Key Points
Ponatinib induces durable responses regardless of baseline BCR-ABL1 mutation status in CP-CML patients.
No single or compound mutant consistently confers primary or secondary resistance to ponatinib in CP-CML.
Abstract
BCR-ABL1 kinase domain mutations can confer resistance to first- and second-generation tyrosine kinase inhibitors (TKIs) in chronic myeloid leukemia (CML). In preclinical studies, clinically achievable concentrations of the third-generation BCR-ABL1 TKI ponatinib inhibit T315I and all other single BCR-ABL1 mutants except T315M, which generates a single amino acid exchange, but requires 2 sequential nucleotide exchanges. In addition, certain compound mutants (containing ≥2 mutations in cis) confer resistance. Initial analyses based largely on conventional Sanger sequencing (SS) have suggested that the preclinical relationship between BCR-ABL1 mutation status and ponatinib efficacy is generally recapitulated in patients receiving therapy. Thus far, however, such analyses have been limited by the inability of SS to definitively identify compound mutations or mutations representing less than ∼20% of total alleles (referred to as “low-level mutations”), as well as limited patient follow-up. Here we used next-generation sequencing (NGS) to define the baseline BCR-ABL1 mutation status of 267 heavily pretreated chronic phase (CP)-CML patients from the PACE trial, and used SS to identify clonally dominant mutants that may have developed on ponatinib therapy (30.1 months median follow-up). Durable cytogenetic and molecular responses were observed irrespective of baseline mutation status and included patients with compound mutations. No single or compound mutation was identified that consistently conferred primary and/or secondary resistance to ponatinib in CP-CML patients. Ponatinib is effective in CP-CML irrespective of baseline mutation status.
Introduction
Tyrosine kinase inhibitors (TKIs) targeting BCR-ABL1 have substantially improved the prognosis of patients with chronic myeloid leukemia (CML).1 However, resistance to TKI therapy, which manifests through both BCR-ABL1–dependent and –independent mechanisms, remains a major challenge.2 The best-characterized mechanism of resistance involves point mutations in the BCR-ABL1 kinase domain (KD) that disrupt TKI binding. A range of point mutations can mediate resistance to first- (imatinib) and second- (dasatinib, nilotinib, and bosutinib) generation TKIs, with the T315I gatekeeper mutation conferring resistance to all 4 agents.3-5 In addition, compound mutants (variants containing ≥2 mutations within the same BCR-ABL1 allele that presumably arise sequentially) can arise under certain selective pressures,6 and a subset is highly TKI resistant.5 Although preclinical data may be used to identify candidate resistance mutations for a particular TKI,7,8 it is the lack of substantial and durable responses in patients who present with particular BCR-ABL1 mutations (primary resistance), coupled with the frequent emergence of those mutations at the time of treatment failure (secondary resistance), that ultimately define such vulnerabilities.9,10
Historically, Sanger sequencing (SS) has been used clinically to identify BCR-ABL1 mutations associated with TKI resistance. However, SS is unable to detect mutations present in less than 10% to 20% of cells and does not allow direct detection of compound mutations, although their presence can be inferred in certain cases (ie, when ≥2 mutations are detected at a combined frequency >100%). Indeed, the use of more sensitive techniques such as mass spectrometry11 or next-generation sequencing (NGS)12 has demonstrated that mutations present at levels below the detection limit of SS (ie, low-level [LL] mutations) can also affect clinical outcomes. For example, mutations that confer resistance to nilotinib or dasatinib have been associated with poor outcomes in patients even when present at low levels.11 In addition, the presence of multiple LL mutations, regardless of their association with TKI resistance, has been shown to identify patients who respond relatively poorly to treatment with dasatinib or nilotinib.13 Importantly, NGS also enables direct detection of BCR-ABL1 compound mutations. However, it has recently been shown that polymerase chain reaction (PCR)-mediated recombination may cause compound mutation frequencies to be overestimated when PCR amplicons are used for NGS.14 Thus, the ability of NGS to reliably detect compound mutations, and therefore their prevalence in CML, remains to be established.
Ponatinib is a third-generation BCR-ABL1 TKI shown to potently inhibit native BCR-ABL1 as well as all single mutants associated with resistance to imatinib and second-generation TKIs, including T315I, in preclinical models.8 However, certain compound mutations, in particular those including T315I, confer resistance to clinically achievable ponatinib concentrations.5,8 In phase 115 and phase 2 (PACE16 ) trials, ponatinib had significant antileukemic activity in heavily pretreated patients with CML (chronic phase [CP], accelerated phase [AP], or blast phase [BP]) or Philadelphia chromosome–positive acute lymphoblastic leukemia (Ph+ ALL), >90% of whom had previously received at least 2 TKIs. Consistent with its preclinical profile, ponatinib demonstrated activity in patients with or without BCR-ABL1 mutation (including T315I) present at baseline, as determined by SS. In addition, analysis of mutations in a subset of patients in the PACE trial who discontinued therapy (after a 15-month median follow-up) failed to identify any single mutations consistently associated with secondary resistance to ponatinib.16 However, in this and a separate study,5 certain compound mutations were associated with secondary resistance, though these were seen predominantly in patients with advanced CML or Ph+ ALL.
The goals of the current study were to characterize the prevalence and nature of LL and compound BCR-ABL1 mutations in heavily pretreated CP-CML patients, and to rigorously explore the relationship between BCR-ABL1 mutation status and primary and secondary ponatinib resistance. To achieve this, we used an optimized NGS analysis to determine the baseline mutation status of all 267 CP-CML patients in the PACE trial (median follow-up, 30.1 months) and performed postbaseline SS on all patients, except those who remained on the trial in continuous response.
Methods
PACE trial and sample analysis
The trial design has been previously described.16 In brief, 449 patients with CML (any phase) or Ph+ ALL with resistance or intolerance to dasatinib or nilotinib, or with a T315I mutation, were treated with ponatinib (initial dose of 45 mg once daily). The analysis described here was limited to the CP-CML patients (N = 267). The primary end point was major cytogenetic response (MCyR) by 12 months, and secondary end points included complete cytogenetic response (CCyR) and major molecular response (MMR) at any time. The data evaluated are as of January 6, 2014; the median follow-up was 30.1 months (range, 0.1-39.3). Patients’ mutation history and prior TKI exposure were collected at enrollment. Patient-recorded daily ponatinib dosing information was also collected.
BCR-ABL1 mutation analysis was conducted on blood samples from all patients at baseline using SS and NGS (Figure 1). Postbaseline analysis using SS was attempted on all patients except those who achieved MCyR and remained on trial in continuous MCyR.
Schematic representation of the BCR-ABL1 mutation analyses performed in this study. BCR-ABL1 mutation analysis (Sanger sequencing [SS] and next-generation sequencing [NGS]) was conducted on baseline samples from all chronic-phase chronic myeloid leukemia (CP-CML) patients in the PACE phase 2 study. Postbaseline analysis (SS) was conducted on any patient who did not achieve major cytogenetic response (MCyR) by 1 year, achieved and then lost MCyR, or achieved MCyR and discontinued while in MCyR. *Postbaseline (post-BL) analysis was not conducted on patients who achieved MCyR and remain on study in continuous MCyR. ^36 patients were not evaluable: 12 did not have a post-BL sample collected, and BCR-ABL1 could not be amplified for sequencing in the remaining 24; 20 of these were associated with low (<1%) BCR-ABL1 transcript levels. Of the 129 patients with evaluable post-BL samples, 102 were collected from patients who did not achieve MCyR by 1 year, 14 from patients who achieved and lost MCyR, and 13 from patients who achieved MCyR and discontinued study while in MCyR.
Schematic representation of the BCR-ABL1 mutation analyses performed in this study. BCR-ABL1 mutation analysis (Sanger sequencing [SS] and next-generation sequencing [NGS]) was conducted on baseline samples from all chronic-phase chronic myeloid leukemia (CP-CML) patients in the PACE phase 2 study. Postbaseline analysis (SS) was conducted on any patient who did not achieve major cytogenetic response (MCyR) by 1 year, achieved and then lost MCyR, or achieved MCyR and discontinued while in MCyR. *Postbaseline (post-BL) analysis was not conducted on patients who achieved MCyR and remain on study in continuous MCyR. ^36 patients were not evaluable: 12 did not have a post-BL sample collected, and BCR-ABL1 could not be amplified for sequencing in the remaining 24; 20 of these were associated with low (<1%) BCR-ABL1 transcript levels. Of the 129 patients with evaluable post-BL samples, 102 were collected from patients who did not achieve MCyR by 1 year, 14 from patients who achieved and lost MCyR, and 13 from patients who achieved MCyR and discontinued study while in MCyR.
Sequencing: Sanger sequencing and next-generation sequencing
SS was conducted as previously described.16 In postbaseline samples, the presence of a compound mutation was inferred by the detection of 2 (or more) mutations with a combined frequency >100% (eg, 100% T315I and 30% F317V).
NGS encompassed the following steps: BCR-ABL1 real-time PCR, random fragmentation of PCR product and size selection of ∼400 base-pair (bp)-sized fragments, sequencing library construction, NGS using IonTorrent PGM (read lengths up to 400 bp). The average sequence read depth for all detected variants was 11 831 reads/base (range, 1000-25 028); variants detected at ≥1% were analyzed. Bases with Phred Score <25 were considered quality control failures and designated as unknown.
A total of 419 sequence variants were detected in ABL1 (amino acids 27-540). All analyses described here focused on the 266 missense mutations detected in the BCR-ABL1 KD (amino acids 237-507). Missense mutations outside the KD were excluded (N = 55) (supplemental Table 1 available on the Blood Web site), as were all polymorphisms (N = 3), deletions (N = 5), frameshift (N = 12), nonsense (N = 18), and synonymous (N = 60) mutations because these are not considered relevant for TKI resistance.17-19
In samples in which multiple BCR-ABL1 missense mutations were detected, the phase (whether present on the same or different alleles) and frequency of all possible mutation combinations were calculated as described in the supplemental Methods.
Identification of false-positive compound mutants
Detailed explanation of the methodology used to eliminate putative false-positive compound mutations is described in the supplemental Methods. In brief, by mixing 2 RNA samples from patients, each positive for a distinct single mutation, a “recombination rate” was calculated based on the frequency at which false compound mutants were generated. A compound mutation was considered true-positive if the observed frequency was above the expected false compound mutation frequency.
Kaplan-Meier estimation of maintenance of response
Duration of MCyR, CCyR, MMR, progression-free (PFS), and overall survival (OS) was estimated using the Kaplan-Meier method. Patients without documented loss of response were censored at their last response assessment. Loss of MCyR and CCyR was confirmed by a second consecutive assessment.
Results
Baseline BCR-ABL1 mutation status determined by NGS
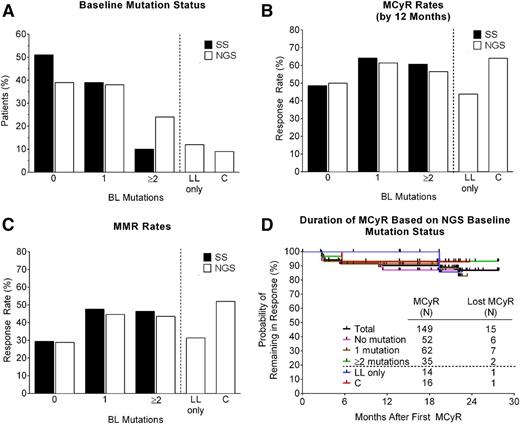
The baseline BCR-ABL1 mutation status of all 267 CP-CML patients in the PACE trial has previously been assessed by SS, with 161 mutations detected in 131 patients (49%).16 To allow detection of LL and compound BCR-ABL1 mutations, the baseline mutation status of all 267 patients was reassessed by NGS (Figure 1). In total, 266 mutations were detected by NGS in 163 patients (61%) (supplemental Table 2 and Figure 2A). NGS identified all 161 mutations detected by SS and 105 additional mutations (ie, LL mutations, which were detected at frequencies ranging from 1%-11%), in 73 patients (27%). Notably, approximately one-third of the LL mutations (34/105; 32%) occurred at amino acids that have not previously been implicated in resistance to imatinib, dasatinib, or nilotinib (supplemental Table 2).4 Twelve percent (32/267) of patients had LL mutations only (Figure 2A), whereas 15% (41/267) of patients had ≥1 high-level mutations (detected by SS) in addition to LL mutations. Overall, 23% (62/267) of patients had >1 mutation detected by NGS (Figure 2A). Thus, consistent with the greater sensitivity of NGS, the percentage of patients with no detectable BCR-ABL1 mutations by NGS was lower than that observed by SS (39% vs 51%), and the percentage of patients with multiple mutations was higher (23% vs 10%) (Figure 2A).
Distribution, response rates, and duration of response according to BCR-ABL1 mutation status at baseline as determined by SS or NGS. (A) Percentage of patients with no mutation (0), one mutation (1), or 2 or more mutations (≥2) at baseline, as determined by SS or NGS. Also shown are subsets of these groups (separated by a dotted line) that had LL mutations only, or compound mutations, as determined by NGS. (B) MCyR rates, (C) MMR rates, and (D) Kaplan-Meier curves indicating the duration of continuous MCyR, according to baseline mutation status. The median follow-up after achievement of MCyR was 27.4 months, with the majority of patients (>80%) remaining in follow-up. BL, baseline; C, compound; LL, low-level; MCyR, major cytogenetic response; MMR, major molecular response.
Distribution, response rates, and duration of response according to BCR-ABL1 mutation status at baseline as determined by SS or NGS. (A) Percentage of patients with no mutation (0), one mutation (1), or 2 or more mutations (≥2) at baseline, as determined by SS or NGS. Also shown are subsets of these groups (separated by a dotted line) that had LL mutations only, or compound mutations, as determined by NGS. (B) MCyR rates, (C) MMR rates, and (D) Kaplan-Meier curves indicating the duration of continuous MCyR, according to baseline mutation status. The median follow-up after achievement of MCyR was 27.4 months, with the majority of patients (>80%) remaining in follow-up. BL, baseline; C, compound; LL, low-level; MCyR, major cytogenetic response; MMR, major molecular response.
Assessment of compound mutant frequency with an optimized NGS analysis
In previous studies using NGS and other techniques, as much as 70% of CML patients with multiple mutations were reported to have compound mutations.12,20 Somewhat surprisingly, in 60% to 80% of cases with compound mutations, both individual mutants were also detected (eg, E255K/T315I detected on the same allele and both E255K and T315I detected on separate alleles). Noting the difficultly in explaining such complexity phylogenetically, Parker et al14 reported the detection of false-positive compound mutations by SS on mixtures of 2 plasmids or cDNAs that each contained a single mutation. To confirm these results and explore strategies to overcome limitations of sequencing protocols when using amplicons, we performed NGS analysis on mixtures of 2 patient-derived RNA samples positive for M244V and F359V to recapitulate all steps of the NGS protocol (supplemental Methods and supplemental Tables 3 and 4). Analyzed individually, each sample only had 1 mutation detected at high prevalence (>95%). However, when the 2 samples, whose mutations were 345 bp apart, were mixed, false-positive compound mutations were detected in ∼11% of the reads. Although several potential mechanisms may contribute (supplemental Figure 1), we were able to estimate a “recombination rate” of ∼15% per 100 bp, which explains the observed 11% frequency of false-positive compound mutations (supplemental Methods and supplemental Table 3).
To correct for this artifact, the percentage of false-positive compound mutations (the product of individual mutation frequencies and the recombination rate) was estimated for 23 patients with a total of 26 “suspect” compound mutations (ie, a compound mutation and both single component mutations were detected) (supplemental Methods). Using this approach, 88% (23/26) of suspect compound mutants were identified as probable false-positives and were excluded, leaving a total of 33 “true” compound mutants (59% [33/56] of those originally called) in 25 patients (supplemental Table 4). Thus, 9% (25/267) of patients overall (Figure 2A and Table 1), and 42% (25/62) of patients with multiple mutations, were found to have true compound mutations at baseline.
Clinical and molecular characteristics of the 25 PACE CP-CML patients harboring compound mutations at baseline
| Pt ID . | Achieved primary end point . | Time on study (d) . | Prior TKI* . | Baseline SS . | Baseline NGS . | Best response‡ . | Time to response (d) . | Response duration (d) . | PADD (mg) . | Reason discontinued . | Postbaseline SS . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation . | %† . | Mutation . | %† . | Mutation . | %† . | Time . | |||||||||
| 191 | N | 8 | I D N | V379I | 100 | V379I | 90 | None | N/A | N/A | 45 | Adverse event | V379I | 100 | EOT |
| V379I/H396P | 3 | ||||||||||||||
| V379I/E459K | 3 | ||||||||||||||
| 264 | N | 11 | I D | H396R | 100 | H396R | 88 | None | N/A | N/A | 45 | Progressive disease | Not evaluable (o postbaseline sample) | ||
| H396R/V299L | 5 | ||||||||||||||
| 49 | N | 27 | I N D | F359C | 100 | F359C/T315I | 76 | None | N/A | N/A | 45 | Progressive disease | Not evaluable (no postbaseline sample) | ||
| T315I | 90 | F359C | 12 | ||||||||||||
| F359C/V299L | 9 | ||||||||||||||
| 263 | N | 226 | I D N | F359V | 90 | F359V | 82 | None | N/A | N/A | 35 | Progressive disease | T315I | 100 | EOT |
| T315I | 3 | ||||||||||||||
| F359V/E255K | 1 | ||||||||||||||
| 149 | N | 245 | I D | F359C | 100 | F359C/F317L | 73 | None | N/A | N/A | 31 | Adverse event | F359C | 100 | EOT |
| F317L | 80 | F359C/T315I | 21 | ||||||||||||
| T315I | 40 | F359C | 3 | ||||||||||||
| 249 | N | 840+ | I D N | Y253H | 100 | Y253H | 94 | None | N/A | N/A | 13 | Ongoing | Y253H | 10 | 1 y |
| G250E/F317L | 1 | ||||||||||||||
| 242 | N§ | 924+ | I D N | T315I | 50 | T315I/F317L | 34 | CCyR‖ | 506 | 417+ | 43 | Ongoing | Not evaluable (no amplification)†† | 1 y | |
| F317L | 40 | T315I | 1 | ||||||||||||
| 35 | N¶ | 1007+ | I D N | F317L | 100 | F317L/H396R | 75 | MMR | 259 | 664+ | 26 | Ongoing | Not evaluable (no amplification)†† | 1y | |
| H396R | 80 | F317L/E279K | 20 | ||||||||||||
| E279K | 10 | F317L/E281K | 2 | ||||||||||||
| F317L | 1 | ||||||||||||||
| 125 | N | 1014+ | I D N | V299L | 100 | H396R/V299L | 97 | None | N/A | N/A | 27 | Ongoing | No mutation detected | 1 y | |
| H396R | 100 | ||||||||||||||
| 262 | Y | 224 | I N D | V299L | 100 | F359V/V299L | 97 | CCyR | 83 | 1# | 26 | Progressive disease | Y253H | 100 | EOT |
| F359V | 100 | F359V | 100 | ||||||||||||
| 247 | Y | 298 | I (D N) B | G250E | 20 | G250E | 27 | CCyR | 175 | 78 | 43 | Death | Not evaluable (no amplification)†† | ||
| G250E/E279V | 1 | ||||||||||||||
| 115 | Y | 594 | I | T315I | 100 | T315I | 95 | MMR | 167 | 433 | 45 | Adverse event | Not evaluable (no amplification)†† | ||
| T315I/I432M | 2 | ||||||||||||||
| T315I/R386M | 1 | ||||||||||||||
| 83 | Y | 848 | I D N | F317L | 100 | F317L/E459K | 95 | MMR | 583 | 82 | 32 | Withdrawal | Not evaluable (no amplification)†† | ||
| E459K | 100 | F317L | 2 | ||||||||||||
| 101 | Y | 875 | I D | T315I | 100 | T315I | 93 | CCyR‖ | 168 | 587 | 29 | Adverse event | Not evaluable (no amplification)†† | ||
| T315I/E275D | 3 | ||||||||||||||
| 7 | Y | 903 | I N D | F317L | 100 | F317L/E450G | 92 | PCyR | 252 | 170 | 5 | Withdrawal | F317L/ E450G | 10 | EOT |
| E450G | 90 | F359V | 6 | ||||||||||||
| 185 | Y | 928+ | I D N | F359I | 84 | MMR | 524 | 322+ | 43 | Ongoing | N/A** | ||||
| F359I | 90 | F359I/T277I | 2 | ||||||||||||
| G250E | 10 | G250E | 1 | ||||||||||||
| E507G | 1 | ||||||||||||||
| 228 | Y | 937+ | I D | V299L | 100 | V299L | 95 | MMR | 55 | 785+ | 44 | Ongoing | N/A** | ||
| V299L/P310L | 2 | ||||||||||||||
| 76 | Y | 944+ | I (D N) B | M244V | 100 | M244V | 89 | MMR | 85 | 841+ | 16 | Ongoing | N/A** | ||
| F359V | 3 | ||||||||||||||
| M244V/G250W | 2 | ||||||||||||||
| 234 | Y | 951+ | I D N | F359V | 100 | F359V | 91 | MMR | 336 | 576+ | 28 | Ongoing | N/A** | ||
| F359V/M244V | 1 | ||||||||||||||
| 113 | Y | 953+ | I D N | F317L | 100 | F317L/E459K | 99 | MMR | 87 | 845+ | 45 | Ongoing | N/A** | ||
| E459K | 100 | ||||||||||||||
| 112 | Y | 967+ | I | T315I | 100 | T315I | 92 | MMR | 81 | 842+ | 44 | Ongoing | N/A** | ||
| C475W | 1 | ||||||||||||||
| T315I/K262N | 1 | ||||||||||||||
| 89 | Y | 992+ | I N B | F359V | 60 | MMR | 84 | 832+ | 31 | Ongoing | N/A** | ||||
| F359V | 90 | T315I | 29 | ||||||||||||
| T315I | 20 | F359V/M237R | 2 | ||||||||||||
| F359V/R362T | 2 | ||||||||||||||
| 36 | Y | 1012+ | I N | F317L | 100 | E459K/F317L | 98 | MMR | 168 | 762+ | 42 | Ongoing | N/A** | ||
| E459K | 100 | ||||||||||||||
| 159 | Y | 1055+ | (D N) I | T315I | 100 | T315I | 65 | MMR | 249 | 759+ | 16 | Ongoing | N/A** | ||
| T315I/M237V | 3 | ||||||||||||||
| 10 | Y | 1119+ | I N | Y253H | 100 | Y253H | 96 | MMR | 83 | 1016+ | 18 | Ongoing | N/A** | ||
| Y253H/A287V | 1 | ||||||||||||||
| Y253H/E282D | 1 | ||||||||||||||
| Y456C | 1 | ||||||||||||||
| Pt ID . | Achieved primary end point . | Time on study (d) . | Prior TKI* . | Baseline SS . | Baseline NGS . | Best response‡ . | Time to response (d) . | Response duration (d) . | PADD (mg) . | Reason discontinued . | Postbaseline SS . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation . | %† . | Mutation . | %† . | Mutation . | %† . | Time . | |||||||||
| 191 | N | 8 | I D N | V379I | 100 | V379I | 90 | None | N/A | N/A | 45 | Adverse event | V379I | 100 | EOT |
| V379I/H396P | 3 | ||||||||||||||
| V379I/E459K | 3 | ||||||||||||||
| 264 | N | 11 | I D | H396R | 100 | H396R | 88 | None | N/A | N/A | 45 | Progressive disease | Not evaluable (o postbaseline sample) | ||
| H396R/V299L | 5 | ||||||||||||||
| 49 | N | 27 | I N D | F359C | 100 | F359C/T315I | 76 | None | N/A | N/A | 45 | Progressive disease | Not evaluable (no postbaseline sample) | ||
| T315I | 90 | F359C | 12 | ||||||||||||
| F359C/V299L | 9 | ||||||||||||||
| 263 | N | 226 | I D N | F359V | 90 | F359V | 82 | None | N/A | N/A | 35 | Progressive disease | T315I | 100 | EOT |
| T315I | 3 | ||||||||||||||
| F359V/E255K | 1 | ||||||||||||||
| 149 | N | 245 | I D | F359C | 100 | F359C/F317L | 73 | None | N/A | N/A | 31 | Adverse event | F359C | 100 | EOT |
| F317L | 80 | F359C/T315I | 21 | ||||||||||||
| T315I | 40 | F359C | 3 | ||||||||||||
| 249 | N | 840+ | I D N | Y253H | 100 | Y253H | 94 | None | N/A | N/A | 13 | Ongoing | Y253H | 10 | 1 y |
| G250E/F317L | 1 | ||||||||||||||
| 242 | N§ | 924+ | I D N | T315I | 50 | T315I/F317L | 34 | CCyR‖ | 506 | 417+ | 43 | Ongoing | Not evaluable (no amplification)†† | 1 y | |
| F317L | 40 | T315I | 1 | ||||||||||||
| 35 | N¶ | 1007+ | I D N | F317L | 100 | F317L/H396R | 75 | MMR | 259 | 664+ | 26 | Ongoing | Not evaluable (no amplification)†† | 1y | |
| H396R | 80 | F317L/E279K | 20 | ||||||||||||
| E279K | 10 | F317L/E281K | 2 | ||||||||||||
| F317L | 1 | ||||||||||||||
| 125 | N | 1014+ | I D N | V299L | 100 | H396R/V299L | 97 | None | N/A | N/A | 27 | Ongoing | No mutation detected | 1 y | |
| H396R | 100 | ||||||||||||||
| 262 | Y | 224 | I N D | V299L | 100 | F359V/V299L | 97 | CCyR | 83 | 1# | 26 | Progressive disease | Y253H | 100 | EOT |
| F359V | 100 | F359V | 100 | ||||||||||||
| 247 | Y | 298 | I (D N) B | G250E | 20 | G250E | 27 | CCyR | 175 | 78 | 43 | Death | Not evaluable (no amplification)†† | ||
| G250E/E279V | 1 | ||||||||||||||
| 115 | Y | 594 | I | T315I | 100 | T315I | 95 | MMR | 167 | 433 | 45 | Adverse event | Not evaluable (no amplification)†† | ||
| T315I/I432M | 2 | ||||||||||||||
| T315I/R386M | 1 | ||||||||||||||
| 83 | Y | 848 | I D N | F317L | 100 | F317L/E459K | 95 | MMR | 583 | 82 | 32 | Withdrawal | Not evaluable (no amplification)†† | ||
| E459K | 100 | F317L | 2 | ||||||||||||
| 101 | Y | 875 | I D | T315I | 100 | T315I | 93 | CCyR‖ | 168 | 587 | 29 | Adverse event | Not evaluable (no amplification)†† | ||
| T315I/E275D | 3 | ||||||||||||||
| 7 | Y | 903 | I N D | F317L | 100 | F317L/E450G | 92 | PCyR | 252 | 170 | 5 | Withdrawal | F317L/ E450G | 10 | EOT |
| E450G | 90 | F359V | 6 | ||||||||||||
| 185 | Y | 928+ | I D N | F359I | 84 | MMR | 524 | 322+ | 43 | Ongoing | N/A** | ||||
| F359I | 90 | F359I/T277I | 2 | ||||||||||||
| G250E | 10 | G250E | 1 | ||||||||||||
| E507G | 1 | ||||||||||||||
| 228 | Y | 937+ | I D | V299L | 100 | V299L | 95 | MMR | 55 | 785+ | 44 | Ongoing | N/A** | ||
| V299L/P310L | 2 | ||||||||||||||
| 76 | Y | 944+ | I (D N) B | M244V | 100 | M244V | 89 | MMR | 85 | 841+ | 16 | Ongoing | N/A** | ||
| F359V | 3 | ||||||||||||||
| M244V/G250W | 2 | ||||||||||||||
| 234 | Y | 951+ | I D N | F359V | 100 | F359V | 91 | MMR | 336 | 576+ | 28 | Ongoing | N/A** | ||
| F359V/M244V | 1 | ||||||||||||||
| 113 | Y | 953+ | I D N | F317L | 100 | F317L/E459K | 99 | MMR | 87 | 845+ | 45 | Ongoing | N/A** | ||
| E459K | 100 | ||||||||||||||
| 112 | Y | 967+ | I | T315I | 100 | T315I | 92 | MMR | 81 | 842+ | 44 | Ongoing | N/A** | ||
| C475W | 1 | ||||||||||||||
| T315I/K262N | 1 | ||||||||||||||
| 89 | Y | 992+ | I N B | F359V | 60 | MMR | 84 | 832+ | 31 | Ongoing | N/A** | ||||
| F359V | 90 | T315I | 29 | ||||||||||||
| T315I | 20 | F359V/M237R | 2 | ||||||||||||
| F359V/R362T | 2 | ||||||||||||||
| 36 | Y | 1012+ | I N | F317L | 100 | E459K/F317L | 98 | MMR | 168 | 762+ | 42 | Ongoing | N/A** | ||
| E459K | 100 | ||||||||||||||
| 159 | Y | 1055+ | (D N) I | T315I | 100 | T315I | 65 | MMR | 249 | 759+ | 16 | Ongoing | N/A** | ||
| T315I/M237V | 3 | ||||||||||||||
| 10 | Y | 1119+ | I N | Y253H | 100 | Y253H | 96 | MMR | 83 | 1016+ | 18 | Ongoing | N/A** | ||
| Y253H/A287V | 1 | ||||||||||||||
| Y253H/E282D | 1 | ||||||||||||||
| Y456C | 1 | ||||||||||||||
B, bosutinib; CCyR, complete cytogenetic response; D, dasatinib; EOT, end of treatment; I, imatinib; MMR, major molecular response; N, nilotinib; N/A, not applicable; PADD, ponatinib average daily dose; PCyR, partial cytogenetic response; Pt, patient; TKI, tyrosine kinase inhibitor.
Patients who did not achieve the primary end point are listed first, with each subgroup then arranged by time on study. Bold font denotes exact or alternate (also underlined) substitutions at amino acids previously associated with TKI resistance4 ; nonbolded font denotes substitutions at amino acids not previously associated with TKI resistance.
The order of TKIs from left to right indicates the sequence of TKI treatment in the patient’s history (first to last); parentheses indicate that the treatment order is unknown.
The percentage indicates the mutation allele frequency in the patient sample.
Best response does not include hematologic response.
Cytogenetic response for this patient was only assessed at 9 months (no MCyR) and 18 months (CCyR).
In this patient, MMR was achieved at 9 months; however, cytogenetic response was not assessed between months 3 and 12; SS was conducted at 1 year and no amplification of the transcript was achieved.
Patients with atypical transcripts in whom molecular response could not be assessed.
1-day duration indicates patient had one assessment at which criteria for response were met and no further evaluable cytogenetic assessments.
Postbaseline mutation status was not evaluated for patients who achieved and remain on study in continuous MCyR.
Patients are in CCyR and/or <1% BCR-ABL1 transcript levels at the time of sample analysis.
Compound mutations were highly heterogeneous. Only 3 compound mutations were observed in multiple patients: F317L/E459K in 3 patients, V299L/H396R in 2 patients, and T315I/F359C in 2 patients (Table 1). Compound mutations invariably involved at least 1 mutation previously implicated in TKI resistance (Table 1). Notably, T315I-inclusive compound mutations were observed in only 3% of all patients (7/267) despite enrichment for T315I-positive patients in the PACE trial. In summary, whereas a majority (61%) of PACE CP-CML patients had at least 1 BCR-ABL1 mutation detected by NGS at baseline, and 23% had >1 mutation, only 9% had compound mutations.
Robust and durable major cytogenetic responses to ponatinib irrespective of baseline mutation status
Previous analyses based on SS revealed high response rates in PACE CP-CML patients, irrespective of the number of BCR-ABL1 mutations detected at baseline (ie, 0, 1, or ≥2), with MCyR rates by 1 year ranging from 49% to 64% and MMR rates at any time ranging from 29% to 48% (Figure 2B-C).16 Response rates in patients with 0, 1, or ≥2 BCR-ABL1 mutations at baseline based on NGS were similar to those based on SS (50%-61% MCyR by 1 year and 29%-45% MMR at any time; Figure 2B-C). Moreover, response rates in patients with LL mutants only (43% MCyR and 31% MMR) were similar to those with no mutations, and response rates in patients with compound mutations (64% MCyR and 52% MMR) (Figure 2B-C) were similar to those in patients with ≥1 mutations. Importantly, responses were durable regardless of baseline mutation status (Figure 2D and supplemental Figure 2). Among patients who achieved MCyR or MMR, the estimated rates of a sustained response of at least 2 years were 87% and 65%, respectively, for the total population, and 90% and 92% for patients with compound mutations. Baseline mutation status also had no significant impact on rates of CCyR, PFS, or OS, estimated to be 79%, 68%, and 86%, respectively, for the total population at 2 years (supplemental Figure 2). In summary, robust and durable responses were observed irrespective of baseline NGS mutation status, including in patients with compound mutants.
No single or compound mutant as a major driver of primary resistance to ponatinib
To further investigate the relationship between BCR-ABL1 mutation status and sensitivity to ponatinib, we evaluated responses according to the specific single or compound mutation present at baseline as determined by NGS. Evidence of efficacy was observed against all 20 single mutants present in at least 2 patients (Table 2). Ten mutations were present in at least 5 patients at baseline. Response rates were high (20%-68% MCyR and 25%-54% MMR) in patients with H396R, V299L, E459K, F317L, F359V, M244V, E255K, G250E, and T315I. Response rates were lower in patients with F359C (14% MCyR and 0% MMR). Of the 7 patients with F359C present at baseline, 6 had a postbaseline sample available for analysis. In 2 patients, F359C was still detectable by SS, whereas in 4 it was not (data not shown), indicating that the mutant clone had not expanded during therapy. Although patient numbers were small, response rates were relatively low in patients with E255V, the mutation with the lowest in vitro sensitivity to ponatinib of those tested (IC50 = 16 nM; 1/4 and 0/4 achieved MCyR and MMR, respectively). Overall, no association between response rates and IC50 was observed (Table 2).
Response rates according to baseline BCR-ABL1 mutation for the 20 mutants detected by NGS in at least 2 patients
| Unique NGS baseline mutations . | Patients (NGS) . | Patients (SS) . | Patients (LL) . | IC50 (nM)* . | MCyR rate (%)† . | MMR rate (%)† . |
|---|---|---|---|---|---|---|
| T315I | 74 | 64 | 10 | 6 | 50/74 (68) | 40/74 (54) |
| F317L | 29 | 22 | 7 | 4 | 13/29 (45) | 12/29 (41) |
| F359V | 20 | 13 | 7 | 4 | 11/20 (55) | 8/20 (40) |
| E255K | 13 | 8 | 5 | 6 | 8/13 (61.5) | 6/13 (46) |
| G250E | 12 | 8 | 4 | 5 | 8/12 (67) | 4/12 (33) |
| M244V | 9 | 5 | 4 | 3 | 4/9 (56) | 3/9 (33) |
| V299L | 8 | 5 | 3 | 4 | 3/8 (37.5) | 2/8 (25) |
| F359C | 7 | 4 | 3 | 6 | 1/7 (14) | 0/7 (0) |
| E459K | 7 | 3 | 4 | 5 | 3/7 (43) | 3/7 (43) |
| H396R | 5 | 5 | 0 | 4 | 1/5 (20)‡ | 2/5 (40) |
| E255V | 4 | 2 | 2 | 16 | 1/4 | 0/4 |
| F359I | 4 | 4 | 0 | 11 | 3/4 | 1/4 |
| L248V | 2 | 2 | 0 | 4 | 1/2 | 1/2 |
| Q252H | 2 | 1 | 1 | ND | 2/2 | 2/2 |
| Y253H | 2 | 2 | 0 | 5 | 1/2 | 1/2 |
| E279K | 2 | 1 | 1 | ND | 0/2§ | 1/2 |
| M351T | 2 | 1 | 1 | 4 | 1/2 | 1/2 |
| E355A | 2 | 2 | 0 | 7 | 1/2 | 1/2 |
| H396P | 2 | 1 | 1 | ND | 1/2 | 0/2 |
| M437I | 2 | 0 | 2 | ND | 1/2 | 1/2 |
| Unique NGS baseline mutations . | Patients (NGS) . | Patients (SS) . | Patients (LL) . | IC50 (nM)* . | MCyR rate (%)† . | MMR rate (%)† . |
|---|---|---|---|---|---|---|
| T315I | 74 | 64 | 10 | 6 | 50/74 (68) | 40/74 (54) |
| F317L | 29 | 22 | 7 | 4 | 13/29 (45) | 12/29 (41) |
| F359V | 20 | 13 | 7 | 4 | 11/20 (55) | 8/20 (40) |
| E255K | 13 | 8 | 5 | 6 | 8/13 (61.5) | 6/13 (46) |
| G250E | 12 | 8 | 4 | 5 | 8/12 (67) | 4/12 (33) |
| M244V | 9 | 5 | 4 | 3 | 4/9 (56) | 3/9 (33) |
| V299L | 8 | 5 | 3 | 4 | 3/8 (37.5) | 2/8 (25) |
| F359C | 7 | 4 | 3 | 6 | 1/7 (14) | 0/7 (0) |
| E459K | 7 | 3 | 4 | 5 | 3/7 (43) | 3/7 (43) |
| H396R | 5 | 5 | 0 | 4 | 1/5 (20)‡ | 2/5 (40) |
| E255V | 4 | 2 | 2 | 16 | 1/4 | 0/4 |
| F359I | 4 | 4 | 0 | 11 | 3/4 | 1/4 |
| L248V | 2 | 2 | 0 | 4 | 1/2 | 1/2 |
| Q252H | 2 | 1 | 1 | ND | 2/2 | 2/2 |
| Y253H | 2 | 2 | 0 | 5 | 1/2 | 1/2 |
| E279K | 2 | 1 | 1 | ND | 0/2§ | 1/2 |
| M351T | 2 | 1 | 1 | 4 | 1/2 | 1/2 |
| E355A | 2 | 2 | 0 | 7 | 1/2 | 1/2 |
| H396P | 2 | 1 | 1 | ND | 1/2 | 0/2 |
| M437I | 2 | 0 | 2 | ND | 1/2 | 1/2 |
ND, not determined.
