Abstract
Evidence of human acute myeloid leukemia stem cells (AML LSCs) was first reported nearly 2 decades ago through the identification of rare subpopulations of engrafting cells in xenotransplantation assays. These AML LSCs were shown to reside at the apex of a cellular hierarchy that initiates and maintains the disease, exhibiting properties of self-renewal, cell cycle quiescence, and chemoresistance. This cancer stem cell model offers an explanation for chemotherapy resistance and disease relapse and implies that approaches to treatment must eradicate LSCs for cure. More recently, a number of studies have both refined and expanded our understanding of LSCs and intrapatient heterogeneity in AML using improved xenotransplant models, genome-scale analyses, and experimental manipulation of primary patient cells. Here, we review these studies with a focus on the immunophenotype, biological properties, epigenetics, genetics, and clinical associations of human AML LSCs and discuss critical questions that need to be addressed in future research.
Introduction
Acute myeloid leukemia (AML) is a rapidly progressing hematopoietic malignancy, characterized by the accumulation of clonal myeloid progenitor cells arrested in their ability to differentiate into mature blood cells that is accompanied by multilineage cytopenias.1 Indeed, it is the lack of normal cells, including red blood cells, platelets, and granulocytes, that eventually contributes to morbidity in this disease. While reductions in leukemic blasts can be achieved initially with cytarabine and anthracycline chemotherapy in the majority of patients, long-term outcomes have not improved significantly for over 3 decades, with 5-year overall survival rates for patients <60 years ranging from 35% to 40% and median overall survival of ∼1 year.1,2 Many samples from patients with AML show evidence of a hierarchical cellular organization, with a minor fraction of self-renewing leukemia stem cells (LSCs) at the apex of this hierarchy that can maintain the disease long-term in immunodeficient mice. LSCs are defined as cells that are capable of initiating the disease when transplanted into immunodeficient animals and can self-renew by giving rise to leukemia in serial transplantations and also partially differentiate into non-LSC bulk blasts that resemble the original disease but are unable to self-renew. The earliest conceptual idea of leukemia being organized in a hierarchical manner traces back to studies performed to identify clonogenic AML progenitors in vitro.3,4 Dick and colleagues later demonstrated that somewhat similar to normal hematopoiesis, AML is organized in this hierarchical fashion in vivo, being driven long-term by LSCs.5-7
From a clinical perspective, the cancer stem cell model implies that in order to eradicate the disease and achieve long-term remissions, treatment courses must eliminate the LSC population.8 While this ultimate goal is yet to be realized, detailed characterization of AML LSCs has demonstrated their properties of self-renewal, relative quiescence, resistance to apoptosis, and increased drug efflux that likely render them less susceptible to conventional therapies aimed at the bulk proliferative disease.9 Recently, high-throughput genome sequencing and DNA methylation profiling has increased our knowledge of the genomic and epigenomic landscape of this disease,10,11 but merging these data with in vivo biology of LSCs is still in its infancy. The application of genomic methodologies has also led to the identification of preleukemic hematopoietic stem cells (HSCs) in AML patients that do not generate leukemia in vivo but carry early competitive driver mutations, typically in epigenetic regulator genes.12-14 In this article, we review the immunophenotype, biological properties, epigenetics, genetics, and clinical associations of human AML LSCs and highlight themes for further investigation.
Past perspectives on leukemia heterogeneity and stem cells
While hematopathologists had long noted morphological differences in leukemic cells both between and within patients, the first in vivo evidence of intraleukemic heterogeneity came directly from AML patients injected with tritiated thymidine.15 Marked differences in the proliferation kinetics of individual leukemic cells could be distinguished, anticipating the existence of a rare leukemic population that cycled very slowly and showed resistance to antiproliferative therapies.16 Unexpectedly, AML cells did not proliferate more extensively than normal hematopoietic cells in patients, and a consistent population of quiescent cells could be distinguished. These observations were later followed by efforts to identify AML progenitors in vitro3,4,17,18 and inspired an interest in conceptualizing AML in a hierarchical organization, akin to what was being observed in normal hematopoiesis at that time. Isolated primary cells that could give rise to leukemic blast colonies (∼1%) in semisolid media were termed AML colony-forming cells (CFCs) and showed wide variation between patients; however, they did not correlate with clinical outcome and exhibited very limited replating efficiency.18 Improvements in culture conditions using normal human bone marrow or mouse fibroblast feeder cell layers and recombinant human cytokines allowed measurement of rarer cells that could give rise to AML CFCs for more than 5 weeks in vitro, termed long-term culture-initiating cells (LTC-IC), with a frequency 5- to 300-fold lower than CFCs.19 In vivo evidence for LSCs was later demonstrated by serial xenotransplantation experiments in severe combined immunodeficiency (SCID) mice,6 and then later in nonobese diabetic (NOD)/SCID mice,7 using fluorescence-activated cell sorting (FACS) to isolate engrafting subpopulations from a number of AML samples. These robust but scarce leukemic cells, termed leukemia-initiating cells (LICs), comprised roughly 1 per 1 × 106 leukemic blasts and had 2 important properties: (1) they gave rise to leukemic engraftment that could be propagated for multiple serial transplants (self-renewal), and (2) they produced non-LSC progeny unable to engraft. In the most rigorous definition, cells that meet both these criteria are termed LSCs, although not all studies in the literature that use this designation perform functional in vivo or serial transplantation experiments.20
Immunophenotype of LSCs in human AML
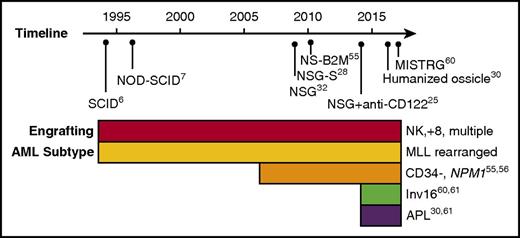
The critical step in the identification of human AML LSCs is the establishment of a xenotransplantation assay that facilitates engraftment of patient samples. Over the years, a number of immunodeficient mouse strains have been developed to facilitate engraftment of normal and malignant human hematopoietic cells, including AML (reviewed in Goyama et al21). However, what is often not apparent from reported studies is that many primary AML samples do not engraft at all when transplanted into these models. Engraftment has historically been defined as human leukemic chimerism >0.1% in the mouse bone marrow, a notably low bar compared with clinical disease. Using this criterion, a review of historical studies of diagnostic samples of AML reporting both engrafting and nonengrafting samples found that in aggregate, 23 out of 49 samples (47%) engrafted.22-25 A recent study reported results of engraftment of 56 AML samples, and found that 45 (80%) engrafted in xenotransplantation assays, although it should be noted that more than half of these were from relapsed or refractory patients.26 Recently, a humanized ossicle xenotransplantation model was reported in which 13 out of 15 diagnostic AML samples (87%) engrafted to high levels of bone marrow chimerism.27 Apart from engraftment, the frequency of LICs varied greatly between samples and models, indicating that identification and frequency of LSCs is highly dependent on the model used (Figure 1). Indeed, to what extent immunological and interspecies differences in specific xenotransplant models have dictated the observed biological properties of LSCs is an important and open question.
Timeline of human AML engraftment models. Historical timeline is indicated with immunodeficient mouse models used for AML engraftment. The panel below indicates the subtypes of AML exhibiting engraftment in the models along with the relevant references. Note that as the immunodeficient mouse models improved the engraftment of more AML subtypes was observed. APL, acute promyelocytic leukemia; NK, normal karyotype; MISTR-G, RAG2-IL2R-γ null with human M-CSF, IL-3, GM-CSF, SIRPA, and TPO; MLL, mixed-lineage leukemia; NSG, NOD-SCID-IL2R-γ null; NS-B2M, NOD-SCID-beta2-microglobulin null; NSG-S, NSG expressing human IL-3, GM-CSF, and stem cell factor.
Timeline of human AML engraftment models. Historical timeline is indicated with immunodeficient mouse models used for AML engraftment. The panel below indicates the subtypes of AML exhibiting engraftment in the models along with the relevant references. Note that as the immunodeficient mouse models improved the engraftment of more AML subtypes was observed. APL, acute promyelocytic leukemia; NK, normal karyotype; MISTR-G, RAG2-IL2R-γ null with human M-CSF, IL-3, GM-CSF, SIRPA, and TPO; MLL, mixed-lineage leukemia; NSG, NOD-SCID-IL2R-γ null; NS-B2M, NOD-SCID-beta2-microglobulin null; NSG-S, NSG expressing human IL-3, GM-CSF, and stem cell factor.
CD34+ AML
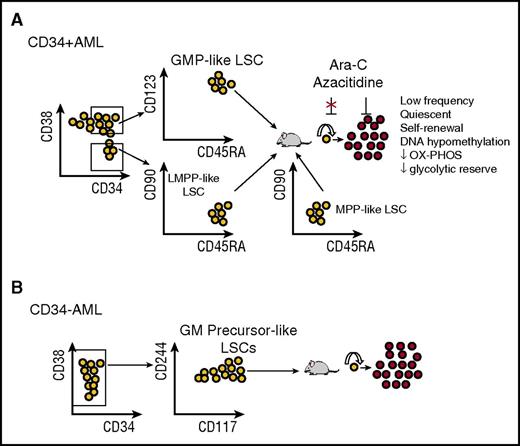
The majority of AML samples (∼75%) are positive for expression of CD34, defined as present on >10% of blasts, and nearly all studies of AML LSCs have focused on this subgroup. In the initial studies, LSCs were shown to be positive for expression of CD34 and negative for expression of CD38 as demonstrated by engraftment in NOD/SCID mice7 and LTC-IC activity in vitro.19,28 Further work by multiple laboratories,22,23,29,30 using both intrafemoral and intravenous transplantation and more permissive models, including NOD/SCID/IL2R-γ null (NSG) mice, showed that for CD34+ AML, LICs predominantly reside in the CD34+CD38− fraction. However, in more than half of the samples, LSCs were also present in at least one other subpopulation, usually the CD34+CD38+ fraction and sometimes in the CD34− fraction.20 To further examine the relationship and plasticity between these subpopulations, one group performed detailed immunophenotypic analysis of 100 CD34+ samples coupled with engraftment studies in 12 cases. Consistent with prior work, they demonstrated the coexistence of at least 2 distinct CD34+ LSC populations in these patients: a CD34+CD38− fraction resembling normal lymphoid-primed multipotent progenitors (LMPP-like LSCs; Lin−CD34+CD38−CD90−CD45RA+) and a CD34+CD38+ fraction resembling granulocyte-macrophage progenitors (GMP-like LSCs; Lin−CD34+CD38+CD123+CD45RA+) (Figure 2A).22 Interestingly, in 80% of patients studied, both populations coexisted with evidence of a hierarchical relationship in which LMPP-like LSCs gave rise to the GMP-like population, but not the converse (Figure 3). It will be important to see if such a hierarchical relationship between these 2 LSC populations is consistently recapitulated in more humanized mouse models. Finally, in 10% to 15% of cases, a dominant population resembling multipotent progenitors was observed (MPP-like, Lin−CD34+CD38−CD90−CD45RA−). In this and other studies, the CD34+CD38− AML populations exhibited a higher LSC frequency compared with the CD34+CD38+ fraction, consistent with higher self-renewal potential of the more immature cells.29,30 Global gene expression profiling showed that these LSC populations were molecularly distinct and, in keeping with their immunophenotype, resembled normal progenitors more than normal stem cells.22 This suggests, but does not prove, that LSCs arise from a progenitor that has acquired self-renewal properties rather than a direct HSC origin for AML.
Immunophenotype and biological characteristics of CD34+and CD34−AML LSCs. (A) AML LSCs in CD34+ AML are primarily detected within LMPP-like (CD34+CD38−CD90−CD45RA+) and GMP-like (CD34+CD38+CD123+CD45RA+) subpopulations when engrafted into NSG mice. Less frequently they are detected in a dominant MPP-like (CD34+CD38−CD90−CD45RA−) subpopulation. Nevertheless, the stability of these markers after chemotherapy has not been rigorously tested, and there is likely to be considerable plasticity between these populations. LSCs are resistant to chemotherapy and give rise to non-LSC leukemic cells. OX-PHOS, oxidative phosphorylation. (B) AML LSCs in CD34− AML are primarily detected within a precursor GM-like (CD34−CD117+CD244+/−) subpopulation based on recent studies. Upon engraftment, these cells give rise to non-LSC leukemic cells. These findings have yet to be validated in additional studies but indicate a striking contrast with CD34+ AML and possibly a more mature myeloid cell of origin that has acquired self-renewal properties.
Immunophenotype and biological characteristics of CD34+and CD34−AML LSCs. (A) AML LSCs in CD34+ AML are primarily detected within LMPP-like (CD34+CD38−CD90−CD45RA+) and GMP-like (CD34+CD38+CD123+CD45RA+) subpopulations when engrafted into NSG mice. Less frequently they are detected in a dominant MPP-like (CD34+CD38−CD90−CD45RA−) subpopulation. Nevertheless, the stability of these markers after chemotherapy has not been rigorously tested, and there is likely to be considerable plasticity between these populations. LSCs are resistant to chemotherapy and give rise to non-LSC leukemic cells. OX-PHOS, oxidative phosphorylation. (B) AML LSCs in CD34− AML are primarily detected within a precursor GM-like (CD34−CD117+CD244+/−) subpopulation based on recent studies. Upon engraftment, these cells give rise to non-LSC leukemic cells. These findings have yet to be validated in additional studies but indicate a striking contrast with CD34+ AML and possibly a more mature myeloid cell of origin that has acquired self-renewal properties.
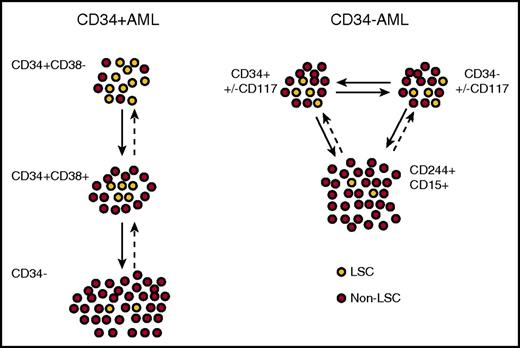
Hierarchical relationships between LSCs in CD34+and CD34−AML at diagnosis. Xenograft models suggest that CD34+ AML is arranged in a semihierarchical structure resembling normal hematopoiesis (left). In many patients, there is an increased LSC frequency in the CD34+CD38− population, which in turn gives rise to a CD34+CD38+ population with a reduced LSC frequency, but reversible plasticity is likely to be present in many cases. In infrequent cases, CD34− cells also contain rare LSCs. In CD34− AML, CD34+ and CD34− populations have similar LSC frequencies and gene expression profiles that most resemble precursor GM cells (right). The CD34− LSCs may express progenitor markers such as CD117 and myeloid antigens such as CD15 and CD244, and both in turn give rise to a non-LSC population with a mature myeloid immunophenotype.
Hierarchical relationships between LSCs in CD34+and CD34−AML at diagnosis. Xenograft models suggest that CD34+ AML is arranged in a semihierarchical structure resembling normal hematopoiesis (left). In many patients, there is an increased LSC frequency in the CD34+CD38− population, which in turn gives rise to a CD34+CD38+ population with a reduced LSC frequency, but reversible plasticity is likely to be present in many cases. In infrequent cases, CD34− cells also contain rare LSCs. In CD34− AML, CD34+ and CD34− populations have similar LSC frequencies and gene expression profiles that most resemble precursor GM cells (right). The CD34− LSCs may express progenitor markers such as CD117 and myeloid antigens such as CD15 and CD244, and both in turn give rise to a non-LSC population with a mature myeloid immunophenotype.
Detailed characterization of the surface immunophenotype of AML LSCs and mapping them back to their normal counterparts have been of great interest, in particular the identification of cell surface markers that are differentially upregulated on LSCs compared with normal HSCs. A number of such markers have been identified, but thus far, no one unique marker has been discovered that is universally expressed on CD34+CD38− LSCs across AML patients, but not on bulk leukemic blasts or on normal hematopoietic stem or progenitor cells (HSPCs). This challenge is partially due to the intrinsic heterogeneity of AML, both between patients and within an individual sample, but it also arises from the fact that LSCs immunophenotypically resemble certain normal hematopoietic progenitor populations. In addition, many lymphoid and myeloid antigens are aberrantly expressed in AML (such as CD7 or CD11b), giving rise to complex leukemia-associated phenotypes that may can change at the time of relapse.
Despite these difficulties, a number of cell surface markers have been identified that are upregulated on CD34+CD38− LSCs compared with normal CD34+CD38− HSPCs, including CD123,31 CD44,32 CD47,33,34 TIM3,35,36 CD96,37 CD99,13,38,39 CLL-1,40,41 CD32,42 CD25,42 IL1RAP,43,44 GPR56,45 and CD93.46 However, most of these markers have not been studied at relapse, so their stability in this setting is not known. In the case of CD123, the frequency of CD123+CD34+CD38− cells consistently increased at relapse.47,48 CD123 also cosegregates with FLT3-internal tandem duplication (ITD) mutation–positive cells within the CD34+CD38− population,49 suggesting it may be a robust LSC marker in FLT3-ITD–mutated AML. Notably, in only a handful of cases have any of these markers been shown to segregate LSC activity between marker-positive and marker-negative leukemic cells.
Antibody and cell therapies directed against LSC antigens
Apart from the refinement and purification of LSCs, these antigens serve as potential targets for immunotherapeutic approaches for the treatment of AML. Indeed, monoclonal antibodies that bind and/or block the function of these antigens have demonstrated antileukemic responses and, in some cases, anti-LSC activity in preclinical models of human AML.50 Monoclonal antibodies or biologics against 3 of these targets have recently entered clinical trials: CD123 (for AML with decitabine; NCT02472145), CD47 (for AML as monotherapy; NCT02678338, NCT02641002, NCT02663518), and IL1RAP (for CML; NCT02842320). Engineered T cells directed against CD123, including T cells with chimeric antigen receptors, have recently entered clinical trials (NCT02623582 and NCT02159495), but toxicity against CD123hi normal HSPCs may be difficult to circumvent.51 Unlike human B-cell populations, in a nonmyeloablative setting, the ultimate success of such approaches depends on stable expression of leukemia-specific antigens that are not present on normal regenerating myeloid progenitors.
CD34− AML
A minor proportion of AML cases (∼25%) lack expression of CD34, defined as present on <10% of blasts, and such cases are enriched for NPM1 mutations52,53 and possibly TET2 mutations.54 Although transplantable LSCs can be found, the majority reside in a CD34− compartment, with some detectable in a smaller CD34+ compartment, indicating a striking difference from the CD34+ AML cases.29,53,55 Rather than one population hierarchically giving rise to the other, both CD34+ and CD34− LSC populations essentially represent the same cells (with aberrant plasticity of CD34 expression) based on transcriptional profiling and a similar frequency of engraftment (Figure 3).54 Thus, CD34 is not a fixed marker of LSCs in CD34− AML, and alternative markers may be more informative in these cases. Indeed, a recent report demonstrated that engrafting LSCs in CD34− AML most resembled normal maturing granulocyte-macrophage precursors rather than CD34+CD38+CD123+CD45RA+ granulocyte-macrophage progenitors, based on immunophenotype, morphology, and gene expression (Figure 2B).54
AML subtypes
AML with translocations t(8;21) and inv16 involving the core binding factor (CBF-AML) account for ∼16% of adult AML cases and are associated with a favorable prognosis, with 10-year overall survival rates >70% using standard chemotherapy regimens.56 Understanding the cellular basis of this long-term remission by characterization of LSCs in this AML subtype has been difficult due to lack of a suitable xenotransplantation model. Engraftment of CBF-AML in immunodeficient mice is extremely low or absent for reasons that are not understood. In vitro studies have shown that leukemic blast colonies from t(8;21) AML can be grown from FACS-purified CD34+CD38−CD90− MPPs, but not from CD34+CD38−CD90+ HSCs,57 in keeping with a progenitor origin for CBF-AML. More recently, several cases of CBF-AML have been shown to robustly engraft in a humanized ossicle model27 and in humanized cytokine knockin mice (macrophage colony-stimulating factor [M-CSF], interleukin-3 [IL-3], GM-CSF, thrombopoietin [TPO]), with evidence that human M-CSF is critical for leukemic growth.58
Acute promyelocytic leukemia (APL) is commonly associated with the t(15;17) PML-RARA translocation resulting in expansion of promyelocytes whose differentiation block can be overcome with retinoic acid. APL blasts have been historically difficult to culture and transplant into immunodeficient mice (Figure 1), resulting in low to no detectable engraftment and an inability to identify and define LSCs in this disease.59,60 Studies from mouse models show that an APL-like disease can be driven by the expression of PML-RARA from a myeloid-lineage restricted promoter, suggesting that the disease initiates in committed progenitors rather than more immature multipotent cells.61,62 Recent xenotransplantation assays using humanized ossicles resulted in extensive engraftment of human APL, and pilot studies suggested that APL LSCs reside in a committed myeloid progenitor-like population.27
Identification of LSCs in mouse models of AML
In human AML, the identification of LSCs is dependent on xenotransplantation assays that introduce potential confounding interspecies mismatches in cytokines, microenvironment, and immune interactions. Thus, studies in mouse models have complemented investigation of the cancer stem cell model for AML. One of the best-studied mouse models involves retroviral transduction of the MLL-AF9 oncogene into mouse bone marrow cells followed by in vivo transplantation into syngeneic hosts. In this model, transduction of mouse HSPCs, purified GMPs, or purified HSCs resulted in LSCs that were enriched in a GMP-like population, but notably, leukemia-initiating activity was also detected in cells expressing more mature myeloid markers.63-65 This model has been extensively studied, yielding significant insights into AML LSC biology. For example, evidence that myeloid differentiation contributes to LSC activity in this model was recently demonstrated in CEBPA knockout mice, where the differentiation block imparted by CEBPA deficiency completely abrogated leukemia development but could be overcome by exogenous administration of myeloid growth factors.66 However, it should be noted that retroviral overexpression of a potent leukemia oncogene has the potential to result in nonphysiological effects. Indeed, in a knockin model in which MLL-AF9 was expressed from endogenous regulatory elements, HSCs, but not GMPs, transplanted disease.67
Apart from retroviral models, AML LSCs have been rigorously identified in few genetically engineered mouse models. Using a knockin mouse model of biallelic CEBPA mutations, lethal AML could be isolated and transplanted only from a myeloid progenitor fraction (Sca1−Mac1locKit+), but not from a more differentiated fraction (Sca1−Mac1hi), or HSC containing fraction (Sca1+).68 In a recent published combinatorial model of TET2 knockout and FLT3-ITD mutation, secondary transplantation of FACS-purified LSK CD48+CD150− MPP-like cells, which are not normally associated with self-renewal, initiated and maintained leukemia.69 In contrast, GMP-like blasts were unable to initiate disease, suggesting the functional LSCs in this model reside within an MPP-like population that can both self-renewal and differentiate into non-LSCs. Notably, expansion of human MPP-like cells in AML is also described in some CD34+ human AML samples.22
Biological properties of human AML stem cells
Cell cycle quiescence
Understanding the biological properties of human AML LSCs, particularly their similarities and differences from normal HSPCs and their heterogeneity between individual patients, is an area of active research, as these properties will be important for the development of AML LSC–targeting therapies. Early in vitro experiments showed that although many clonogenic AML CFCs and LTC-ICs were actively cycling and responded to human IL-3, stem cell factor, GM-CSF, FLT3L, and G-CSF, quiescent progenitors were also detectable in most samples.70 Accordingly, LSCs from the same patient samples capable of engrafting in NOD/SCID mice were virtually always quiescent based on 3H-thymidine labeling and in G0 prior to transplant. Consistent with these findings, a separate study reported that 5-fluorouracil–resistant LSCs robustly engrafted SCID mice,71 and lentiviral tracking of AML samples after serial transplantation identified a rarer population of extremely slowly dividing LSCs that did not emerge until the tertiary recipient.72 More recently, xenotransplantation of AML in NSG mice demonstrated the presence of CD34+CD38− LSCs in the endosteal region of mouse bone marrow,23 which were primarily quiescent with >50% of cells in G0 and resistant to cytarabine chemotherapy; however, these cells could be induced to enter the cell cycle and become sensitized to cytarabine by administration of exogenous G-CSF.42 Notably, G-CSF priming was explored in several large-scale clinical trials with mixed results, although FLAG-IDA, a regimen which includes G-CSF, has become frontline therapy in some centers.2 IL-3 is a critical cytokine involved in myeloid differentiation, and more than 70% of primary AML blasts cycled in vitro in response to IL-3 alone (both autocrine and paracrine), while blocking antibodies to IL-3 receptor α chain (CD123) inhibited the engraftment and growth of LSCs in NOD/SCID mice.31 While the bulk of AML progenitors assayed in vitro did not appear to cycle robustly in response to M-CSF,73 M-CSF may be required for the outgrowth of certain cytogenetic subtypes in vivo.74 Together, these studies suggest LSCs are more likely to be quiescent than bulk AML cells but can still respond to hematopoietic growth factors.
A recent paper identified miR-126 as a critical regulator of LSC quiescence from gene expression profiling of engrafting and nonengrafting AML subpopulations.75 Overexpression of miR-126 expanded LSCs, increased the number of cells in G0 and enhanced resistance to daunorubicin. Multiple components of the phosphatidylinositol 3-kinase/akt/mTOR pathway were repressed by miR-126, consistent with previous mouse studies demonstrating that this pathway critically regulates HSC and LSC self-renewal. For example, expression of a constitutively active form of Akt76 in HSC or loss of the negative regulator PTEN77 was associated with HSC exhaustion but increased self-renewal in LSCs, suggesting targetable differences in the regulation of quiescence between HSCs and LSCs.
Metabolism
Quiescent cells are associated with low energy requirements, and thus LSCs would be predicted to have lower levels of oxidative phosphorylation. Using fluorescent chemical probes that measured mitochondrial reactive oxygen species (ROS), functional LSCs were shown to be quiescent (G0 ∼72%) and have lower levels of ROS and oxygen consumption rates, indicative of a lower oxidative metabolism.78 Furthermore, the same ROS-low cells were impaired in their reserve glycolytic capacity, unlike CD34+ cells from normal bone marrow, making them more dependent on oxidative phosphorylation. Inhibition of the antiapoptotic protein Bcl-2 with ABT-263 or ABT-737 further reduced oxidative phosphorylation and selectively eradicated quiescent LSCs.78 In a separate study, LSCs from patient samples with mutations in either IDH1 or IDH2 were shown to have an increased dependence on Bcl-2 due to inhibition of cytochrome c oxidase by 2-hydroxyglutarate, leading to increased susceptibility of LSCs to ABT-199 in vivo.79 A recently published early phase 1/2 study with the oral Bcl-2 inhibitor ABT-199 in relapsed/refractory AML resulted in complete remission in 4 out of 12 patients with IDH1/2 mutations, but the effect on immunophenotypic or functionally validated LSCs was not determined.80 Metabolic heterogeneity of LSCs was reported in a recent study implicating fatty acid oxidation in promoting LSC quiescence and chemotherapy resistance.81 In a mouse model, LSCs resident in adipose tissue upregulated the fatty acid transporter CD36 and induced local lipolysis associated with higher fatty acid oxidation rate and cellular quiescence. High rates of fatty acid oxidation are noted in normal HSCs and reduce oxidative stress by increasing NADH levels,82 but detailed studies of fatty acid metabolism in human LSCs and somatic mutations have not been reported.
Epigenetic heterogeneity in AML and LSCs
In vivo experiments with primary cells show that the progeny of LSCs are clonally related, downstream leukemic blasts that lack the ability to engraft even though they share a common set of genetic mutations. Therefore, a major implication of the cancer stem cell model is that functional properties that differ between LSCs and non-LSCs are likely driven by epigenetic differences. Thus far, only 1 recent study integrated epigenetic information with AML LSCs by conducting genome-scale DNA methylation analysis for 15 patient samples in FACS-purified CD34+CD38−, CD34+CD38+, and CD34− subpopulations for which the engraftment characteristics were determined.83 Interestingly, a large number of differentially methylated regions were identified between LSC-containing (engrafting) and non–LSC-containing (nonengrafting) fractions, providing the first evidence of epigenetic differences between cancer stem cells and their non–stem cell progeny. These differentially methylated regions were predominantly hypomethylated in LSCs and largely associated with transcriptional upregulation, suggesting that hypomethylating agents, such as azacitidine or decitabine, may not have activity against LSC populations.84 An LSC epigenetic signature consisting of 71 genes that were differentially expressed in LSCs compared with non-LSC subpopulations and inversely correlated with DNA methylation was enriched for a number of HOX genes, underscoring the importance of this gene cluster in LSC regulation and its link to DNA methylation.83,85 Furthermore, by comparing the DNA methylation profiles of LSC subpopulations with normal HSPCs, 2 distinct clusters were revealed: a GMP-like cluster (enriched for FLT3 and NPM1 mutations) and an LMPP-like cluster (enriched for mutations in IDH1 and IDH2). These data may reflect a progenitor cell of origin in many cases of AML, consistent with the flow cytometric and gene expression studies discussed above.22
Histone modifications are also critical determinants of epigenetic states in both leukemia and normal hematopoiesis and may be directly linked to cellular metabolism.86 The mono- and dimethyl lysine demethylase LSD1 (KDM1A) is highly expressed in AML and is associated with transcriptional repression through direct and indirect mechanisms, and differentiation block.87 Pharmacological inhibitors of LSD1 resulted in markedly decreased engraftment in combination with all-trans-retinoic acid,88 suggesting that certain histone marks may be important for maintaining LSC function, as has been suggested in recent studies in mice.89
Pre-LSCs in Human AML
Studies investigating surface antigen expression on AML LSCs compared with normal HSCs90 identified a number of candidates upregulated on CD34+CD38− LSCs compared with normal HSCs, including TIM3, a cell surface mucin-domain–containing molecule.35 TIM3 was found to be an informative marker to prospectively separate residual HSCs from leukemic cells (including LSCs) in AML samples from the time of diagnosis and along with CD99 permitted isolation of residual HSCs from all samples studied.12,13 Genotyping of these residual HSCs found that many patients harbored a population of mutated preleukemic HSCs bearing some, but not all, of the mutations present in the corresponding AML. In parallel, sequencing studies in purified HSCs, HPCs, and mature hematopoietic cells also found evidence of preleukemic HSCs in AML patients both at diagnosis and persisting into remission.14 Upon transplantation into NSG mice, these residual HSCs gave rise to normal lymphoid and myeloid progeny harboring these preleukemic mutations. Remarkably, the proportion of preleukemic HSCs in the residual HSC compartment at the time of diagnosis was associated with poor clinical outcomes, highlighting the clinical significance of these cells. Single-cell analysis demonstrated that clonal evolution in AML proceeds through the stepwise acquisition of mutations in self-renewing preleukemic HSCs and suggested that relapse could possibly occur not only from leukemic clonal or subclonal outgrowth but also from further evolution of preleukemic mutated clones.12,91 Analysis of clonal evolution patterns in AML determined that mutations identified in preleukemic HSCs were enriched in epigenetic modifiers such as TET2, DNMT3A, and IDH1/2 rather than proliferative mutations, underscoring a role for epigenetic dysregulation in establishing a preleukemic state.12 Recently, the existence of clonal hematopoiesis harboring mutations in many of these epigenetic modifiers was discovered in large cohort studies performed in individuals with no history of hematologic malignancies.92,93 This clinical entity, termed clonal hematopoiesis of indeterminate potential, was associated with increased risk of progression to hematologic malignancy, raising a number of critical questions for screening and prevention in AML.94
Genetic heterogeneity in AML LSCs
Genomic analysis of AML patient samples has demonstrated that most cases of AML are associated with multiple subclones based on variant allele frequencies.10,11 The relationship between genetically defined subclones and AML LSCs is only beginning to be explored but has significant implications for understanding the role of heterogeneity in AML (reviewed in Kreso and Dick20 ). The first study to examine this question sequenced patient AML samples before and after engraftment in immunodeficient mice and found that in most cases, subclones variably engrafted and typically only a single subclone established engraftment in an individual mouse.95 A second study examined this deeply through the analysis of engraftment and clonal structure in CD34− AML samples, and found similar results where subclones variably engrafted both in primary and secondary transplants, with most individual mice harboring a single subclone.54 A third study of a large number of cases found both concordant and discordant variant allele frequencies between patient clinical samples and patient-derived xenografts.26 These studies raise the possibility that heterogeneity in engraftment potential may result from different properties of genetically defined subclones and not just from epigenetic differences between LSCs and their non-LSC progeny.
Clinical associations of human AML stem cells
LSCs are predictive of patient survival
Because self-renewal provides LSCs with the capability for long-term disease maintenance, the cancer stem cell model predicts that patient outcome is linked more closely to the properties of LSCs than non-LSCs and that cure must depend on LSC eradication. Initial studies investigated established features of AML LSCs such as flow cytometry enumeration or xenotransplantation for their association with clinical outcomes. A high proportion of CD34+CD38− cells at the time of AML diagnosis were associated with LSC engraftment in NOD/SCID mice, high minimal residual disease after chemotherapy, and poor patient survival.96 Similarly, ALDH bright cells in primary samples correlated with leukemic blast persistence after induction chemotherapy and adverse outcome.97 In addition, patients whose leukemic cells were able to engraft in NOD/SCID mice had a significantly shorter overall survival than those with leukemia cells that did not engraft, indicating that standard LIC assays have prognostic meaning.98
Further evidence for the link between LSCs and clinical outcomes was obtained from 2 studies that investigated the association of LSC gene expression signatures with clinical outcomes in large cohorts of AML patients. In the first report, gene expression profiling from CD34+CD38− LSCs compared with CD34+CD38+ progenitor-like and CD34− blasts was used to define an LSC score that was then found to be independently predictive of overall and relapse-free survival in 4 patient cohorts.99 The second report developed an LSC gene expression signature composed of functionally validated LSC populations from 16 patients.30 Interestingly, certain genes enriched in LSCs were shared with normal HSCs constituting a stemness program that was an independent predictor of patient overall survival. Recently, the same group developed a microRNA signature from functionally validated LSC populations and further demonstrated its independent association with overall survival.75 Finally, an integrated gene expression and DNA methylation analysis of functionally validated LSC populations led to the development of an LSC epigenetic signature that was independently associated with overall survival in multiple cohorts of patients.83 Together, these studies demonstrate that diverse molecular features of AML LSCs are independently associated with clinical outcome, supporting the significance of the cancer stem cell model for human AML.
Treatment responses and LSCs
The cancer stem cell model proposes that refractoriness to chemotherapy and disease relapse are driven by leukemia stem cells, but few prospective studies have measured the frequency, immunophenotype, and biological changes in LSCs at relapse. A clinical study of CD34+ subpopulations in patients receiving epigenetic therapies azacitidine and sodium valproate showed persistence of LMPP-like and GMP-like LSCs measured by flow cytometry of serial bone marrow samples, even in patients that achieved a complete remission.84 In a recent study, LSC frequencies were determined from 7 patients at the time of both diagnosis and relapse after fractionating into 4 subpopulations based on CD34 and CD38 expression.48 A 9- to 90-fold increase in LSC frequency was observed from diagnosis to relapse, together with an extended immunophenotype, such that LSCs after treatment were detectable in immunophenotypic compartments not containing LSCs at diagnosis. This study demonstrates that as proposed by the cancer stem cell model, the LSC compartment expands at relapse, likely contributing to the poor clinical responses and outcomes seen in such patients.
Open questions and future directions
The field of AML stem cell biology has come a long way since the original observations made decades ago. However, there are a number of critical open questions that remain to be addressed. As xenotransplantation models improve, how will this impact our conception of LSCs? Can LSCs be identified and assayed at the single-cell level? How heterogeneous are LSCs at the single-cell level? Will there be different classes of LSCs like HSCs? What is the molecular nature of critical interactions between LSCs and the microenvironment? What are the epigenetic drivers that functionally differentiate between LSCs and their non-LSC progeny? How and why do preleukemic HSCs progress to LSCs, and is there a possibility to screen for or even prevent such progression? Finally, what evidence will be required to show that LSC-targeted therapies are required for cure? Ultimately, the true validation of the cancer stem cell model will require clinical evidence that specific targeting of LSCs (but not non-LSC blasts) leads to improved outcomes and long-term disease-free survival.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the members of the Majeti laboratory for helpful discussion.
D.T. is supported by a CJ Martin Overseas Biomedical Research Fellowship (National Health and Medical Research Council, Australia). R.M. is a New York Stem Cell Foundation Robertson Investigator and Leukemia and Lymphoma Society Scholar. This work was supported by the New York Stem Cell Foundation and National Institutes of Health National Cancer Institute grant R01CA188055 (R.M.).
Authorship
Contribution: D.T. and R.M. wrote the paper.
Conflict-of-interest disclosure: R.M. is an inventor of multiple patents related to CD47 and antibody targeting of human AML stem cells and is founder and consultant to Forty Seven Inc. D.T. declares no competing financial interests.
Correspondence: Ravindra Majeti, Stanford Institute for Stem Cell Biology and Regenerative Medicine, Lokey Stem Cell Building, 265 Campus Dr, Stanford, CA 94305; e-mail: rmajeti@stanford.edu.