Key Points
High NPM1-mutant allele burden at diagnosis is associated with poor clinical outcome in de novo AML.
The adverse effect of high NPM1-mutant allele burden is independent of comutations and clinical variables.
Abstract
Acute myeloid leukemia (AML) with mutated NPM1 is a newly recognized separate entity in the revised 2016 World Health Organization classification and is associated with a favorable prognosis. Although previous studies have evaluated NPM1 in a binary fashion, little is known about the significance of its mutant allele burden at diagnosis, nor has the effect of comutations (other than FLT3) been extensively evaluated. We retrospectively used targeted sequencing data from 109 patients with de novo AML with mutated NPM1 to evaluate the potential significance of NPM1 variant allele frequency (VAF), comutations, and clinical parameters with regard to patient outcomes. We observed that high NPM1 VAF (uppermost quartile) correlated with shortened overall survival (median, 12.1 months vs not reached; P < .0001) as well as event-free survival (median, 7.5 vs 65.44 months; P < .0001) compared with the other NPM1-mutated cases. In both univariate and multivariable analyses, high NPM1 VAF had a particularly adverse prognostic effect in the subset of patients treated with stem-cell transplantation in first remission (P = .0004) and in patients with mutated DNMT3A (P < .0001). Our findings indicate that the prognostic effect of NPM1 mutation in de novo AML may be influenced by the relative abundance of the mutated allele.
Introduction
Acute myeloid leukemia (AML) with mutated nucleophosmin 1 (NPM1) gene is recognized as a separate entity in the revised 2016 World Health Organization (WHO) classification of myeloid neoplasms, after previous inclusion as a provisional entity in the 2008 WHO classification.1 NPM1 is one of the most commonly observed mutations in AML; it is detected in ∼30% of all cases and in 50% to 60% of those with normal cytogenetics.1-6 Although AML with mutated NPM1 is considered a distinct entity,1 NPM1 mutation alone seems to be insufficient for leukemogenesis, and it typically occurs in association with founder mutations such as DNMT3A.7-9 Similarly, internal tandem duplication (ITD) mutations in FLT3 are twice as frequent in NPM1-mutant AML as compared with AML with wild-type NPM1.10-14 NPM1 mutation is generally associated with a favorable clinical outcome; however, FLT3-ITD comutation has been shown to diminish the favorable NPM1 effect,15 particularly in the presence of mutant DNMT3A.7 Conversely, RAS pathway comutations may positively influence outcome of AML with mutated NPM1.16
Among patients with AML with FLT3-ITD, a high mutational burden using a polymerase chain reaction–based assay has been associated with an inferior survival in NPM1-mutated AML.17 However, assessment of the FLT3-ITD–mutated allele burden is not currently performed in many centers. Conversely, the influence of NPM1 mutational load on outcome has yet to be investigated, particularly within the setting of FLT3-ITD and/or DNMT3A comutation. Previous mutational landscape–focused studies have evaluated NPM1 in a binary fashion, without emphasis on quantitative mutational burden at the time of diagnosis.
Patients with NPM1-mutated AML without FLT3-ITD, or with a low FLT3-ITD allelic burden, comprise a prognostically favorable subgroup and are often treated with induction chemotherapy and an intensive chemotherapy-based consolidation regimen rather than allogeneic stem-cell transplantation (SCT). The decision to perform SCT in first complete remission (CR1) is complex, and even NPM1-mutated/FLT3-ITD–low or FLT3 wild-type patients may be referred for SCT based on older age or presence of unfavorable comutations, adverse cytogenetics, or measurable residual disease (MRD).18-20 Nonetheless, outcomes in patients who have detectable NPM1 mutations after chemotherapy are poor, even when treated with subsequent SCT.19-21 It remains unclear whether MRD negativity after induction chemotherapy can obviate the need for SCT in a patient whose disease exhibits unfavorable biology at diagnosis or whether MRD positivity is truly an indication for SCT in a patient with good-risk disease. Moreover, sensitive NPM1 MRD assessment is a specialized test that is not available in most centers at the current time. Thus, additional independent predictive markers at diagnosis may still be useful in AML with mutated NPM1.
We examined patient outcomes in a series of patients with NPM1-mutated de novo AML treated with induction chemotherapy, with or without subsequent SCT, focusing on the influence of commonly occurring comutations and NPM1 variant allele frequency (VAF) at the time of diagnosis.
Methods
Case selection
After institutional review board approval, we identified 109 cases of newly diagnosed de novo AML with mutated NPM1 from the pathology archives of Brigham and Women’s Hospital/Dana-Farber Cancer Institute and Massachusetts General Hospital (2008-2017) that fulfilled 2016 WHO classification criteria for AML with mutated NPM1; thus, we excluded patients who had received prior cytotoxic therapy, carried a prior diagnosis of any myeloid neoplasm, or had WHO-defined recurrent cytogenetic abnormalities. All patients were treated with standard anthracycline-cytarabine induction chemotherapy, with or without subsequent allogeneic SCT.
NGS studies
For all patients meeting inclusion criteria, we reviewed data from targeted sequencing studies performed on bone marrow aspirates (n = 93) or peripheral blood specimens (n = 16) at the time of diagnosis, as previously described.22-24 Target regions of 87 genes (hybrid capture system; Agilent Technologies, Santa Clara, CA) were evaluated in 46 patients, 95 genes (rapid heme panel [RHP]; Illumina TruSeq custom amplicon kit; San Diego, CA) in 56 patients, and 54 genes (Massachusetts General Hospital; SNaPshot; Illumina TruSeq) in 7 patients; genes were selected based on pathogenic involvement in myeloid malignancies. Genes assessed in all platforms were NPM1, FLT3, DNMT3A, IDH1, IDH2, TET2, ASXL1, EZH2, BCOR, SETBP1, BCORL, SH2B3, SETD2, CREBBP, WT1, PHF6, CEBPA, RUNX1, ETV6, STAG2, PDS5B, RAD21, KRAS, NRAS, KIT, CBL, RIT1, PTPN11, NF1, JAK2, U2AF1, ZRSR2, PRPF40b, SRSF2, SF3B1, CSF3R, BRAF, GATA2, and TP53. Variant calls were made using minimum criteria of either 10 variant reads with VAF >2% or 5 to 9 variant reads with VAF >33% and a minimum total read depth of 50. All cases were evaluated for average read count across all amplicons and also for the read count and VAF of NPM1. We classified variants as pathogenic mutations based on mutation type, position, and frequency in publicly available single-nucleotide polymorphism databases. Mutations were additionally segregated by pathway as previously described25 : DNA methylation, epigenetic regulation, transcription factor, cohesin complex, RAS, and spliceosome. FLT3-ITD was detected by either a sizing assay based on polymerase chain reaction amplification followed by fragment analysis capillary electrophoresis or a next-generation sequencing (NGS)–based methodology (RHP).24 FLT3-ITD mutation load was segregated into high (>50% VAF) or low (<50%) level for the subset cases in which these data were available.
Statistical analyses
We performed multivariable linear regression for the following variables with respect to NPM1 VAF: total number of mutations, presence or absence of a subclone, DNMT3A, NRAS/KRAS, PTPN11, TET2, IDH1/IDH2, FLT3 mutations other than ITD, FLT3-ITD and DNMT3A and FLT3-ITD comutations (genes mutated and gene combinations comutated in >10% of the cohort), highest VAF value of all mutations for each case, age, sex, white blood cell (WBC) count, hemoglobin, platelet count, bone marrow cellularity, bone marrow blast percentage, peripheral blood blast percentage, and karyotype (normal vs abnormal).
We evaluated patients’ overall survival (OS) and event-free survival (EFS), defined as previously reported.23,26 Briefly, OS was defined as the time in months from the date of diagnosis to last follow-up or death, and EFS was defined as the time in months from the date of diagnosis to relapse, death, or last follow-up (the latter was used as the end point in those who did not relapse). We examined the following variables: age, sex, WBC count, hemoglobin, platelet count, bone marrow cellularity, bone marrow blast percentage, peripheral blood blast percentage (all as continuous variables), karyotype (normal vs abnormal), SCT in CR1 status, NPM1 VAF assessed in quartiles and as a continuous variable, FLT3-ITD status, and mutation status of any genes mutated or gene combinations comutated in >10% of the cohort. Univariate analysis (log-rank test) followed by multivariable analysis (Cox proportional hazards model) was performed for OS and EFS for the entire cohort as well as for EFS of the entire cohort censoring patients at the time of SCT. The same variables were also analyzed for effect on EFS from the time of diagnosis in the subset of patients undergoing SCT in CR1. Statistical analyses were performed using XLSTAT (version 2017.5) and Prism 7.0c (GraphPad) software packages.
Results
Patient characteristics
We identified 109 patients with AML with NPM1 mutations (male/female ratio, 0.84) who met inclusion criteria, with a median age of 60 years (range, 15-83 years) and median follow-up time of 18.1 months for all patients. All patients were treated with standard induction chemotherapy, and 59 (54%) underwent SCT, including 45 (41%) in CR1, before any AML relapse (Table 1). In the patients undergoing SCT in CR1, the SCT occurred a median of 3.7 months after AML diagnosis (range, 1.4-12.4 months). Clinical reasons for SCT varied and are shown in Table 2. Thirty-nine patients (36%) relapsed, and 73 (67%) were alive at last follow-up. Of note, 57% of the FLT3-ITD+ patients received FLT3-inhibitor therapy before any relapse (sorafenib, n = 14; midostaurin, n = 7; quizartinib, n = 1; gilteritinib, n = 1; crenolanib, n = 1).7
Characteristics of patients with de novo AML with mutated NPM1 (N = 109)
| . | All patients (N = 109) . | NPM1 VAF ≤ 0.43 (n = 85) . | NPM1 VAF ≥ 0.44 (n = 24) . |
|---|---|---|---|
| Patient characteristics | |||
| Median age (range), y | 60 (15-83) | 60 (19-75) | 63 (15-83) |
| Male/female ratio | 0.84 | 0.79 | 1 |
| Clinical parameters | |||
| Median WBC (range), ×109/L | 24.0 (0.8-340) | 22.8 (0.8-340) | 44.3 (1.5-309) |
| Median PB blasts (range), % | 26 (0-98) | 26 (0-97) | 48 (1-98) |
| Median BM blasts (range), % | 71 (21-96) | 73 (21-95) | 70 (29-96) |
| Abnormal cytogenetics, N (%) | 14 (13) | 10 (12) | 4 (17) |
| Median NPM1 VAF (range) | 0.39 (0.04-0.54) | 0.38 (0.04-0.43) | 0.46 (0.44-0.54) |
| FLT3-ITD positive, N (%) | 42 (39) | 31 (36) | 11 (46) |
| Comutations by pathway, N (%) | |||
| DNA methylation | |||
| DNMT3A | 55 (50) | 45 (53) | 10 (42) |
| IDH1 | 24 (22) | 17 (20) | 7 (29) |
| IDH2 | 12 (11) | 11 (13) | 1 (4) |
| TET2 | 29 (27) | 26 (31) | 3 (13) |
| Epigenetic regulation | |||
| ASXL1 | 2 (2) | 2 (2) | 0 (0) |
| EZH2 | 0 (0) | 0 (0) | 0 (0) |
| BCOR | 2 (2) | 2 (2) | 0 (0) |
| SETBP1 | 0 (0) | 0 (0) | 0 (0) |
| BCORL | 1 (1) | 1 (1) | 0 (0) |
| SH2B3 | 2 (2) | 1 (1) | 1 (4) |
| SETD2 | 0 (0) | 0 (0) | 0 (0) |
| CREBBP | 1 (1) | 1 (1) | 0 (0) |
| Transcriptional regulation | |||
| WT1 | 7 (7) | 5 (6) | 2 (9) |
| PHF6 | 1 (1) | 1 (1) | 0 (0) |
| CEBPA | 5 (5) | 2 (2) | 3 (13) |
| RUNX1 | 1 (1) | 1 (1) | 0 (0) |
| ETV6 | 0 (0) | 0 (0) | 0 (0) |
| Cohesin complex | |||
| STAG2 | 2 (2) | 1 (1) | 1 (4) |
| PDS5B | 2 (2) | 1 (1) | 1 (4) |
| RAD21 | 4 (4) | 3 (4) | 1 (4) |
| RAS pathway | |||
| KRAS | 4 (4) | 4 (5) | 0 (0) |
| NRAS | 26 (24) | 23 (27) | 3 (13) |
| FLT3 (non-ITD) | 26 (24) | 19 (23) | 7 (29) |
| KIT | 1 (1) | 0 (0) | 1 (4) |
| CBL | 3 (3) | 2 (2) | 1 (4) |
| RIT1 | 4 (4) | 3 (4) | 1 (4) |
| PTPN11 | 26 (24) | 23 (27) | 3 (13) |
| NF1 | 3 (3) | 3 (4) | 0 (0) |
| JAK2 | 0 (0) | 0 (0) | 0 (0) |
| Spliceosome | |||
| U2AF1 | 0 (0) | 0 (0) | 0 (0) |
| ZRSR2 | 0 (0) | 0 (0) | 0 (0) |
| PRPF40b | 0 (0) | 0 (0) | 0 (0) |
| SRSF2 | 5 (5) | 4 (5) | 1 (4) |
| SF3B1 | 0 (0) | 0 (0) | 0 (0) |
| Other | |||
| CSF3R | 2 (2) | 1 (1) | 1 (4) |
| BRAF | 0 (0) | 0 (0) | 0 (0) |
| GATA2 | 1 (1) | 0 (0) | 1 (4) |
| TP53 | 0 (0) | 0 (0) | 0 (0) |
| Outcome, N (%) | |||
| Relapsed | 39 (36) | 24 (29) | 15 (63) |
| Alive at last follow-up | 73 (67) | 62 (74) | 10 (42) |
| Underwent SCT, N (%) | 59 (54) | 49 (58) | 10 (42) |
| SCT in CR1 | 45 (41) | 38 (45) | 7 (29) |
| Conditioning (all SCTs) | |||
| Reduced intensity | 36 (61) | 32 (65) | 4 (40) |
| Myeloablative | 23 (39) | 17 (35) | 6 (60) |
| SCT type (all SCTs), N (%) | |||
| Matched related donor | 18 (31) | 14 (29) | 4 (40) |
| Matched unrelated donor | 31 (53) | 27 (55) | 4 (40) |
| Mismatched unrelated donor | 3 (5) | 3 (6) | 0 (0) |
| Haploidentical | 5 (8) | 3 (6) | 2 (20) |
| Cord blood | 2 (3) | 2 (4) | 0 (0) |
| . | All patients (N = 109) . | NPM1 VAF ≤ 0.43 (n = 85) . | NPM1 VAF ≥ 0.44 (n = 24) . |
|---|---|---|---|
| Patient characteristics | |||
| Median age (range), y | 60 (15-83) | 60 (19-75) | 63 (15-83) |
| Male/female ratio | 0.84 | 0.79 | 1 |
| Clinical parameters | |||
| Median WBC (range), ×109/L | 24.0 (0.8-340) | 22.8 (0.8-340) | 44.3 (1.5-309) |
| Median PB blasts (range), % | 26 (0-98) | 26 (0-97) | 48 (1-98) |
| Median BM blasts (range), % | 71 (21-96) | 73 (21-95) | 70 (29-96) |
| Abnormal cytogenetics, N (%) | 14 (13) | 10 (12) | 4 (17) |
| Median NPM1 VAF (range) | 0.39 (0.04-0.54) | 0.38 (0.04-0.43) | 0.46 (0.44-0.54) |
| FLT3-ITD positive, N (%) | 42 (39) | 31 (36) | 11 (46) |
| Comutations by pathway, N (%) | |||
| DNA methylation | |||
| DNMT3A | 55 (50) | 45 (53) | 10 (42) |
| IDH1 | 24 (22) | 17 (20) | 7 (29) |
| IDH2 | 12 (11) | 11 (13) | 1 (4) |
| TET2 | 29 (27) | 26 (31) | 3 (13) |
| Epigenetic regulation | |||
| ASXL1 | 2 (2) | 2 (2) | 0 (0) |
| EZH2 | 0 (0) | 0 (0) | 0 (0) |
| BCOR | 2 (2) | 2 (2) | 0 (0) |
| SETBP1 | 0 (0) | 0 (0) | 0 (0) |
| BCORL | 1 (1) | 1 (1) | 0 (0) |
| SH2B3 | 2 (2) | 1 (1) | 1 (4) |
| SETD2 | 0 (0) | 0 (0) | 0 (0) |
| CREBBP | 1 (1) | 1 (1) | 0 (0) |
| Transcriptional regulation | |||
| WT1 | 7 (7) | 5 (6) | 2 (9) |
| PHF6 | 1 (1) | 1 (1) | 0 (0) |
| CEBPA | 5 (5) | 2 (2) | 3 (13) |
| RUNX1 | 1 (1) | 1 (1) | 0 (0) |
| ETV6 | 0 (0) | 0 (0) | 0 (0) |
| Cohesin complex | |||
| STAG2 | 2 (2) | 1 (1) | 1 (4) |
| PDS5B | 2 (2) | 1 (1) | 1 (4) |
| RAD21 | 4 (4) | 3 (4) | 1 (4) |
| RAS pathway | |||
| KRAS | 4 (4) | 4 (5) | 0 (0) |
| NRAS | 26 (24) | 23 (27) | 3 (13) |
| FLT3 (non-ITD) | 26 (24) | 19 (23) | 7 (29) |
| KIT | 1 (1) | 0 (0) | 1 (4) |
| CBL | 3 (3) | 2 (2) | 1 (4) |
| RIT1 | 4 (4) | 3 (4) | 1 (4) |
| PTPN11 | 26 (24) | 23 (27) | 3 (13) |
| NF1 | 3 (3) | 3 (4) | 0 (0) |
| JAK2 | 0 (0) | 0 (0) | 0 (0) |
| Spliceosome | |||
| U2AF1 | 0 (0) | 0 (0) | 0 (0) |
| ZRSR2 | 0 (0) | 0 (0) | 0 (0) |
| PRPF40b | 0 (0) | 0 (0) | 0 (0) |
| SRSF2 | 5 (5) | 4 (5) | 1 (4) |
| SF3B1 | 0 (0) | 0 (0) | 0 (0) |
| Other | |||
| CSF3R | 2 (2) | 1 (1) | 1 (4) |
| BRAF | 0 (0) | 0 (0) | 0 (0) |
| GATA2 | 1 (1) | 0 (0) | 1 (4) |
| TP53 | 0 (0) | 0 (0) | 0 (0) |
| Outcome, N (%) | |||
| Relapsed | 39 (36) | 24 (29) | 15 (63) |
| Alive at last follow-up | 73 (67) | 62 (74) | 10 (42) |
| Underwent SCT, N (%) | 59 (54) | 49 (58) | 10 (42) |
| SCT in CR1 | 45 (41) | 38 (45) | 7 (29) |
| Conditioning (all SCTs) | |||
| Reduced intensity | 36 (61) | 32 (65) | 4 (40) |
| Myeloablative | 23 (39) | 17 (35) | 6 (60) |
| SCT type (all SCTs), N (%) | |||
| Matched related donor | 18 (31) | 14 (29) | 4 (40) |
| Matched unrelated donor | 31 (53) | 27 (55) | 4 (40) |
| Mismatched unrelated donor | 3 (5) | 3 (6) | 0 (0) |
| Haploidentical | 5 (8) | 3 (6) | 2 (20) |
| Cord blood | 2 (3) | 2 (4) | 0 (0) |
BM, bone marrow; PB, peripheral blood.
Clinical rationale for SCT in CR1 (n = 45)
| Clinical rationale . | N of patients (%) . |
|---|---|
| Age >60 y | 17 (38) |
| FLT3-ITD | 25 (56) |
| Abnormal cytogenetics | 3 (7) |
| Unfavorable molecular profile | 6 (13) |
| Extramedullary disease | 3 (7) |
| Clinical rationale . | N of patients (%) . |
|---|---|
| Age >60 y | 17 (38) |
| FLT3-ITD | 25 (56) |
| Abnormal cytogenetics | 3 (7) |
| Unfavorable molecular profile | 6 (13) |
| Extramedullary disease | 3 (7) |
Some cases had multiple clinical rationales.
Cytogenetic and molecular characteristics
Most patients (95 [87%] of 109) had a normal karyotype; the karyotype abnormalities are listed in Table 3. The NPM1 read count (total depth) was >100 in 98% of cases, with 2 cases having <100 reads (64 and 81 total reads, respectively). The median for average coverage across all tested loci was 1222 (range, 161-4606). The median VAF for NPM1 was 0.39 (range, 0.04-0.54; supplemental Table 1, available on the Blood Web site). We did not identify any effect of NPM1 read count on NPM1 VAF (P = .86). There was no significant difference in NPM1 VAF measured between the 3 testing platforms (median, 0.39 [n = 46], 0.395 [n = 56], and 0.42 [n = 7] for hybrid capture, RHP, and SNaPshot, respectively; Kruskall-Wallis P = .71) or between blood (median, 0.395 [n = 16]) and bone marrow (median, 0.39 [n = 93]) samples (Mann-Whitney P = .45). NPM1 VAF exhibited no significant correlation with patient age (P = .37) but correlated positively with WBC (r = 0.26; P = .005), peripheral blast percentage (r = 0.29; P = .002), and percentage of marrow blasts (r = 0.25; P = .008).
Cases with abnormal cytogenetics (n = 14)
| Case . | Karyotype . |
|---|---|
| 1 | 46,XX, t(1;13)(q42;q14)[19]/46,XX[1] |
| 2 | 47,XX,+mar[13]/48,idem,+21[7] |
| 3 | 47,XY,+8[13]/46,XY[7] |
| 4 | 46,XY,der(1)t(1;2)(p34;q1?3)[5]/45,idem,10[10]/46,XY[5] |
| 5 | 47,XY,+4[3]/46,XY[17] |
| 6 | 46,XY,del(3)(q21),add(9)(p22)[2]/46,XY[18] |
| 7 | 48,XX,+4,+8[17]/46,XX[3] |
| 8 | 47,XX,+4[19]/46,XX[1] |
| 9 | 46,XY,-6,+r[cp17]/46,XY[3] |
| 10 | 47,XY,+8[7]/48,idem,+5[2]/46,XY[cp11] |
| 11 | 46,XX,i(21)(q10)[6]/46,XX,idem,?inv(20)(p11.2q12)[6]/46,XX[8] |
| 12 | 46,XY,t(3;18)(q26;q21),del(6)(q13q26)[18]/46,XY[2] |
| 13 | 46,XX,del(9)(q12q22)[20] |
| 14 | 47,XX,+8 [20] |
| Case . | Karyotype . |
|---|---|
| 1 | 46,XX, t(1;13)(q42;q14)[19]/46,XX[1] |
| 2 | 47,XX,+mar[13]/48,idem,+21[7] |
| 3 | 47,XY,+8[13]/46,XY[7] |
| 4 | 46,XY,der(1)t(1;2)(p34;q1?3)[5]/45,idem,10[10]/46,XY[5] |
| 5 | 47,XY,+4[3]/46,XY[17] |
| 6 | 46,XY,del(3)(q21),add(9)(p22)[2]/46,XY[18] |
| 7 | 48,XX,+4,+8[17]/46,XX[3] |
| 8 | 47,XX,+4[19]/46,XX[1] |
| 9 | 46,XY,-6,+r[cp17]/46,XY[3] |
| 10 | 47,XY,+8[7]/48,idem,+5[2]/46,XY[cp11] |
| 11 | 46,XX,i(21)(q10)[6]/46,XX,idem,?inv(20)(p11.2q12)[6]/46,XX[8] |
| 12 | 46,XY,t(3;18)(q26;q21),del(6)(q13q26)[18]/46,XY[2] |
| 13 | 46,XX,del(9)(q12q22)[20] |
| 14 | 47,XX,+8 [20] |
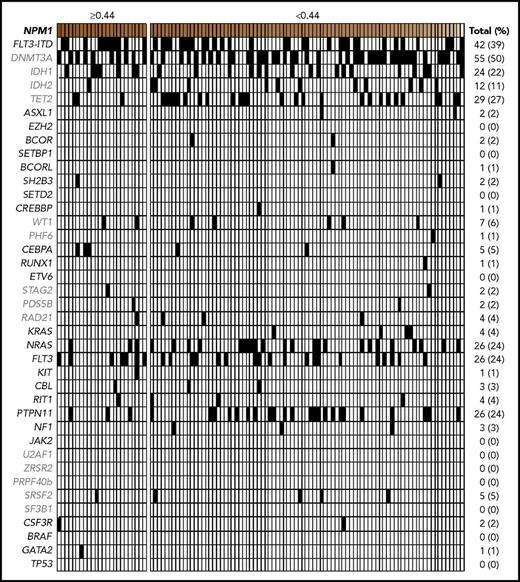
The total mutation count per case ranged from 1 to 8 (including the NPM1 mutations, which were present in all cases), with a median of 4. NPM1 was the sole mutation in a small subset of cases (2 [1.9%] of 109). The most common cooccurring mutations included DNMT3A (50%), FLT3-ITD (39%), TET2 (27%), FLT3 non-ITD (24%), NRAS (24%), and PTPN11 (24%), similar to previous reports.7 DNA methylation pathway mutations were overall the most frequent (78%), followed by RAS pathway mutations including FLT3-ITD (74%), other RAS pathway mutations (44%), and epigenetic regulation (8%), cohesin complex (7%), transcriptional regulation (7%), and spliceosome (5%) pathway mutations (Figure 1; Table 1).
Comutational profiles for all patients with de novo AML with mutated NPM1. All cases of de novo AML with mutated NPM1 evaluated (n = 109). Each column represents an individual patient. Intensity gradient corresponds to NPM1 VAF, and patients in highest VAF quartile (≥0.44) are shown. All comutations, including FLT3-ITD status, provided in binary format.
Comutational profiles for all patients with de novo AML with mutated NPM1. All cases of de novo AML with mutated NPM1 evaluated (n = 109). Each column represents an individual patient. Intensity gradient corresponds to NPM1 VAF, and patients in highest VAF quartile (≥0.44) are shown. All comutations, including FLT3-ITD status, provided in binary format.
Clinical and comutational variables
In univariate analyses for OS, older patient age was significantly associated with shorter OS (P = .03), and SCT in CR1 was associated with longer OS (P = .009; Figure 2A). A DNMT3A mutation was significantly associated with shorter OS (P = .04). In univariate analyses for EFS, SCT in CR1 was associated with longer EFS (P = .0002; Figure 2B). In univariate analysis for EFS censoring patients at the time of any SCT in CR1, older age was significantly associated with shorter EFS (P = .015). Stratification of the cohort at the median for total mutational number (>4 vs ≤4) revealed no significant difference in OS between these groups (P = .93). There was also no significant effect of mutation number as a continuous variable on OS (P = .27), although there was borderline association of higher mutation number as a continuous variable with shorter EFS (P = .07). FLT3-ITD (P = .043) and FLT3-ITD plus DNMT3A mutations (P = .029; Figure 2C) were associated with shorter EFS, whereas NRAS/KRAS mutations were associated with longer EFS (P = .044; Figure 2D) in the analysis that censored patients at the time of SCT. There were no significant associations of any variables with EFS in the 45 patients receiving SCT in CR1. No statistically significant associations with OS, EFS, or EFS censored at the time of SCT were identified with peripheral blood blast percentage or comutations in epigenetic regulation, transcription factor, spliceosome, or cohesin complex pathways or specifically with TET2, non-ITD FLT3, or PTPN11 mutations (genes mutated in >10% of the total cohort). Ten percent of patients harbored mutations associated with secondary AML,27 and there was no association of this subset with OS, EFS, EFS censored at the time of SCT, or EFS of patients undergoing SCT in CR1.
Kaplan-Meier curves for effects of SCT and selected comutations. (A) Effect of SCT in CR1 (n = 45) on OS (35.4 months vs not reached; P = .009). (B) Effect of SCT in CR1 on EFS (16.1 months vs not reached; P = .0002). (C) Effect of combined FLT3-ITD and DNMT3A mutations (n = 22) on EFS in comparison with cases with FLT3-ITD−/DNTM3A+, FLT3-ITD+/DNMT3A−, or FLT3-ITD−/DNMT3A− (6.6 vs 25.8 months; P = .029), censoring patients at the time of SCT. (D) Effect of either KRAS or NRAS mutation (n = 30) on EFS (16.3 vs 32.4 months; P = .044), censoring patients at the time of SCT.
Kaplan-Meier curves for effects of SCT and selected comutations. (A) Effect of SCT in CR1 (n = 45) on OS (35.4 months vs not reached; P = .009). (B) Effect of SCT in CR1 on EFS (16.1 months vs not reached; P = .0002). (C) Effect of combined FLT3-ITD and DNMT3A mutations (n = 22) on EFS in comparison with cases with FLT3-ITD−/DNTM3A+, FLT3-ITD+/DNMT3A−, or FLT3-ITD−/DNMT3A− (6.6 vs 25.8 months; P = .029), censoring patients at the time of SCT. (D) Effect of either KRAS or NRAS mutation (n = 30) on EFS (16.3 vs 32.4 months; P = .044), censoring patients at the time of SCT.
Features of high NPM1 VAF
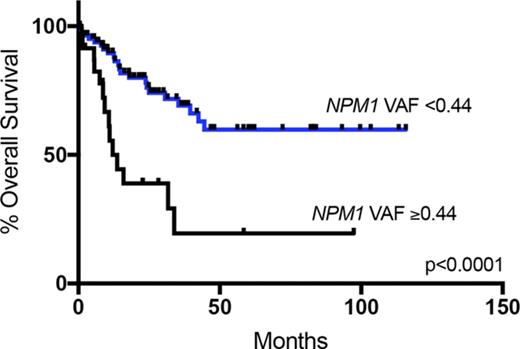
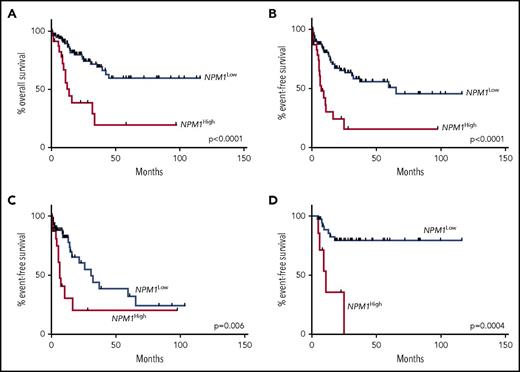
NPM1 VAF showed significant association with shorter OS and EFS, when analyzed as a continuous variable (P = .042 and P = .026, respectively), stratified at the median (≥0.40 or <0.40; P = .0015) and as quartile groupings (VAF, <0.36, 0.36-0.39, 0.40-0.43, and ≥0.44; P = .0004 for both OS [supplemental Figure 1] and EFS). Patients in the uppermost quartile (high NPM1 VAF, ≥0.44; n = 24 patients) had shortened OS (median, 12.1 months vs not reached; P < .0001) and EFS (median, 7.5 vs 65.44 months; P < .0001; Figure 3A-B). High NPM1 VAF correlated with shortened OS both in patients age >60 years (n = 51; P = .01) and in those age ≤60 years (n = 58; P = .006). High NPM1 VAF was also significantly associated with shorter EFS in patients censored at the time of any SCT (median, 6.7 vs 31.0 months; P = .006; Figure 3C). In the 45 patients treated with SCT in CR1, NPM1 VAF as a continuous variable was associated with shortened EFS (P = .02), and high NPM1 VAF (n = 7) was also associated with shortened EFS (median, 11.0 months vs not reached; P = .0004; Figure 3D). These results remained significant on exclusion of the 2 cases with NPM1 read counts of <100 (data not shown).
Kaplan-Meier curves showing effect of NPM1 VAF. (A) Effect of high NPM1 VAF on OS in the entire cohort (12.1 months vs not reached; P < .0001). (B) Effect of high NPM1 VAF on EFS in the entire cohort (7.5 vs 65.44 months; P < .0001). (C) Effect of high NPM1 VAF on EFS in patients censored at the time of any SCT in CR1 (6.7 vs 31.0 months; P = .006). (D) Effect of high NPM1 VAF (n = 7) on EFS in patients treated with SCT in CR1 (n = 45; 11.0 months vs not reached; P = .0004).
Kaplan-Meier curves showing effect of NPM1 VAF. (A) Effect of high NPM1 VAF on OS in the entire cohort (12.1 months vs not reached; P < .0001). (B) Effect of high NPM1 VAF on EFS in the entire cohort (7.5 vs 65.44 months; P < .0001). (C) Effect of high NPM1 VAF on EFS in patients censored at the time of any SCT in CR1 (6.7 vs 31.0 months; P = .006). (D) Effect of high NPM1 VAF (n = 7) on EFS in patients treated with SCT in CR1 (n = 45; 11.0 months vs not reached; P = .0004).
High NPM1 VAF status was not significantly associated with the presence of comutations in FLT3-ITD (P = .48), FLT3 non-ITD (P = .59), DNMT3A (P = .36), TET2 (P = .12), or PTPN11 (P = .18) and was borderline associated with a lack of KRAS/NRAS mutations (Fisher’s exact test P = .07). High NPM1 VAF correlated with shortened EFS in both FLT3-ITD+ (5.9 months vs not reached; P = .018) and FLT3-ITD− (10.6 vs 37.4 months; P = .002) patients and in both DNMT3A-mutated (5.7 vs 37.4 months; P < .0001) and DNMT3A wild-type (11.0 months vs not reached; P = .04) patients (Figure 4A-B). High NPM1 VAF borderline correlated with shortened OS (12.1 months vs not reached; P = .052) in the FLT3-ITD+ patients and significantly correlated with shortened OS in patients who harbored DNMT3A mutation (10.0 vs 44.5 months; P < .0001). In the 22 patients who harbored both FLT3-ITD and DNMT3A mutations, high NPM1 VAF (n = 5) trended toward a correlation with shortened OS (P = .071).
