMXqsBbXAmRG8PEUY2WkesTXwleQQZVp2ZgcTOd~Ju4nmgyDtVVo1pzZpdLvDdSRC9uwF1zifjw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal