Key Points
In pediatric T-ALL, oncogenetic markers, MRD, and WBC count are independent predictors of outcome.
These factors should be used together for individual treatment stratification.
Abstract
Risk stratification in childhood T-cell acute lymphoblastic leukemia (T-ALL) is mainly based on minimal residual disease (MRD) quantification. Whether oncogenetic mutation profiles can improve the discrimination of MRD-defined risk categories was unknown. Two hundred and twenty FRALLE2000T-treated patients were tested retrospectively for NOTCH1/FBXW7/RAS and PTEN alterations. Patients with NOTCH1/FBXW7 (N/F) mutations and RAS/PTEN (R/P) germ line (GL) were classified as oncogenetic low risk (gLoR; n = 111), whereas those with N/F GL and R/P GL mutations or N/F and R/P mutations were classified as high risk (gHiR; n = 109). Day 35 MRD status was available for 191 patients. Five-year cumulative incidence of relapse (CIR) and disease-free survival were 36% and 60% for gHiR patients and 11% and 89% for gLoR patients, respectively. Importantly, among the 60% of patients with MRD <10−4, 5-year CIR was 29% for gHiR patients and 4% for gLoR patients. Based on multivariable Cox models and stepwise selection, the 3 most discriminating variables were the oncogenetic classifier, MRD, and white blood cell (WBC) count. Patients harboring a WBC count ≥200 × 109/L, gHiR classifier, and MRD ≥10−4 demonstrated a 5-year CIR of 46%, whereas the 58 patients (30%) with a WBC count <200 × 109/L, gLoR classifier, and MRD <10−4 had a very low risk of relapse, with a 5-year CIR of only 2%. In childhood T-ALL, the N/F/R/P mutation profile is an independent predictor of relapse. When combined with MRD and a WBC count ≥200 × 109/L, it identifies a significant subgroup of patients with a low risk of relapse.
Medscape Continuing Medical Education online

In support of improving patient care, this activity has been planned and implemented by Medscape, LLC and the American Society of Hematology. Medscape, LLC is jointly accredited by the Accreditation Council for Continuing Medical Education (ACCME), the Accreditation Council for Pharmacy Education (ACPE), and the American Nurses Credentialing Center (ANCC), to provide continuing education for the healthcare team.
Medscape, LLC designates this Journal-based CME activity for a maximum of 1.00 AMA PRA Category 1 Credit(s)™. Physicians should claim only the credit commensurate with the extent of their participation in the activity.
All other clinicians completing this activity will be issued a certificate of participation. To participate in this journal CME activity: (1) review the learning objectives and author disclosures; (2) study the education content; (3) take the post-test with a 75% minimum passing score and complete the evaluation at http://www.medscape.org/journal/blood; and (4) view/print certificate. For CME questions, see page 374.
Disclosures
Laurie Barclay, freelance writer and reviewer, Medscape, LLC, owns stock, stock options, or bonds from Alnylam, Biogen, and Pfizer. Associate Editor Jorge Cortes and the authors declare no competing financial interests.
Learning objectives
Upon completion of this activity, participants will be able to:
Compare outcomes among oncogenetic low-risk vs oncogenetic high-risk patients with childhood T-cell acute lymphoblastic leukemia (T-ALL).
Assess outcome prediction among patients with childhood T-ALL based on a combination of oncogenetic classifier, minimal residual disease, and other factors.
Determine the clinical implications of these findings regarding outcome prediction for patients with childhood T-ALL based on a combination of oncogenetic classifier, minimal residual disease, and other factors.
Release date: January 18, 2018; Expiration date: January 18, 2019
Introduction
Childhood T-cell acute lymphoblastic leukemia (T-ALL) accounts for ∼15% of pediatric ALL. Despite improvements in treatment by using more intensive therapies, childhood T-ALL still has an inferior prognosis compared with B-cell precursor ALL, with an overall survival (OS) of ∼80%.1,2 White blood cell (WBC) count at diagnosis or early response-based parameters such as prednisone response (PR) at day 8 have been used as prognostic factors for risk stratification.3,4 Postremission minimal residual disease (MRD) is currently the best predictor of outcome and is used to drive therapeutic decisions, even if the optimal time point is still contentious.5-9 However, MRD is not always strictly correlated with outcome. Most importantly, it fails to detect some patients with an apparently optimal molecular response who nevertheless relapse. It is therefore important to identify additional risk factors.
T-ALLs represent expansions of precursor T cells arrested at specific stages of thymic development,10,11 with the underlying genetic abnormality often determining the stage of maturation arrest.12 Recognized T-ALL oncogenic pathways include proto-oncogene activation, tumor suppressor gene deletion, and activation of the Notch1 pathway by NOTCH1/FBXW7 (N/F) mutations,13,14 leading to various combinations of gene alterations and complex oncogenic networks.11,15-18 N/F mutations involve either the heterodimerization domain (probably facilitating cleavage of the NOTCH1 receptor) and/or the negative regulatory PEST domain, increasing the half-life of intracellular NOTCH1.19 An alternative mechanism of constitutive Notch1 pathway activation involves loss-of-function mutations of FBXW7, leading to the inhibition of ubiquitin-mediated degradation of activated NOTCH1.20 The GRAALL (Group for Research on Adult Acute Lymphoblastic Leukemia) group reported a significantly better outcome in adult T-ALL patients harboring N/F mutations than in nonmutated cases.14 Although pediatric studies also suggest better overall outcome, the impact of N/F mutations in childhood T-ALL remains more controversial.21-24 The pro-proliferative Ras/Raf/MEK/extracellular signal-regulated kinase and phosphatidylinositol 3-kinase/PTEN/Akt/mTOR pathways have also been reported to be deregulated and associated with poor outcome in limited numbers of pediatric T-ALL.25,26 Recently, the GRAALL group demonstrated that the presence of N/F mutations in the absence of RAS or PTEN abnormalities predicts a very good outcome in almost 50% of adult T-ALL. Conversely, the absence of N/F or the presence of RAS/PTEN (R/P) alterations identifies the remaining cohort of patients with a poor prognosis.27 When combined with postremission induction MRD, N/F and R/P alterations are independent predictors of outcome in adult ALL.28
The FRALLE2000 T guidelines (French Acute Lymphoblastic Leukaemia Study Group) were Berlin-Frankfurt-Münster (BFM) inspired and were used from 2000 to 2010 to treat children with T-ALL, using historical factors and MRD to stratify treatment between standard-risk and high-risk arms. In order to identify new prognostic factors, a retrospective study was conducted among 220 children with T-ALL, treated according to these guidelines, to test whether oncogenetic features identified in the GRAALL cohort could improve the detection of patients at risk of relapse.
Patients and methods
Treatments
From 2000 to 2010, 405 T-ALL patients aged 1 to 14 years were treated in 16 French pediatric FRALLE study group hematology departments according to the FRALLE2000 T guidelines (as briefly described in supplemental Figure 1 and supplemental Tables 1 and 2, available on the Blood Web site). Fourteen patients died before reaching complete remission (CR). After induction, 368 reached CR. Among the 23 patients who failed, 16 achieved CR after salvage treatment, resulting in a total of 384 patients (94.8%) in CR (supplemental Figure 2). Forty-one patients received allogeneic stem cell transplantation in the first CR. Patient outcomes were updated in 2015. The 5-year OS, disease-free survival (DFS), and cumulative incidence of relapse (CIR) were 76.6%, 72.8%, and 24.6%, respectively, with a median follow-up of 5 years (supplemental Figure 3).
Study cohort
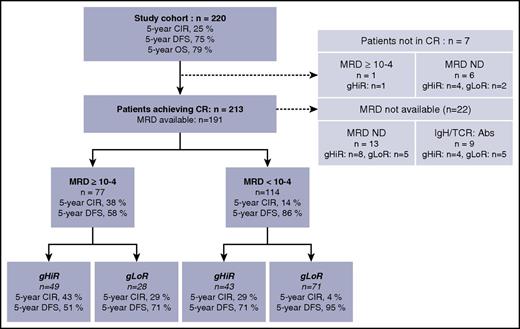
The study cohort comprised the 220 patients for whom diagnostic DNA and/or complementary DNA was available to perform genetic analysis (Figure 1). These patients were representative of the whole FRALLE2000T population, although slightly more patients in this study cohort were male (supplemental Table 3; supplemental Figure 4). Informed consent for data registration was provided according to the Declaration of Helsinki. This study was approved by the Leukemias Committee of the National Scientific Committee of the SFCE (Société Française des Cancers et Hémopathies de l’Enfant et de l’adolescent) and by the ethics committee of each participating center. MRD monitoring was based on quantitative detection of leukemic clone-specific T-cell receptor (TCR) and/or immunoglobulin gene rearrangements according to EuroMRD guidelines.29 NOTCH1, FBXW7, and K-RAS/N-RAS mutations and PTEN alterations (major deletion or mutation) were identified by direct sequencing and/or comparative genomic hybridization array as described previously.27,30 We did not search for microdeletions.31 In brief, the oncogenetic low-risk (gLoR) group was defined by the presence of an N/F mutation and the absence of an R/P mutation, and the oncogenetic high-risk (gHiR) group was defined by N/F germ line (GL) and R/P GL or altered N/F mutations and R/P altered, as for adult GRAALL patients.27
Patient flowchart. Abs, absence; IgH/TCR, immunoglobulin and T-cell receptor markers; ND, not done.
Patient flowchart. Abs, absence; IgH/TCR, immunoglobulin and T-cell receptor markers; ND, not done.
MRD status and outcome
MRD analysis was conducted with a sensitivity threshold of 10−4, as recommended for ALL assessment.32 Among the 213 patients achieving CR after induction, MRD quantification of immunoglobulin/TCR gene rearrangements was available for 191 patients. Evaluation was not performed in 22 patients (including 9 patients with an absence of an immunoglobulin/TCR marker). MRD levels were <10−4 for 114 patients (60%) and ≥10−4 for 77 patients (40%). Patients with MRD ≥10−4 showed a significantly higher risk of relapse than patients with MRD <10−4, with a 5-year CIR and DFS for MRD ≥10−4 vs <10−4 of 37.6% and 58.4% vs 13.6% and 86.4%, respectively (Figure 2). The cause-specific hazard of relapse (CSHR) for CIR was 3.11 (95% confidence interval [CI], 1.74-5.57) for patients with MRD ≥10−4 (P = .00013).
CIR and DFS according to MRD. (A) CIR in patients with MRD ≥10−4 vs MRD <10−4. (B) DFS in patients with MRD ≥10−4 vs MRD <10−4.
CIR and DFS according to MRD. (A) CIR in patients with MRD ≥10−4 vs MRD <10−4. (B) DFS in patients with MRD ≥10−4 vs MRD <10−4.
Statistical methods
Patient characteristics and CR rates were compared using either the Fisher’s exact test or the Mann-Whitney U test. OS was calculated from the date of prephase initiation until the date of death. CIR and DFS were calculated from the date of CR achievement until the date of relapse or death free of relapse. OS and DFS were estimated using the Kaplan-Meier method and then compared using the log-rank test.33 Multivariable regressions were performed using the Cox model.34 CIR was estimated taking into account death in first CR as a competing risk and then compared using the Gray test. Multivariable models used the cause-specific hazard Cox model. CSHRs and hazard ratios (HRs) were given with 95% CIs. Interactions were assessed by the Gail and Simon statistics. All statistical tests were 2 sided, with P ≤ .05 denoting statistical significance. All analyses were performed on R 2.14 (http://www.R-project.org) statistical software. Tabulated summary statistics are frequency (percentages) for discrete variables and medians (interquartile range) for continuous variables, unless otherwise specified.
Results
NOTCH1, FBXW7, PTEN and K/N-RAS gene status in pediatric T-ALL
All 220 patients were explored for NOTCH1, FBXW7, PTEN, and K/N-RAS gene status. N/F mutations were identified in 134 patients (61%), including 94 with NOTCH1 mutation alone, 12 with FBXW7 mutation alone, and 28 with both. PTEN alterations (mutation and/or major deletion) were identified in 30 patients, including 16 patients with N/F mutation. K/N-RAS mutations were found in 18 patients, including 2 patients with PTEN alteration and 7 patients with N/F mutations. Overall, 163 patients (74%) harbored at least 1 mutation/deletion in NOTCH1, FBXW7, PTEN, or K/N-RAS gene. Clinical features, genotype subsets, and response to treatment were analyzed according to N/F, PTEN, and K/N-RAS status (Table 1). T-ALLs with PTEN alteration were more frequently associated with higher median WBC counts (P = .027) and with SIL-TAL positivity (P = .002), as described previously.27 Regarding early response to treatment, patients with PTEN alteration were less responsive to prednisone at day 8. Patients harboring N/F mutations had better response to induction therapy in terms of good PR (65% vs 50%, P = .03) and MRD <10−4 at day 35 (61% vs 37%, P = .0006).
Characteristics of patients according to their N/F, PTEN, and RAS status
| . | All patients . | N/F . | PTEN . | K/N-RAS . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (N = 220) . | Mut, 134 (61%) . | WT, 86 (39%) . | P* . | Altered, 30 (14%) . | Not altered 190 (86%) . | P* . | Mut 18 (8%) . | WT 202 (92%) . | P* . | |
| Genotype subsets analyzed | 134 | 88 | 46 | — | 22 | 112 | — | 14 | 120 | — |
| CALM-AF10 | 5 (4) | 2 (2) | 3 (6) | NS | 2 (9) | 3 (3) | NS | 0 | 5 (4) | NS |
| SIL-TAL1 | 18 (13) | 9 (10) | 9 (20) | NS | 8 (36) | 10 (9) | .002 | 1 (7) | 17 (14) | NS |
| TLX1 | 9 (7) | 7 (8) | 2 (4) | NS | 1 (4) | 8 (7) | NS | 1 (7) | 8 (7) | NS |
| TLX3 | 35 (26) | 24 (27) | 11 (24) | NS | 2 (10) | 33 (29) | NS | 7 (50) | 28 (23) | .049 |
| None of above | 67 (50) | 46 (52) | 21 (46) | NS | 9 (41) | 58 (52) | NS | 5 (36) | 62 (52) | NS |
| N/Fmutated | 134 (61) | 134 (100) | 0 | — | 16 (53) | 118 (62) | NS | 7 (39) | 127 (63) | NS |
| PTENaltered | 30 (14) | 16 (12) | 14 (16) | NS | 30 (100) | 0 | — | 2 (11) | 28 (14) | NS |
| K/N-RASmutated | 18 (8) | 7 (5) | 11 (13) | NS | 2 (7) | 16 (8) | NS | 18 (100) | 0 | — |
| Clinical features | ||||||||||
| Age, median (range), y | 9.5 (1.1-19.5) | 9.8 | 8.9 | NS | 8.7 | 9.5 | NS | 9.3 | 9.5 | NS |
| Median WBC, ×109/L | 100 | 101.5 | 93.9 | NS | 162.5 | 79.8 | .027 | 82 | 97 | NS |
| WBC count ≥200 × 109/L | 55 (25) | 27 (20) | 28 (32) | NS | 12 (40) | 43 (23) | NS | 4 (22) | 51 (25) | NS |
| CNS involvement | 18 (8) | 11 (8) | 7 (8) | NS | 2 (7) | 16 (8) | NS | 0 | 18 (9) | NS |
| Day 8 GPR | 125 (60) | 84 (65) | 41 (50) | .03 | 11 (37) | 114 (60) | .014 | 10 (59) | 115 (60) | NS |
| Day 21 CHs | 186 (87) | 117 (90) | 69 (83) | NS | 28 (93) | 158 (83) | NS | 14 (82) | 172 (87) | NS |
| CR | 213 (97) | 132 (98) | 81 (94) | NS | 30 (100) | 183 (96) | NS | 15 (83) | 191 (95) | NS |
| MRD <10−4 | 114/192 (59) | 82/118 (69) | 32/74 (43) | .0006 | 15/26 (58) | 99/166 (60) | NS | 6/15 (40) | 108/177 (61) | NS |
| . | All patients . | N/F . | PTEN . | K/N-RAS . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (N = 220) . | Mut, 134 (61%) . | WT, 86 (39%) . | P* . | Altered, 30 (14%) . | Not altered 190 (86%) . | P* . | Mut 18 (8%) . | WT 202 (92%) . | P* . | |
| Genotype subsets analyzed | 134 | 88 | 46 | — | 22 | 112 | — | 14 | 120 | — |
| CALM-AF10 | 5 (4) | 2 (2) | 3 (6) | NS | 2 (9) | 3 (3) | NS | 0 | 5 (4) | NS |
| SIL-TAL1 | 18 (13) | 9 (10) | 9 (20) | NS | 8 (36) | 10 (9) | .002 | 1 (7) | 17 (14) | NS |
| TLX1 | 9 (7) | 7 (8) | 2 (4) | NS | 1 (4) | 8 (7) | NS | 1 (7) | 8 (7) | NS |
| TLX3 | 35 (26) | 24 (27) | 11 (24) | NS | 2 (10) | 33 (29) | NS | 7 (50) | 28 (23) | .049 |
| None of above | 67 (50) | 46 (52) | 21 (46) | NS | 9 (41) | 58 (52) | NS | 5 (36) | 62 (52) | NS |
| N/Fmutated | 134 (61) | 134 (100) | 0 | — | 16 (53) | 118 (62) | NS | 7 (39) | 127 (63) | NS |
| PTENaltered | 30 (14) | 16 (12) | 14 (16) | NS | 30 (100) | 0 | — | 2 (11) | 28 (14) | NS |
| K/N-RASmutated | 18 (8) | 7 (5) | 11 (13) | NS | 2 (7) | 16 (8) | NS | 18 (100) | 0 | — |
| Clinical features | ||||||||||
| Age, median (range), y | 9.5 (1.1-19.5) | 9.8 | 8.9 | NS | 8.7 | 9.5 | NS | 9.3 | 9.5 | NS |
| Median WBC, ×109/L | 100 | 101.5 | 93.9 | NS | 162.5 | 79.8 | .027 | 82 | 97 | NS |
| WBC count ≥200 × 109/L | 55 (25) | 27 (20) | 28 (32) | NS | 12 (40) | 43 (23) | NS | 4 (22) | 51 (25) | NS |
| CNS involvement | 18 (8) | 11 (8) | 7 (8) | NS | 2 (7) | 16 (8) | NS | 0 | 18 (9) | NS |
| Day 8 GPR | 125 (60) | 84 (65) | 41 (50) | .03 | 11 (37) | 114 (60) | .014 | 10 (59) | 115 (60) | NS |
| Day 21 CHs | 186 (87) | 117 (90) | 69 (83) | NS | 28 (93) | 158 (83) | NS | 14 (82) | 172 (87) | NS |
| CR | 213 (97) | 132 (98) | 81 (94) | NS | 30 (100) | 183 (96) | NS | 15 (83) | 191 (95) | NS |
| MRD <10−4 | 114/192 (59) | 82/118 (69) | 32/74 (43) | .0006 | 15/26 (58) | 99/166 (60) | NS | 6/15 (40) | 108/177 (61) | NS |
Values represent n (%) of patients unless otherwise indicated.
CH, chemosensitivity (defined by blasts ≤5% in bone marrow); CNS, central nervous system; GPR, good PR; Mut, mutated; NA, not applicable; NS, not significant; WT, wild-type.
Student t test or Fisher’s exact test where appropriate.
Oncogenetic classifier in pediatric T-ALL
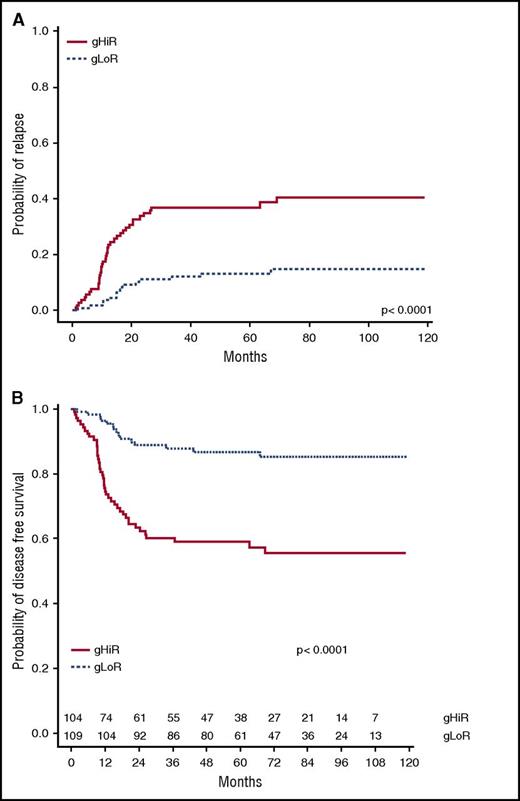
As shown in adults with T-ALL, the favorable prognostic significance of N/F mutations was essentially restricted to pediatric patients without K/N-RAS mutations or PTEN alterations (R/P mutations) and had no effect in N/F GL cases, demonstrating the importance of including oncogenic synergy in evaluation of prognostic impact (supplemental Figure 5). In the group of patients with N/F mutations, 5-year CIR and DFS were respectively 49.8% (95% CI, 27.2-68.8) and 45.9% (95% CI, 29.2-72.2) for patients with R/P mutations and 13.3% (95% CI, 7.6-20.5) and 86.7% (95% CI, 80.5-93.5) for patients without R/P mutations (P < .0001). By using the oncogenetic classifier previously described,27 111 of the 220 patients (50%) were classified as gLoR and 109 as gHiR. At diagnosis, this classifier was not associated with significant differences in age, sex, WBC count, WBC ≥200 ×109/L, or central nervous system status (supplemental Table 4). A good PR at day 8 was significantly lower for gHiR patients (P = .049), but this difference disappeared after adjusting on baseline WBC levels (P = .11). The classifier alone was unable to predict CR status (supplemental Table 4) because CR was 98.2% (95% CI, 93.6-99.8) for gLoR and 95.4% (95% CI, 89.6-98.5) for gHiR patients. Among the 213 patients achieving CR after induction, 109 were gLoR and 104 were gHiR. 5-year CIR and DFS were respectively 36.3% (95% CI, 27.2-44.8) and 60.4% (95% CI, 51.0-71.6) for the gHiR group and 11.4% (95% CI, 4.8-17.2) and 88.6% (95% CI, 82.4-95.2) for the gLoR group (P < .0001) (Figure 3).
CIR and DFS according to oncogenetic classifier. (A) CIR in gHiR vs gLoR patients. (B) DFS in gHiR vs gLoR patients.
CIR and DFS according to oncogenetic classifier. (A) CIR in gHiR vs gLoR patients. (B) DFS in gHiR vs gLoR patients.
Oncogenetic classifier in patients with MRD ≥10−4
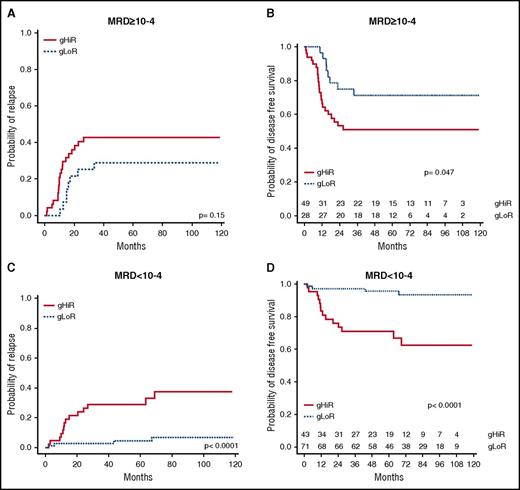
The oncogenetic classifier was then combined with MRD status to determine whether it could provide additional information regarding relapse risk. gHiR patients were significantly associated with MRD ≥10−4 (P = .0004) and a higher risk of relapse (P = .00002) (supplemental Table 5). Among the 77 patients with MRD ≥10−4, gHiR worsened the risk of relapse. 5-year CIR and DFS were respectively 42.8% (95% CI, 28.9-56.7) and 50.9% (95% CI, 38.4-67.6) for gHiR patients and 28.9% (95% CI, 11.7-46.1) and 71.1% (95% CI, 56.0-90.2) for gLoR patients (Figure 4). These data show that among high-risk patients with MRD ≥10−4, gHiR status has an additional negative impact on outcome.
CIR and DFS according to oncogenetics combined with MRD. (A) CIR in patients with MRD ≥10−4, comparing gHiR and gLoR patients. (B) DFS in patients with MRD ≥10−4, comparing gHiR and gLoR patients. (C) CIR in patients with MRD <10−4, comparing gHiR and gLoR patients. (D) DFS in patients with MRD <10−4, comparing gHiR and gLoR patients.
CIR and DFS according to oncogenetics combined with MRD. (A) CIR in patients with MRD ≥10−4, comparing gHiR and gLoR patients. (B) DFS in patients with MRD ≥10−4, comparing gHiR and gLoR patients. (C) CIR in patients with MRD <10−4, comparing gHiR and gLoR patients. (D) DFS in patients with MRD <10−4, comparing gHiR and gLoR patients.
Oncogenetic classifier in patients with MRD <10−4
Among the 104 patients with MRD <10−4, 18 patients (16%) relapsed, leading to 9 deaths, including 8 related to leukemia. Interestingly, among these patients, the oncogenetic classifier identified patients with a higher risk of relapse. Indeed, relapse occurred in 14 of the 42 patients with MRD <10−4 and gHiR (including 10 relapses before 2 years), leading to 6 deaths (5 leukemia related), compared with 4 relapses in the 71 patients with MRD <10−4 and gLoR. 5-year-CIR and DFS were respectively 28.9% (95% CI, 15.0-42.8) and 71.0% (95% CI, 58.4-86.3) in the gHiR group and 4.4% (95% CI, 0-9.2) and 95.5% (95% CI, 90.7-1.00) in the gLoR group (Figure 4). Oncogenetic status therefore improved the detection of patients at risk of relapse, especially in those with MRD <10−4 after induction.
Prognostic analyses and clinical classification
Prognostic values of conventional and new oncogenetic risk factors were then analyzed together (Table 2). By univariate analyses, in addition to the classifier, other factors that were identified to predict relapse were male gender, WBC ≥200 × 109/L, chemoresistance at day 21, and MRD ≥10−4 at the end of induction. Based on a stepwise selection procedure, the 3 variables retained were the classifier, WBC count ≥200 × 109/L, and MRD (supplemental Table 6). The CSHR was 3.22 (95% CI, 1.65-6.29) for oncogenetic gHiR vs gLoR (P = .0006), 2.30 (95% CI, 1.26-4.20) for MRD ≥10−4 vs MRD <10−4 (P = .0070), and 1.85 (95% CI, 1.01-3.37) for WBC ≥200 × 109/L vs <200 × 109/L (P = .0456) (supplemental Table 6; Table 2). Based on these 3 parameters, 8 subsets of patients were defined according to the estimated 5-year CIR (Table 3). The 58 patients (30%) with a WBC count <200 × 109/L, classifier gLoR, and MRD<10−4 were at very low risk of relapse (with a 5-year estimated CIR of 1.7%). Patients harboring at least 1 of the parameters WBC count ≥200 × 109/L, classifier gHiR, or MRD ≥10−4 acquired an increased cumulative risk of relapse (up to a 5-year CIR of 45.8% if all 3 parameters were associated). Taken together, the oncogenetic classifier improves prediction of the risk of relapse for patients with a WBC count ≥ 200 × 109/L at diagnosis and/or MRD ≥10−4 at end of induction. Interestingly, the oncogenetic classifier efficiently discriminated the risk of relapse in patients with MRD <10−4 and identified a subset of patients at very low risk of relapse when associating gLoR, MRD <10−4, and WBC count <200 × 109/L.
Cause-specific hazards of relapse in the 213 patients with CR
| . | No. of patients . | No. of relapses . | Univariable analysis (213 patients, 54 relapses) . | Multivariable analysis (191 patients, 46 events) . | ||||
|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |||
| Age, y | 213 | — | 1.02 | (0.96-1.08) | .51 | — | — | — |
| Sex | ||||||||
| Female | 37 | 4 | Ref | — | — | — | — | — |
| Male | 176 | 50 | 2.98 | (1.08-8.25) | .036 | — | — | — |
| CNS involvement | ||||||||
| No | 197 | 51 | Ref | — | — | — | — | — |
| Yes | 16 | 3 | 0.62 | (0.19-2) | .42 | — | — | — |
| WBC count, ×109/L | ||||||||
| <200 | 162 | 36 | Ref | — | — | — | — | — |
| ≥200 | 51 | 18 | 1.93 | (1.1-3.4) | .023 | 1.85 | (1.01-3.37) | .0456 |
| Day 21 CHr | ||||||||
| Yes | 184 | 42 | Ref | — | — | — | — | — |
| No | 26 | 12 | 2.43 | (1.28-4.61) | .007 | — | — | — |
| Oncogenetics | ||||||||
| gLoR | 109 | 15 | Ref | |||||
| gHiR | 104 | 39 | 3.49 | (1.92-6.34) | <.0001 | 3.22 | (1.64-6.28) | .0006 |
| MRD | ||||||||
| <10−4 | 114 | 18 | Ref | — | — | — | — | — |
| ≥10−4 | 77 | 28 | 2.84 | (1.57-5.15) | .0006 | 2.30 | (1.20-4.02) | .0007 |
| . | No. of patients . | No. of relapses . | Univariable analysis (213 patients, 54 relapses) . | Multivariable analysis (191 patients, 46 events) . | ||||
|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |||
| Age, y | 213 | — | 1.02 | (0.96-1.08) | .51 | — | — | — |
| Sex | ||||||||
| Female | 37 | 4 | Ref | — | — | — | — | — |
| Male | 176 | 50 | 2.98 | (1.08-8.25) | .036 | — | — | — |
| CNS involvement | ||||||||
| No | 197 | 51 | Ref | — | — | — | — | — |
| Yes | 16 | 3 | 0.62 | (0.19-2) | .42 | — | — | — |
| WBC count, ×109/L | ||||||||
| <200 | 162 | 36 | Ref | — | — | — | — | — |
| ≥200 | 51 | 18 | 1.93 | (1.1-3.4) | .023 | 1.85 | (1.01-3.37) | .0456 |
| Day 21 CHr | ||||||||
| Yes | 184 | 42 | Ref | — | — | — | — | — |
| No | 26 | 12 | 2.43 | (1.28-4.61) | .007 | — | — | — |
| Oncogenetics | ||||||||
| gLoR | 109 | 15 | Ref | |||||
| gHiR | 104 | 39 | 3.49 | (1.92-6.34) | <.0001 | 3.22 | (1.64-6.28) | .0006 |
| MRD | ||||||||
| <10−4 | 114 | 18 | Ref | — | — | — | — | — |
| ≥10−4 | 77 | 28 | 2.84 | (1.57-5.15) | .0006 | 2.30 | (1.20-4.02) | .0007 |
CHr, chemoresistance (defined by blasts >5% in bone marrow); CNS, central nervous system; Ref, reference.
Estimated CIR according to classifier, WBC count, and MRD
| WBC count (×109/L) . | Classifier . | MRD ≥10−4 . | Prevalence, n (%) . | 5-y CIR (%) . | Clinical risk classification . |
|---|---|---|---|---|---|
| <200 | gLoR | No | 58/191 (30.3) | 1.7 | Low |
| ≥200 | gLoR | No | 13/191 (6.8) | 17.5 | Intermediate |
| <200 | gHiR | No | 30/191 (15.7) | 24.8 | Intermediate |
| ≥200 | gHiR | No | 13/191 (6.8) | 38.4 | High |
| <200 | gLoR | Yes | 21/191 (11.0) | 28.9 | Intermediate |
| ≥200 | gLoR | Yes | 7/191 (3.7) | 28.6 | Intermediate |
| <200 | gHiR | Yes | 33/191 (17.3) | 41.5 | High |
| ≥200 | gHiR | Yes | 16/191 (8.4) | 45.8 | High |
| WBC count (×109/L) . | Classifier . | MRD ≥10−4 . | Prevalence, n (%) . | 5-y CIR (%) . | Clinical risk classification . |
|---|---|---|---|---|---|
| <200 | gLoR | No | 58/191 (30.3) | 1.7 | Low |
| ≥200 | gLoR | No | 13/191 (6.8) | 17.5 | Intermediate |
| <200 | gHiR | No | 30/191 (15.7) | 24.8 | Intermediate |
| ≥200 | gHiR | No | 13/191 (6.8) | 38.4 | High |
| <200 | gLoR | Yes | 21/191 (11.0) | 28.9 | Intermediate |
| ≥200 | gLoR | Yes | 7/191 (3.7) | 28.6 | Intermediate |
| <200 | gHiR | Yes | 33/191 (17.3) | 41.5 | High |
| ≥200 | gHiR | Yes | 16/191 (8.4) | 45.8 | High |
Patients were classified into 3 clinical risk groups according to 5-year CIR.
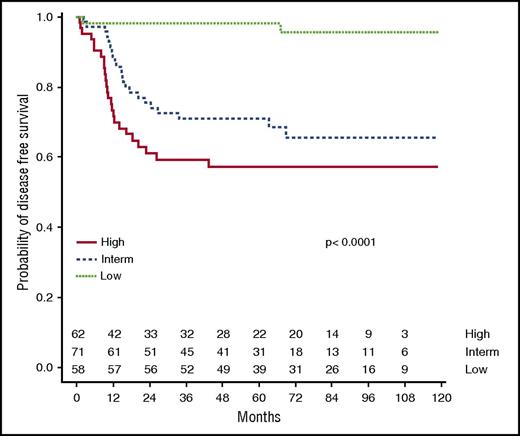
Because it is not realistic for clinicians to design a therapeutic approach based on 8 risk subsets, a simplified “clinical classification” into 3 groups was then tested (Table 3; Figure 5). The clinical low-risk group corresponded to gLoR patients with a WBC count <200 × 109/L at diagnosis and MRD <10−4, leading to a 5-year CIR and DFS at 1.7% (95% CI, 0-5.1) and 98.3% (95% CI, 95.0-100). The clinical high-risk group combined patients with MRD ≥10−4 and gHiR (regardless of WBC count) or patients with a WBC count >200 × 109/L and gHiR (regardless of MRD status), leading to a 5-year CIR and DFS of 37.7% (95% CI, 25.3-50.1) and 57.3% (95% CI, 45.9-71.6). Alternatively stated, the clinical high-risk group was defined by gHiR in association with a WBC count >200 × 109/L and/or MRD ≥10−4. The clinical intermediate-risk group included all other patients, with a 5-year CIR and DFS of 29.1% (95% CI, 18.3-39.7) and 70.9% (95% CI, 61.0-82.5), respectively. This classification divides patients into roughly equal subgroups (30% at low risk, 33% at intermediate risk, and 37% at high risk), which is easily transferable to clinical practice, including therapeutic stratification.
DFS according to clinical classification. Using the clinical classification integrating oncogenetics, MRD, and WBC count, 5-year DFS was estimated at 98.3%, 70.9%, and 57.3% in the clinical low-risk group, intermediate-risk group, and high-risk group, respectively.
DFS according to clinical classification. Using the clinical classification integrating oncogenetics, MRD, and WBC count, 5-year DFS was estimated at 98.3%, 70.9%, and 57.3% in the clinical low-risk group, intermediate-risk group, and high-risk group, respectively.
Discussion
The objective of this study was to test whether new genetic factors, when combined with historical parameters, can improve outcome prediction in pediatric T-ALL.
Although the MRD value (measured by immunoglobulin/TCR clone-specific real-time quantitative polymerase chain reaction) is a well-established factor predicting relapse in children with precursor B-cell ALL,35-37 it has been less widely studied in pediatric T-ALL.5-7 In the present cohort, which was treated uniformly with FRALLE2000T guidelines, 191 patients had available MRD data, representing the second largest cohort of pediatric T-ALL with available MRD data quantified by immunoglobulin/TCR clone-specific real-time quantitative polymerase chain reaction. In the BFM and Italian Association of Pediatric Hematology and Oncology (AIEOP) study of 464 de novo pediatric T-ALL, MRD quantified prospectively at day 33 and day 78 was used for risk stratification.7 Patients were treated with identical chemotherapy during the first 9 weeks but then treated slightly differently in the AIEOP and BFM arms. MRD negativity at day 33 (sensitivity ≤10−4) was the most favorable prognostic factor, and the patients who were MRD ≥10−3 on day 33 but negative on day 78 (32%) had an excellent outcome, with a 7-year CIR of 8.5%, leading to the conclusion that day 33 MRD was prognostically irrelevant whereas MRD ≥10−3 at day 78 (21% of patients) constituted the most important factor predicting relapse.7 Day 78 MRD was not performed sufficiently frequently in the FRALLE2000T to allow assessment of this parameter, but it would be interesting to know the oncogenic status of day 33–positive/day 78–negative AIEOP/BFM patients. In the FRALLE2000T cohort, MRD status at the time of CR (around day 35), using a threshold of 10−4, was a strong independent risk factor predicting relapse. MRD was <10−4 for 60% of patients (114), with a 5-year CIR of 13.6%, which was similar to the 58% of samples <10−4 by 8-color flow cytometry in the AALL0434 COG study38 but different from the 16% of MRD-negative BFM/AEIOP T-ALL patients, who had a 7-year CIR of 7.5%, suggesting that positivity <10−4 may be significant.7 The arrival of novel techniques for potentially more sensitive MRD detection, such as next-generation sequencing and multidimensional flow cytometry, will help address these issues.39 Analysis of the impact of MRD at the end of induction for 1144 T-ALL patients treated in the AALL0434 COG study showed that MRD ≥10−4 on day 29 was associated with inferior event-free survival (EFS) (76.3% vs 89.0%; P = .0001) and OS (86.6% vs 93.8%; P = .0008),38 in keeping with the positive impact of an early MRD time point identified here. Although this study was not powered to evaluate the value of our oncogenetic classifier relative to the combined evaluation of MRD at the end of induction and day 78 or the end of consolidation, these data should allow comparison of the place of oncogenic markers, WBC count, and MRD, as performed by the aforementioned cooperative groups. Because a significant proportion of MRD ≥10−4 patients relapse (16% of patients in FRALLE2000T), we sought to identify the diagnostic parameters that could usefully complement MRD assessment.
Several oncogenetic aberrations have been reported to detect high-risk pediatric T-ALL patients. One of the most interesting and promising aberrations, NOTCH1 gain-of-function mutations, has raised hope for a “targetable” genetic hit. N/F activating mutations had a favorable clinical impact in some studies,23,24,40 but not others.21,22,41 PTEN alterations can modulate this prognostic impact.42,43 Having identified the prognostic value of this 4-gene classifier in adult T-ALL,27,28 we now show that this also applies to pediatric T-ALLs treated according to the FRALLE2000T guidelines. The incidence of N/F (61%), PTEN (14%) and K/N RAS (8%) aberrations were similar to other pediatric T-ALL series.22,24,40,41,43,44 Recently, 145 pediatric T-ALLs treated on the UKALL2003 trial were tested for PTEN/K/N-RAS and N/F mutations. Neither PTEN nor K/N-RAS genotype significantly impacted outcome, and there was no evidence that they changed the highly favorable outcome of patients with double N/F mutations.44 Application of the N/F/R/P classifier presented here to this cohort did, however, show a trend towards an inferior OS outcome, but not an inferior relapse-free survival outcome. The reasons for this discordance merit closer examination but potentially include minor treatment variables, the relatively small size of the UKALL cohort, and/or different molecular techniques and/or interpretation, because the incidence of PTEN abnormalities was slightly higher in the UKALL2003 cohort (22%) than the FRALLE2000 cohort (14%).
In addition to the intrinsic prognostic value of the 4-gene classifier, we show that it provides added information to both the WBC count at diagnosis and MRD at the end of induction, particularly within the large group of MRD <10−4 patients. Patients with an elevated WBC count (≥100 × 109/L) are more likely to have certain genetic aberrations, such as SIL-TAL1 fusions,45-47 and have a different response to treatment, with more frequent poor PR and slower MRD clearance, although the prognostic value of the WBC count often loses independent discrimination on multivariate analysis with MRD.5,7,35,47 This was not the case here, when a WBC count ≥200 × 109/L at diagnosis proved to be an independent, albeit weaker, risk factor, in addition to MRD and the oncogenetic classifier.
By combining WBC count with the 2 other prognostic factors, we identified 30% of patients (n = 58) with a very low risk of relapse. Interestingly, in the AALL0434 COG study, a WBC count >200 × 109/L (27% of patients) was associated with inferior EFS and OS in non–early T-cell precursor and near–early T-cell precursor ALL, but not in early T-cell precursor ALL.38 In the CCG-1961 protocol, pediatric T-ALL with a WBC count ≥200 × 109/L had no difference in outcome compared with T-ALL patients with a WBC count <200 × 109/L, with an EFS of 72.0% ± 4.3% vs 73.7% ± 2.7%, although this could be explained by a preferential response to treatment intensification in patients with a WBC count >200 × 109/L. Indeed, T-lineage patients with a WBC count >200 × 109/L treated with standard intensity had a DFS of 63.1% ± 8.8% compared with 81.3% ± 7.2% when treated with augmented-intensity regimens (P = .1134).48
Because the development of massive genetic screening of leukemic samples promises to improve the prediction and outcome of ALL patients, we propose prospective testing of a new simple stratification for pediatric T-ALL based on a WBC count ≥200 × 109/L, MRD at end of induction with a 10−4 (0.01%) cutoff, and a 4-gene oncogenetic classifier including NOTCH1, FBXW7, N/K-RAS, and PTEN. If confirmed prospectively in current protocols, it will allow treatment reduction for the 30% of low-risk children with T-ALL. Second, it will help allow for the judicious use of stem cell or innovative therapy in the 30% of high-risk cases, potentially also guided by the results of extensive molecular screening. Because the best time point for MRD remains controversial, it will be interesting to evaluate the impact of 2 time points (end of induction and around day 78) on outcome within these 3 groups. It will also be interesting to apply this WBC count/MRD/4-gene classifier to other BFM-based pediatric protocols in order to determine which minor therapeutic modifications could explain potential differential prognostic value.
Presented at the 58th annual meeting of the American Society of Hematology, San Diego, CA, 3-6 December 2016.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Marie-Françoise Auclerc for her assistance with FRALLE data collection. The authors also thank the SFCE and the investigators of the 16 SFCE-involved centers: Nathalie Aladjidi, Anne Auvrignon, Claire Berger, Pascale Blouin, Benoit Brethon, Hervé Chambost, Pascal Chastagner, Gérard Couillault, Jean-Hugues Dalle, Marianne Debre, Catherine Devoldere, Catherine Dollfus, Jean Donadieu, Cécile Dumesnil De Maricourt, Sylvie Fasola, Fanny Fouyssac, Claire Galambrun, Virginie Gandemer, Stéphanie Haouy, Mathilde Jehanne, Charlotte Jubert, Justyna Kanold, Thierry Leblanc, Odile Lejars, Philippe Le Moine, Aude Marie-Cardine, Françoise Mechinaud, Claire Oudin, Caroline Oudot, Catherine Paillard, Brigitte Pautard, Yves Perel, Fanny Rialland, Claudine Schmitt, Pascale Schneider, Jean-Louis Stephan, Marie-Dominique Tabone, Caroline Thomas, and Jean-Pierre Vannier.
This study was supported by Association Laurette Fugain and Enfants et Santé.
Authorship
Contribution: A.B. and V.A. conceived the study; A.P., P.B., J.-M.C., N.G., B.B., H.L., C.S., S.T., G.M., C.P., J.S., J.L.-P., G.L., and A.B. provided study materials or patients; A. Trinquand, A. Touzard, E.M., and V.A. performed molecular analyses; A. Trinquand, S.C., and V.A. collected and assembled data; S.C. performed statistical analysis; A.P., V.A., A.B., and S.C. analyzed and interpreted data; A.P., A.B., E.M., and V.A wrote the manuscript; all authors approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the members of the French Acute Lymphoblastic Leukemia Study Group appears in “Appendix.”
Correspondance: Vahid Asnafi, Hôpital Necker-Enfants Malades, Laboratoire d’onco-hématologie, 149 rue de Sèvres, 75015 Paris, France; e-mail: vahid.asnafi@nck.aphp.fr; and André Baruchel, Hopital Robert Debré, Service d’hématologie Pédiatrique, 48 Blvd Serrurier, 75019 Paris, France; e-mail: andre.baruchel@aphp.fr.
Appendix: study group members
The members of the French Acute Lymphoblastic Leukemia Study Group are: Nathalie Aladjidi, Anne Auvrignon, André Baruchel, Claire Berger, Pascale Blouin, Benoit Brethon, Hervé Chambost, Gérard Couillault, Jean-Hugues Dalle, Marianne Debre, Catherine Devoldere, Catherine Dollfus, Jean Donadieu, Cécile Dumesnil De Maricourt, Christine Edan, Sylvie Fasola, Fanny Fouyssac, Claire Galambrun, Virginie Gandemer, Stéphanie Haouy, Mathilde Jehanne, Charlotte Jubert, Justyna Kanold, Judith Landman-Parker, Thierry Leblanc, Odile Lejars, Philippe Le Moine, Guy Leverger, Aude Marie-Cardine, Geneviève Margueritte, Françoise Mechinaud, Gérard Michel, Claire Oudin, Caroline Oudot, Catherine Paillard, Brigitte Pautard, Yves Perel, Arnaud Petit, Christophe Piguet, Anne-France Ray-Lunven, Fanny Rialland, Claudine Schmitt, Pascale Schneider, Jean-Louis Stephan, Marie-Dominique Tabone, Caroline Thomas, and Jean-Pierre Vannier.
References
Author notes
A.P. and A. Trinquand contributed equally to this study.