Kaplan-Meier curves showing effect of NPM1 VAF within subpopulations defined by DNMT3A comutation. (A) Effect of high NPM1 VAF (n = 10) on EFS in DNTM3A-mutated patients (n = 55; 5.7 vs 37.4 months; P < .0001). (B) Effect of high NPM1 VAF (n = 14) on EFS in DNMT3A wild-type patients (n = 54; 11.0 months vs not reached; P = .04).
Kaplan-Meier curves showing effect of NPM1 VAF within subpopulations defined by DNMT3A comutation. (A) Effect of high NPM1 VAF (n = 10) on EFS in DNTM3A-mutated patients (n = 55; 5.7 vs 37.4 months; P < .0001). (B) Effect of high NPM1 VAF (n = 14) on EFS in DNMT3A wild-type patients (n = 54; 11.0 months vs not reached; P = .04).
The NPM1 variant had the highest VAF of all mutated genes in 26 cases (24%). Of note, there was no association between the highest VAF among all mutated genes in each case and EFS in the entire cohort (P = .41). Among the 55 patients with DNMT3A mutation, higher DNMT3A VAF was borderline associated with shorter EFS (P = .093), and DNMT3A VAF was positively correlated with NPM1 VAF (r = 0.356; P = .008). Among the 22 FLT3-ITD cases in which the mutant level could be determined, 10 had high FLT3-ITD and 12 had low FLT3-ITD; there was no significant difference in NPM1 VAF between FLT3-ITD–high and FLT3-ITD–low cases (Mann-Whitney P = .94). Using previously published criteria for defining subclones,28 we identified NPM1 as subclonal in 29 cases (27%). Using the log-rank test, there were no significant differences in OS (P = .44) or EFS (P = .53) based on whether or not the NPM1 mutation was subclonal.
In multivariable linear regression analyses to evaluate for variables associated with NPM1 VAF, the final model showed that higher peripheral blood blasts (P < .005) was the only variable significantly associated with higher NPM1 VAF, whereas the presence of DNMT3A mutation (P = .12) and presence of FLT3 mutations other than ITD (P = .09) were borderline associated with higher NPM1 VAF.
In multivariable analyses with respect to outcome in the entire patient cohort, high NPM1 VAF and DNMT3A mutation were each independently associated with OS, whereas high NPM1 VAF, DNMT3A mutation, and SCT in CR1 were each independently associated with EFS (Table 4). When censoring at the time of any SCT, age and combined FLT3-ITD and DNMT3A comutation were independently associated with EFS (Table 4). DNMT3A VAF was not independently associated with OS or EFS in any of the models, either when considered as a continuous variable or when using cutoffs at the median or highest quartile. In the cohort of patients treated with SCT in CR1, high NPM1 VAF (P = .002) and NPM1 VAF as a continuous variable (P = .021) were each significantly associated with EFS in multivariable analyses; other tested variables were not significant in this patient cohort.
Multivariable analysis of factors influencing OS, EFS, and EFS censoring patients at the time of SCT
| Variable . | P . | Hazard ratio . | 95% CI . |
|---|---|---|---|
| OS of all patients | |||
| High (≥0.44) NPM1 VAF | <.0001 | 4.043 | 2.037-8.024 |
| DNMT3A mutation | .017 | 2.381 | 1.165-4.869 |
| EFS of all patients | |||
| High (≥0.44) NPM1 VAF | <.0001 | 3.777 | 1.946-7.333 |
| DNMT3A mutation | .023 | 2.088 | 1.107-3.939 |
| SCT in CR1 | .003 | 0.365 | 0.188-0.711 |
| EFS of all patients censored at time of SCT | |||
| Age (continuous, per year) | .01 | 1.044 | 1.010-1.078 |
| Combined FLT3-ITD and DNMT3A mutation | .02 | 2.71 | 1.171-6.273 |
| Variable . | P . | Hazard ratio . | 95% CI . |
|---|---|---|---|
| OS of all patients | |||
| High (≥0.44) NPM1 VAF | <.0001 | 4.043 | 2.037-8.024 |
| DNMT3A mutation | .017 | 2.381 | 1.165-4.869 |
| EFS of all patients | |||
| High (≥0.44) NPM1 VAF | <.0001 | 3.777 | 1.946-7.333 |
| DNMT3A mutation | .023 | 2.088 | 1.107-3.939 |
| SCT in CR1 | .003 | 0.365 | 0.188-0.711 |
| EFS of all patients censored at time of SCT | |||
| Age (continuous, per year) | .01 | 1.044 | 1.010-1.078 |
| Combined FLT3-ITD and DNMT3A mutation | .02 | 2.71 | 1.171-6.273 |
CI, confidence interval.
Discussion
In this study, we investigated the potential effects of clinical variables, comutations, and NPM1 mutational burden at diagnosis in de novo AML with mutated NPM1. The frequencies of various comutations in our cohort were similar to those reported by other groups.7 Our results confirm previous findings that older patient age as well as concomitant FLT3-ITD or DNMT3A mutations may offset the otherwise favorable effect of NPM1 mutation; of note, the frequent use of FLT3 inhibitor therapy in the FLT3-ITD–mutated cases may have dampened the negative effect of the latter variable in our cohort.29,30 We also found that patients in our cohort with either KRAS or NRAS mutations experienced longer relapse-free survival in a univariate analysis that censored for SCT.16 As previously reported, we found that the combination of FLT3-ITD and DNMT3A mutations conferred a negative effect on outcome when censoring for SCT,7 but these mutations did not influence the outcome of patients treated with SCT in CR1.