The mutations are sorted by number of patients with the corresponding mutant as detected by NGS, and then by ascending amino acid sequence.
IC50 values are based on ponatinib potency in Ba/F3 cells harboring the corresponding BCR-ABL1 mutation.29
MCyR rate by 12 months or MMR rate at any time is reported based on patients with mutations detected by NGS in this study; percent of MCyR or MMR is shown when the number of evaluable patients was ≥5. Bold font denotes exact substitutions at amino acids previously associated with TKI resistance4 ; nonbolded font denotes a substitution at an amino acid not previously associated with TKI resistance.
One patient achieved MMR at 12 months but did not have documented evidence of MCyR by 12 months because CyR was not assessed between 3 and 12 months; therefore, this patient was not included as achieving MCyR.
In one patient, MMR was achieved at 9 months; however, cytogenetic response was not assessed between months 3 and 12.
Of the 25 patients found to harbor compound mutations at baseline, 16 achieved MCyR by 1 year (Table 1). Of the 9 patients who did not, 1 with 3 compound mutations (patient 35; F317L/H396R, F317L/E279K, and F317L/E281K) achieved MMR at 9 months but did not have a cytogenetic assessment between months 3 and 12, and 1 with a T315I/F317L compound mutation (patient 242) subsequently achieved CCyR and remains on study. Of the remaining 7 patients, 5 did not have compound mutations detected by SS in postbaseline samples, indicating that these compound mutants did not expand during therapy (Table 1, patients 191, 263, 149, 249, and 125); the remaining 2 could not be evaluated. In summary, with the possible exception of F359C, no recurrent single or compound mutation conferring primary resistance to ponatinib was detected at baseline in the 267 CP-CML patients from the PACE trial.
No single or compound mutant as a major driver of secondary resistance to ponatinib
To evaluate whether single or compound mutants undetectable by NGS at baseline emerged and expanded during ponatinib therapy, postbaseline samples were analyzed by SS. Postbaseline sequencing was attempted on all treated patients, except those who achieved MCyR by 1 year and remain on the trial in continuous MCyR (Figure 1). In total, postbaseline samples from 129 patients were evaluable. Of these patients, 24 had ceased ponatinib for at least 1 month before the postbaseline mutation analysis (34-239 days). Previous studies have demonstrated that some mutants can be rapidly deselected in the absence of kinase inhibition.21-23 Therefore, some mutations may have become undetectable by SS at the time of the postbaseline analysis. Of the 129 patients evaluated, 8 harbored mutations that were not detected at baseline by NGS (Table 3). E255V was detected in 1 patient at the time of MCyR loss (patient 248) and was associated with a low average daily dose of ponatinib (13 mg). T315I was detected in 3 patients: in 2 at the time of MCyR loss (patients 29 and 155) and in 1 after 1 year of treatment in a patient (patient 121) with a low average daily dose of ponatinib (6 mg) and a history of T315I before commencing ponatinib. Finally, compound mutations were detected at the end of treatment in 4 patients—Y253H/F359V in 2 patients, and T315I/M351T and T315I/F359V each in 1 patient (in each case 1 mutation was detected at a frequency of 100% and the second was detected at 40% to 100%, using SS). Notably, there was no evidence of F359C being newly acquired in any patient. Thus, at a median follow-up of 30.1 months, emergence of previously undetected single and compound mutants during ponatinib therapy is a rare event.
Clinical and molecular characteristics of patients with postbaseline mutations that were not identified by NGS at baseline
| Patient ID . | Prior TKI* . | Baseline NGS (frequency) . | Postbaseline SS (frequency) . | Achieved primary end point . | Lost primary end point . | Reason for discontinuation . | Sample analysis at . | IC50 (nM)† . | Average daily dose (mg) . | Comment . |
|---|---|---|---|---|---|---|---|---|---|---|
| 248 | I D N | G250E (1%) | E255V (100%) | Y | Y | Adverse event | EOT | 16 | 13 | Low drug exposure |
| 29 | I N | None | T315I (100%) | Y | Y | Progressive disease | EOT | 6 | 43 | Possible BCR-ABL–independent resistance |
| 155 | I D | None | T315I (100%) | Y | Y | Withdrawal by subject | EOT | 6 | 44 | Possible BCR-ABL–independent resistance |
| 121 | I N | E355A (81%) | T315I (10%) | N | N/A | On study | 1 y | 6 | 6 | Low drug exposure and re-emergence of T315I |
| E355A (10%) | 7 | |||||||||
| 262 | I D N | V299L/F359V (97%) | Y253H/F359V (100%/100%) | Y | N‡ | Progressive disease | EOT | 5 | 26 | Compound mutant at EOT |
| 158 | I D N | F359V (93%) | Y253H/F359V (100%/100%) | N | N/A | Other | EOT | 5 | 34 | Compound mutant at EOT |
| M244V (4%) | ||||||||||
| 142 | B | T315I (41%) | T315I/M351T (100%/40%) | N | N/A | Other | EOT | 27 | 45 | Compound mutant at EOT |
| 167 | I D | None | T315I/F359V (100%/90%) | Y | N‡ | Adverse event | EOT | 5 | 30 | Compound mutant at EOT |
| Patient ID . | Prior TKI* . | Baseline NGS (frequency) . | Postbaseline SS (frequency) . | Achieved primary end point . | Lost primary end point . | Reason for discontinuation . | Sample analysis at . | IC50 (nM)† . | Average daily dose (mg) . | Comment . |
|---|---|---|---|---|---|---|---|---|---|---|
| 248 | I D N | G250E (1%) | E255V (100%) | Y | Y | Adverse event | EOT | 16 | 13 | Low drug exposure |
| 29 | I N | None | T315I (100%) | Y | Y | Progressive disease | EOT | 6 | 43 | Possible BCR-ABL–independent resistance |
| 155 | I D | None | T315I (100%) | Y | Y | Withdrawal by subject | EOT | 6 | 44 | Possible BCR-ABL–independent resistance |
| 121 | I N | E355A (81%) | T315I (10%) | N | N/A | On study | 1 y | 6 | 6 | Low drug exposure and re-emergence of T315I |
| E355A (10%) | 7 | |||||||||
| 262 | I D N | V299L/F359V (97%) | Y253H/F359V (100%/100%) | Y | N‡ | Progressive disease | EOT | 5 | 26 | Compound mutant at EOT |
| 158 | I D N | F359V (93%) | Y253H/F359V (100%/100%) | N | N/A | Other | EOT | 5 | 34 | Compound mutant at EOT |
| M244V (4%) | ||||||||||
| 142 | B | T315I (41%) | T315I/M351T (100%/40%) | N | N/A | Other | EOT | 27 | 45 | Compound mutant at EOT |
| 167 | I D | None | T315I/F359V (100%/90%) | Y | N‡ | Adverse event | EOT | 5 | 30 | Compound mutant at EOT |
B, bosutinib; D, dasatinib; EOT, end of treatment; I, imatinib; N, nilotinib; N/A, not applicable.
Mutations detected postbaseline that were not detected at baseline are bolded; if such mutations had been detected before baseline (in the patient’s history), they are also underlined.
The order of TKIs from left to right indicates the sequence of TKI treatment in the patient’s history (first to last).
IC50 values are based on in vitro ponatinib activity against cells harboring bolded BCR-ABL1 mutations.29
Patient achieved primary end point; however, loss of response before discontinuation was not documented.
Discussion
BCR-ABL1 KD mutations are a major mechanism of CML resistance to TKIs, and the BCR-ABL1 genotype is used to rationalize selection of salvage therapy after first-line TKI failure.4 KD mutants present at low levels in specimens obtained before switching (“switch samples”) can be selected on therapy if not covered by the second-generation TKI chosen for salvage therapy.11 In contrast, we show that the genotype of the switch sample, whether assessed by SS or NGS, has no impact on cytogenetic or molecular responses of CP-CML patients treated with ponatinib. These data are consistent with the in vitro profile of ponatinib that predicts activity against all single mutants previously associated with resistance to imatinib, including the T315I, F317L, Y253H, F359V/C, E255K/V, and V299L mutants associated with resistance to second-generation TKIs.2,5 Response rates were notably lower in the 7 patients with the F359C mutation. Based on its in vitro ponatinib IC50 of 6 nM, F359C should be responsive to the ∼64 nM trough plasma concentrations achieved with the 45-mg daily initial ponatinib dose used in the PACE study.15 The fact that F359C was undetectable in 4 of 6 patients with evaluable follow-up samples argues against a causative role of F359C for resistance, although deselection remains possible in 1 patient whose last dose of ponatinib predated the mutation analysis sample by 85 days. Responses were generally durable regardless of baseline mutation status: overall 87%, 79%, and 66% of patients were estimated to remain in MCyR, CCyR, and MMR, respectively, for 2 years, with similar durations observed across the various subgroups (compound, LL only, 0, 1, ≥2 mutants). Consistent with this, baseline mutation status had no impact on PFS and OS.
BCR-ABL1 compound mutations, particularly those including T315I, have recently been shown to confer high-level resistance against all approved TKIs, including ponatinib, raising the question of whether preexisting compound mutants may be selected on therapy.5 Although SS cannot distinguish between polyclonal and compound mutations, unless the combined mutant allele burden clearly exceeds 100%, NGS identifies compound mutations as long as the average read length exceeds the distance between the 2 single nucleotide variations. With an average read length of 400 bp, our assay is expected to detect the vast majority of compound mutations, although some long-distance combinations may be missed. Recent reports have drawn attention to “recombination” events that can cause false-positive compound mutation calls.14 In our analysis, a substantial percentage of compound mutation calls appear to be false-positives (41%; 23/56), which increased to 88% (23/26) when a compound mutation and both single mutations were detected in the same sample. The precise mechanisms responsible for these recombination events remain to be defined (supplemental Figure 1). Based on a direct measurement of a false-positive “recombination rate” between 2 mutations separated by a known distance, we developed a simple algorithm that predicts the frequency with which false-positive compound mutants are observed with remarkable precision (supplemental Methods). Still, after eliminating all suspect compound mutation calls, we observed no correlation between compound mutation detection at baseline and cytogenetic and molecular response to ponatinib. This surprising result may be explained by the very low frequency of compound mutations containing T315I, and compound mutants without T315I remaining sensitive to ponatinib.5 Conversely, in the 4 patients with compound mutations at end of treatment, NGS failed to detect the respective compound mutations at baseline, suggesting they were below the detection limit or were acquired on therapy.
Thus, our data show that the increased sensitivity of mutation detection afforded by NGS is of no clinical utility in terms of predicting response to ponatinib in patients with CP-CML. However, because current guidelines3 suggest using ponatinib in patients with the T315I mutation, and low-level detection of T315I is associated with failure of dasatinib and nilotinib, it is conceivable that NGS, similarly to targeted approaches such as mass spectrometry,11,13 may identify patients likely to benefit from ponatinib. Importantly, relapse rates were much higher in PACE patients with AP-CML or BP-CML compared with CP-CML, and compound mutations were detected by SS in many AP- and BP-CML patients at the time of discontinuation,5 suggesting that NGS may have predictive utility in these more advanced cases.
Although 161 mutations detected by SS occurred at amino acids previously implicated in TKI resistance, 34 of the 105 LL mutations (31%) occurred at amino acids that were not.4 The fact that none of these “novel” mutants was found to expand during ponatinib therapy to become detectable by SS strongly suggests they are not relevant to resistance. Similarly, responses to ponatinib were independent of the number of BCR-ABL1 mutations detected at baseline by NGS (0, 1, or ≥2; Figure 2B-C) and substantially exceeded the 26% MCyR and 3% MMR rates observed in these patients on their prior line of therapy.16 Interestingly, response rates in patients with LL mutants only (43% and 31% MCyR by 1 year and MMR at any time, respectively) were more similar to those with no mutations (50% and 29%) than to patients with compound mutations (64% and 52%) or with ≥1 mutations (57%-64% and 44%-48%). We speculate that the relatively low response rates in patients with no BCR-ABL1 mutations may reflect the presence of BCR-ABL1–independent mechanisms of resistance, and that this may also be the case in patients with exclusively LL mutations. BCR-ABL1–independent resistance may also account for the fact that ∼30% of T315I patients did not respond to ponatinib, although this mutant should be sensitive to ponatinib plasma concentrations expected with 45-mg daily dosing. Low drug exposure as a result of pharmacokinetic factors and/or dose reductions caused by adverse events may be contributing factors. Inducing rapid and deep reductions in disease burden may ultimately reduce the probability of randomly acquiring additional resistance mutations and achieve better outcomes.24-26
Acquired resistance to imatinib and second-generation TKIs is frequently associated with the emergence of new BCR-ABL1 mutations. We analyzed postbaseline samples from 129 patients (all except those who remain on trial in continuous MCyR at a median follow-up of 30.1 months). In total, 8 of 129 patients were found to have mutations that were not detectable at baseline: 3 of 102 who did not achieve MCyR by 1 year, 3 of 14 who achieved and lost MCyR, and 2 of 13 who achieved MCyR and discontinued study while in MCyR. In 4 of the patients, a single mutation emerged (T315I in 3 patients and E255V in 1 patient with relatively low exposure to ponatinib [average dose = 13 mg]). High response rates were seen in patients with T315I (MCyR 68%), suggesting that BCR-ABL1 kinase–independent mechanisms are responsible or at least contributed to resistance in the T315I cases. Although the number of patients with E255V at baseline was small, the response rate was low (MCyR 25%), which suggests that E255V may contribute more directly to ponatinib resistance. However, 2 of 2 patients with AP-CML in the PACE study who had an E255V mutation at baseline achieved MCyR,16 demonstrating that E255V does not confer uniform resistance to ponatinib. Compound mutants emerged in the remaining 4 patients. Y253H/F359V was observed in 2 patients previously successively treated with imatinib, dasatinib, and nilotinib, and T315I/M351T and T315I/F359V were observed in 1 patient each. T315I/M351T and T315I/F359V confer significant resistance to ponatinib (IC50 ∼100 nM), consistent with a causal role for clinical resistance in these patients, whereas Y253H/F359V has thus far not been characterized.5 Overall, however, mutational escape is rare in CP-CML patients treated with ponatinib, despite the fact that many of these patients were pretreated with multiple TKIs. In contrast, certain compound mutants have been found to emerge frequently in BP-CML and Ph+ ALL patients treated with ponatinib or other TKIs.5,16 Compound mutants that arise through sequential mutation of a single amino acid residue have also been observed (eg, T315M, which can arise from a T315-I315, followed by an I315-M315 mutation event).5 Higher levels of genomic instability and greater clonal diversity are likely to contribute to the higher frequency of compound mutations in advanced BCR-ABL1–positive leukemias. In our study, NGS was limited to the baseline samples, and it is unknown whether ponatinib exposure changes the frequency and spectrum of LL mutations—for example, by improving genetic stability through profound inhibition of BCR-ABL1 kinase activity.
Ponatinib is the most potent BCR-ABL1 TKI, inducing rapid, deep, and durable responses in CP-CML patients; however, ponatinib treatment is associated with considerable cardiovascular toxicity that may be dose-dependent—as described in a post hoc multivariate analysis of the PACE trial.27 The results discussed in this study are based on the analysis of patients treated with a starting dose of 45 mg ponatinib, though the average daily dose (over the median follow-up of 30.1 months) was only ∼30 mg owing to dose reductions. As strategies to lower average dose intensity are explored, it will be important to determine whether this results in an increase in mutation-based resistance. Although preliminary analyses of PACE patients suggest that responses are maintained in most patients after dose reductions to 15 or 30 mg, more follow-up is required to maximize the benefit/risk ratio in a rational manner.28
In summary, we show that pretherapeutic BCR-ABL1 mutation profiles, whether obtained by SS or by NGS, have little impact on ponatinib response, and no single or compound mutant has been identified as a major driver of primary and secondary resistance to ponatinib in CP-CML patients. The role of NGS in this setting may be to identify patients with LL T315I who are unlikely to derive lasting benefit from second-generation TKIs, but have a high likelihood of achieving durable cytogenetic and molecular responses to ponatinib, an important factor for balancing risks and benefits of salvage therapy selection.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all the patients, their families and caregivers, the site investigators, and research personnel for their participation in the PACE trial. They acknowledge their colleagues at ARIAD Pharmaceuticals, Inc. and in the CML community for their contributions. They also thank Thihan Padukkavidana (ARIAD Pharmaceuticals, Inc.) for scientific writing and editorial assistance.
This study was funded by ARIAD Pharmaceuticals, Inc.
Authorship
Contribution: M.W.D., S.B., V.M.R., and J.G.H. designed experiments, performed research, analyzed data, and wrote the manuscript; S.L. performed statistical analyses and contributed to writing the manuscript; N.P.S., J.E.C., D.-W.K., F.E.N., M.T., M.B., M.C.M., J.L., W.T.P., F.G., H.M.K., S.S., A.H., and T.P.H. analyzed data and contributed to writing the manuscript; and T.C. and F.G.H. reviewed the manuscript, provided critical feedback, and contributed to writing the manuscript.
Conflict-of-interest disclosure: M.W.D. researched research funding from BMS, Novartis, Celgene, Genzyme, and Gilead, and is on the advisory board and consultant for BMS, ARIAD, Novartis, Incyte, and Pfizer. J.G.H., S.L., T.C., F.G.H., and V.M.R. are employees and hold equity ownership at ARIAD Pharmaceuticals Inc. N.P.S. received research funding from ARIAD and BMS. J.E.C. is a consultant for ARIAD, BMS, Novartis, and Pfizer, and received research funding from ARIAD, BMS, Novartis, Pfizer, and Teva. D.-W.K. is a consultant for BMS and Novartis, and received honoraria and research funding from BMS, Novartis, and Wyeth. F.E.N. is a consultant for Novartis, BMS, and ARIAD; received research funding from Novartis; and received honoraria from Novartis, BMS, and ARIAD. M.T. received research funding from ARIAD, BMS, Sanofi, Incyte, and Pfizer. M.B. is a consultant for ARIAD, Novartis, and BMS, and received honoraria and is on the speakers bureau for ARIAD, BMS, Novartis, Pfizer, and Teva. M.C.M. is a consultant to and received research funding and honoraria from ARIAD, BMS, and Novartis. J.L. is an employee of MolecularMD, Inc. F.G. received honoraria from ARIAD. H.M.K. received research funding from ARIAD. S.S. is a consultant for ARIAD, Novartis, and BMS. A.H. received honoraria and research funding from ARIAD, Novartis, BMS, and Pfizer. T.P.H. and S.B. received honoraria and research funding from ARIAD, Novartis, BMS. W.T.P. declares no competing financial interests.
Correspondence: Michael Deininger, Division of Hematology and Hematologic Malignancies, Huntsman Cancer Institute, The University of Utah, Salt Lake City, UT 84112; e-mail: michael.deininger@hci.utah.edu.

![Figure 1. Schematic representation of the BCR-ABL1 mutation analyses performed in this study. BCR-ABL1 mutation analysis (Sanger sequencing [SS] and next-generation sequencing [NGS]) was conducted on baseline samples from all chronic-phase chronic myeloid leukemia (CP-CML) patients in the PACE phase 2 study. Postbaseline analysis (SS) was conducted on any patient who did not achieve major cytogenetic response (MCyR) by 1 year, achieved and then lost MCyR, or achieved MCyR and discontinued while in MCyR. *Postbaseline (post-BL) analysis was not conducted on patients who achieved MCyR and remain on study in continuous MCyR. ^36 patients were not evaluable: 12 did not have a post-BL sample collected, and BCR-ABL1 could not be amplified for sequencing in the remaining 24; 20 of these were associated with low (<1%) BCR-ABL1 transcript levels. Of the 129 patients with evaluable post-BL samples, 102 were collected from patients who did not achieve MCyR by 1 year, 14 from patients who achieved and lost MCyR, and 13 from patients who achieved MCyR and discontinued study while in MCyR.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/6/10.1182_blood-2015-08-660977/4/m_703f1.jpeg?Expires=1765885225&Signature=V01I7pIN4tBBOTqVqKe6-2yiFfjj5~EXYekGRZNuwZhi7ciDNkwBFJHiF8XeuDJwd5V3mZ~oJjikMIfr6~LJQO0ba98X3SWlrGeJf5QTUmcsmvOsfuBUfxCeTpM7~sefbofFhVyXPam2D5Cerjp10KoRjy-sJW-Lwz1cg1ajq2hNUZ~LIe4tihIkOHMTiFqNRgoM7xcaSFmBf61Y9V8D~tcdZzh8zO1lNE1gQ4vT26tnKEKJwwi8UsAPtoiE7TBMiiydxOgXPMHRHwiBlqXGwviqxfL-MPFL17lbIwo0v8Dn3dtoKahp~zYMYs3gJSQHg40TTQhOsTd-rjXPLfDCZQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)