Interestingly, we identified a powerful negative effect on survival of high NPM1 mutational burden at diagnosis. Although high NPM1 VAF was correlated with higher WBC and blast percentages, other variables classically associated with higher-risk disease, its effect on prognosis was independent of these factors. We did not observe differential outcomes based on our VAF-based mathematical approximation of the position of NPM1 within the clonal hierarchy (ie, clone vs subclone); however, we are limited in our ability to validate this finding in the absence of single-cell sequencing data. Because NPM1 mutation has been shown to be present in nonblast maturing hematopoietic cells in NPM1-mutated AML,31 the VAF may in fact capture the true disease burden more effectively than blast percentage in blood or bone marrow. Importantly, the prognostic effect of NPM1 VAF was observed across our entire cohort, including in subset analyses of patients whose samples were run on either the hybrid capture (n = 46) or amplicon-based (RHP; n = 56) NGS platforms (data not shown). The effect was also seen in subset analyses of patients with and without FLT3-ITD comutation but was especially prominent in patients with DNMT3A comutation and in patients who underwent SCT in CR1. Although the high NPM1 VAF variable showed only borderline association with OS in subset analyses of FLT3-ITD–mutated and FLT3-ITD/DNMT3A comutated cases (possibly because of diminishing sample sizes), these findings nevertheless raise the possibility that DNMT3A comutation may potentiate ability of high-burden NPM1-mutated hematopoiesis to persist after induction chemotherapy. Higher NPM1 mutant allele burden may be less amenable to eradication by induction chemotherapy, resulting in a higher likelihood of MRD, which has been associated with relapse in patients treated with SCT.32 Alternatively, high NPM1-mutant allele burden may indicate the presence of disease in hematopoietic cell populations that are resistant to chemotherapy and foster relapse after SCT. Unlike detection of low-level MRD after therapy, which requires specialized high-sensitivity testing that is not currently available in most laboratories, NPM1 VAF information is readily available from most NGS testing platforms. In our cohort, NGS was performed in only a subset of patients posttreatment; 2 of 4 cases with high NPM1 VAF and 9 of 17 cases without high NPM1 VAF had persistent NPM1 mutations identified after induction therapy.
Although we attempted to control for all potential effect-bearing variables in multivariable analyses, our study is limited by its retrospective nature, the relatively small number of examined cases, and the variable application of SCT in our patient cohort, reflecting the controversial nature of the need for SCT in the setting of NPM1-mutated AML. Although the NPM1 VAF values were derived from more than one molecular testing platform and included testing of both blood and bone marrow samples, we did not observe significant differences in the VAF based on the platform used or sample type. VAF may be subject to variability because of skewed amplification, particularly in cases with low read numbers; however, it should be noted that such variability is inherent in any quantitative prognostic marker, such as blast count strata, which are used routinely to risk stratify myelodysplastic syndromes. We recognize that although our data suggested a VAF cutoff of ≥0.44, this value likely represents an approximation of a true biologic cutoff, given variation among assays and testing methodologies. We also acknowledge that the VAF cutoff of 0.44 is based on a relatively small series of patients and should be evaluated in other patient cohorts. Until now, NPM1 VAF has not generally been investigated as a risk factor in AML; we feel that the results of our study could stimulate additional investigation of this variable in terms of its effect on outcome in other independent patient cohorts with NPM1-mutated AML. Further study in larger patient cohorts and using different testing platforms will be necessary to optimize a VAF cutoff indicating higher-risk disease.
In summary, we have shown in this cohort of patients with AML with mutated NPM1, diagnosed according to current WHO criteria, that high NPM1-mutant allele burden at diagnosis is an independent predictor of unfavorable clinical outcomes, particularly in patients treated with SCT and in the subset of patients with DNMT3A comutation. These findings raise the possibility that the biology of this leukemia subtype might differ based on NPM1 clone size. Although these results require validation in a larger patient cohort, they suggest that routine quantification of NPM1 mutational burden at diagnosis may provide important prognostic information for patients with de novo AML and help guide subsequent management.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.S.P., R.P.H., and O.K.W. analyzed clinical databases, performed statistical analyses, and wrote the manuscript; F.C.K., C.J.G., and V.N. performed select data analyses; M.W., R.J.S., D.P.S., R.M.S., E.P.A., D.J.D., Y.-B.A.C., A.T.F., A.M.B., and T.A.G. provided patients and contributed critical clinical insight; and all authors reviewed and approved the final manuscript.
Conflict-of-interest disclosure: D.P.S. has the following relevant relationships: safety monitoring committee (Janssen, Onconova), equity (Acceleron), consulting (H3 Biosciences, Celgene, Amphivena, Tesaro). A.T.F. has the following relevant relationships: consulting (Seattle Genetics, Celgene), advisory boards (Celgene, Agios, Jazz Pharmaceuticals). Y.-B.A.C. has the following relevant relationships: consulting (Takeda Pharmaceuticals, Magenta Therapeutics), advisory boards (Takeda Pharmaceuticals, Incyte Diagnostics). R.J.S. has the following relevant relationships: consulting (Juno, NMDP, GSK, Merck, Gilead), management board (Kiadis). R.P.H. has the following relevant relationship: consulting (Celgene). The remaining authors declare no competing financial interests.
Correspondence: Robert P. Hasserjian, Department of Pathology, Massachusetts General Hospital, 55 Fruit St, Warren 244, Boston, MA 02114-2696; e-mail: rhasserjian@partners.org.
References
Author notes
R.P.H. and O.K.W. contributed equally to this work.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal