TO THE EDITOR:
Children diagnosed with Down syndrome (DS) have a 150-fold increased risk of developing a unique acute myeloid leukemia (ML-DS) within the first 4 years of life.1,2 ML-DS is preceded by a fetal/neonatal myeloproliferative disorder, transient abnormal myelopoiesis (TAM). ML-DS and TAM require mutations in the X-chromosome encoded erythro-megakaryocyte transcription factor GATA1.3-7 In normal human GATA1-expressing cells, 2 GATA1 isoforms are detected; a full-length 414-amino-acid protein (GATA1fl) and an N-terminal truncated 331-amino-acid protein known as GATA1s. These arise by either alternative splicing or use of alternate translational start sites from the full-length transcript. In TAM and ML-DS. GATA1 mutations abrogate GATA1fl production, leading to exclusive production of GATA1s. The morphology and immunophenotypic profile of TAM and ML-DS blasts8-11 are not absolutely specific, but can be shared with normal immature blast cells in neonates with DS.7 Consequently, the current standard assay for a specific diagnosis of TAM or ML-DS requires detection of GATA1s mutations by DNA sequencing7 or immunofluorescence to detect exclusive production of GATA1s in blast cells.12 GATA1s mutations marking TAM and ML-DS blasts can also serve to monitor measurable residual disease (MRD) following treatment6,13,14 by targeted next-generation sequencing (NGS) with a sensitivity of 0.3%.7 NGS methods are robust but require technical expertise, expensive equipment,15 and are not usually a same-day test. Therefore, a simple, cost-effective method to identify and track GATA1s mutations is needed. Here, we describe a highly sensitive intracellular flow cytometry (iFC)-based method to identify GATA1s cells within the CD45low CD117+ gate in TAM and ML-DS. The novel iFC method can diagnose TAM or ML-DS.
Parents gave written informed consent in accordance with the Declaration of Helsinki, and the study was approved by the Thames Valley Research Ethics Committee (06MRE12-10; NIHR portfolio no. 6362). Neonates with TAM were (1) diagnosed within 3 months of birth (majority younger than 14 days of age), (2) positive for a GATA1 variant by deep sequencing/conventional Sanger sequencing, and (3) evidence of subsequent clinical and/or GATA1 mutation resolution. ML-DS samples were allocated if ≥3 months from birth and positive for GATA1 variant by the criteria given previously. Subject characteristics are in supplemental Table 1 on the Blood Web site.
DNA was isolated using the DNeasy Blood and Tissue Kit (Qiagen). Fifty nanograms of genomic DNA was used for polymerase chain reaction (PCR) amplification using FastStart High Fidelity PCR System (Roche, Burgess Hill, UK). Five GATA1 primer pairs with CS1 and CS2 tags (supplemental Table 2) were used. PCR products were pooled and diluted 1:50 before barcoding with the Access Array Barcode library (Fluidigm, Cambridge, UK; PN 100-4876) as recommended. Barcoded products were pooled proportionately and purified using Beckman Coulter Agencourt AMPure XP beads (Beckman Coulter, High Wycombe, UK). Products were diluted to 4 nM and sequenced on an Illumina MiSeq using 300 bp-phycoerythrin sequencing. Mapping was done by the Burrows-Wheeler16 algorithm and variants were called using Varscan.17 Called variants were inspected in the Integrative Genomics Viewer. Variant allele frequency (VAF%) was determined using the UNIX command “grep” on fastq files as previously published.7
Cryopreserved mononuclear cells were washed and stained for flow cytometry with antibodies against human CD117, CD45, CD34, CD36, CD235A, CD41, and CD7 (supplemental Table 3 for antibody details). To discriminate dead cells, LIVE/DEAD Fixable (Thermo Fisher Scientific, Hemel Hempstead, UK) was used. Cells were fixed and permeabilized using the Transcription-Factor-Buffer-Set by BD Pharmingen (Becton Dickinson, Oxford, UK). To detect GATA1fl and GATA1s, cells were immunostained with rabbit anti-human GATA1 D52H6 phycoerythrin-conjugated and D24E4, (Cell Signaling, London, UK) conjugated to AF647 (Zenon AlexaFluor-647 Rabbit IgG Labeling Kit, Thermo Fisher Scientific). Samples were run on a BD FACSymphony flow cytometer (Becton Dickinson) and analyzed in FlowJo-10.6.0 (Becton Dickinson). A population was defined as containing ≥10 events, unless otherwise stated. Statistical analysis was performed with Prism v.10 (GraphPad, San Diego, CA).
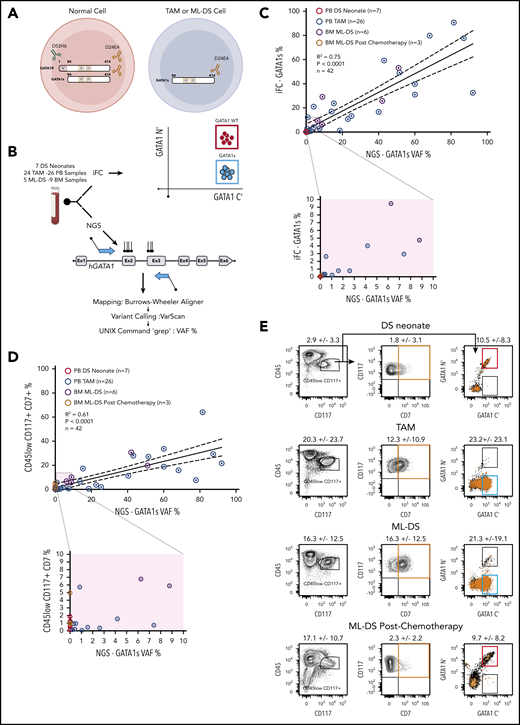
The iFC method uses 2 anti-GATA1 antibodies: 1 that detects the N-terminus present only in GATA1fl and 1 that detects the C terminus present in GATA1s and GATAfl. Immunostaining with both antibodies identifies normal cells expressing both GATA1 protein isoforms (GATA1 wild-type [GATA1WT] cells) and TAM or ML-DS cells that express only GATA1s (GATA1s+ cells) (Figure 1A). The full gating strategy and flow cytometric controls for iFC detection of GATA1 protein isoforms is in supplemental Figure 1. We compared detection of the GATA1s mutation by NGS to the iFC method using either peripheral blood (PB) or bone marrow (BM) samples from 7 DS neonates (7 samples), 24 DS patients with TAM (26 samples), and 5 DS patients with ML-DS (9 samples) (Figure 1B). To determine if the iFC method can be used as an alternative to NGS to identify TAM and ML-DS cells, we compared the GATA1s VAF% with the percent of GATA1s+ cells with either GATA1s mutation (26 PB-TAM, 6 BM-ML-DS) or 10 control samples with no detectable GATA1s mutations by NGS (7 PB-DS samples, 3 postchemotherapy BM-ML-DS samples) (Figure 1C). Linear regression analysis revealed a significant concordance (R2 = 0.75; P < .0001) between iFC and NGS methods.
Intracellular flow cytometric detection of GATA1s cells in TAM and ML-DS is comparable to NGS. (A) Antibody directed against GATA1 N terminus (green) detects only GATA1fl. Antibody directed against GATA1 C terminus (brown) detects both GATA1fl and GATA1s. Left, a normal cell (red circle) expresses both GATA1fl and GATA1s. Right, TAM or ML-DS cells (blue circle) only express GATA1s. (B) PB and BM samples were subject to both iFC (top) and NGS sequencing of GATA1 exon 2 and 3 (bottom). Top right, schematic of iFC plot of wild-type cells (red gate) and GATA1s-only expressing cells (blue gate) immunostained with anti GATA1 N′ terminus (y-axis) and anti-GATA1 C′ terminus (x-axis). (C) Linear regression analysis showing correlation between GATA1s VAF (x-axis) by NGS and percentage of GATA1s+ cells detected by iFC, as a percentage of live cells (y-axis). Samples tested: PB DS neonates screened by NGS and found to have no GATA1 mutations (red open circles), PB TAM (blue open circles), ML-DS BM (purple open circles), and ML-DS postchemotherapy treatment BM (brown open circles). Dotted lines show 95% confidence intervals. R2 = 0.75; P < .0001; n = 42. For samples in which more than 1 mutation was identified, the biggest VAF value was used. Bottom, samples with 1% to 10% VAF and 1% to 10% GATA1s+ cells are magnified to illustrate data more clearly. (D) Linear regression analysis showing correlation between GATA1s VAF (x-axis) by NGS and percentage of cells in the live, CD45lowCD117+CD7+ gate (y-axis) by flow cytometry. Dotted lines show 95% confidence intervals. R2 = 0.61; P < .0001; n = 42. The rest of the panel is as in panel C. (E) Representative flow cytometry plots of PB DS neonate, PB TAM, and BM ML-DS before and after chemotherapy. Cells within the CD45low CD117+ (left) were plotted in the CD117 vs CD7 (middle) or in the GATA1 N terminus (GATA1 N′) vs GATA1 C terminus (GATA1 C′) (left). CD117+CD7+ cells (orange box) were overlaid (as orange dots) on the GATA1 N′ and GATA1 C′ axis. Figures indicate the mean percentage ± 1 SD of cells within the gates. Number of samples tested: 7 PB DS neonate samples, 26 PB TAM samples, and 6 and 3 BM ML-DS samples before and after chemotherapy, respectively.
Intracellular flow cytometric detection of GATA1s cells in TAM and ML-DS is comparable to NGS. (A) Antibody directed against GATA1 N terminus (green) detects only GATA1fl. Antibody directed against GATA1 C terminus (brown) detects both GATA1fl and GATA1s. Left, a normal cell (red circle) expresses both GATA1fl and GATA1s. Right, TAM or ML-DS cells (blue circle) only express GATA1s. (B) PB and BM samples were subject to both iFC (top) and NGS sequencing of GATA1 exon 2 and 3 (bottom). Top right, schematic of iFC plot of wild-type cells (red gate) and GATA1s-only expressing cells (blue gate) immunostained with anti GATA1 N′ terminus (y-axis) and anti-GATA1 C′ terminus (x-axis). (C) Linear regression analysis showing correlation between GATA1s VAF (x-axis) by NGS and percentage of GATA1s+ cells detected by iFC, as a percentage of live cells (y-axis). Samples tested: PB DS neonates screened by NGS and found to have no GATA1 mutations (red open circles), PB TAM (blue open circles), ML-DS BM (purple open circles), and ML-DS postchemotherapy treatment BM (brown open circles). Dotted lines show 95% confidence intervals. R2 = 0.75; P < .0001; n = 42. For samples in which more than 1 mutation was identified, the biggest VAF value was used. Bottom, samples with 1% to 10% VAF and 1% to 10% GATA1s+ cells are magnified to illustrate data more clearly. (D) Linear regression analysis showing correlation between GATA1s VAF (x-axis) by NGS and percentage of cells in the live, CD45lowCD117+CD7+ gate (y-axis) by flow cytometry. Dotted lines show 95% confidence intervals. R2 = 0.61; P < .0001; n = 42. The rest of the panel is as in panel C. (E) Representative flow cytometry plots of PB DS neonate, PB TAM, and BM ML-DS before and after chemotherapy. Cells within the CD45low CD117+ (left) were plotted in the CD117 vs CD7 (middle) or in the GATA1 N terminus (GATA1 N′) vs GATA1 C terminus (GATA1 C′) (left). CD117+CD7+ cells (orange box) were overlaid (as orange dots) on the GATA1 N′ and GATA1 C′ axis. Figures indicate the mean percentage ± 1 SD of cells within the gates. Number of samples tested: 7 PB DS neonate samples, 26 PB TAM samples, and 6 and 3 BM ML-DS samples before and after chemotherapy, respectively.
TAM and ML-DS cells variably express erythroid and megakaryocytic markers, as well as CD45, CD117, and CD7. Expression of these 3 markers has been used to identify TAM and ML-DS cells.8,11 Thus, we compared the GATA1s VAF% with the % of live cells with CD45lowCD117+CD7+ expression in the same set of samples. Linear regression analysis showed less concordance between GATA1s VAF% and percentage of live cells with CD45lowCD117+CD7+ expression (R2 = 0.61; P < .0001) (Figure 1D). The reason for this is that both control and GATA1s mutant samples have a population of CD45lowCD117+ cells, with CD7 expression, irrespective of GATA1s mutation status (Figure 1E). As shown in Figure 1E, there is a small population of CD45lowCD117+CD7+ cells (1.8% ± 3.1% in DS neonates without GATA1 mutation and 2.3% ± 2.2% in ML-DS postchemotherapy) that express GATA1 N terminus and GATA1 C terminus and thus are WT cells. This confounds use of surface CD7 expression to identify GATA1s mutant cells.
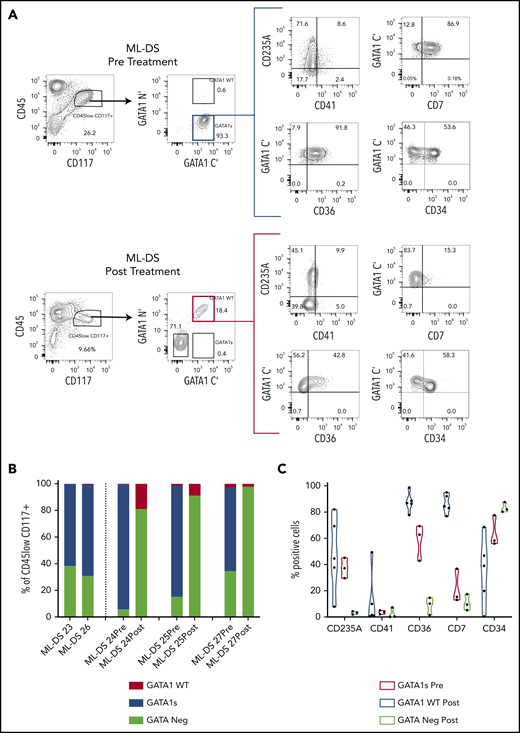
We noted that most GATA1s+ cells colocated in the CD45lowCD117+ gate (supplemental Figure 2A). Linear regression analysis revealed a significant concordance between percentage of live GATA1s+ cells measured by iFC and live cells in the CD45lowCD117+ gate (R2 = 0.82; P < .0001) (supplemental Figure 2B). We then wanted to define the immunophenotype of ML-DS cells and hemopoietic reconstitution before and after chemotherapy for ML-DS (Figure 2A-C; supplemental Figure 2C). Before treatment, GATA1s+ cells within the CD45lowCD117+ gate in ML-DS are characterized by variable CD235A, CD34, and CD41 expression, and high CD7 and CD36 expression. After treatment, cells in the CD45lowCD117+ gate are GATA1WT with reduced CD36 and CD7 expression.
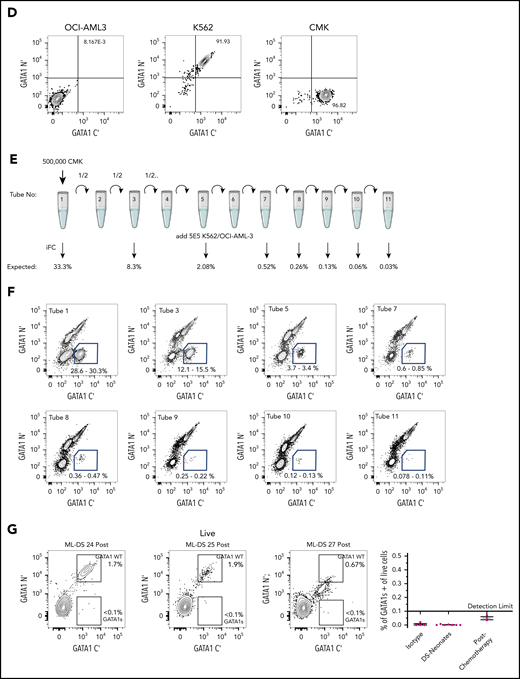
Intracellular flow detection of GATA1s+ cells is highly sensitive and can be used to measure MRD in ML-DS. (A) Representative flow cytometry panels of ML-DS samples before (top) and after chemotherapy (bottom) in which additional markers previously described as aberrant (CD235A, CD41, CD7, CD36, and CD34) were examined in GATA1s+ cells (blue gate) and GATA1WT (red gate). (B) Bar graph showing the proportion of GATA1s+ (blue), GATA1WT (red) and GATA1 negative (light green) cells within the CD45lowCD117+ gate in 5 prechemotherapy and 3 postchemotherapy ML-DS samples. (C) Violin plots showing the percent of cells positive for CD235A, CD41, CD36, CD34, and CD7 in GATA1s+ cells before chemotherapy (blue), GATA1WT after chemotherapy (red), and GATA1− cells after chemotherapy (light green). (D) Flow cytometry plots of dual immunostaining with antibodies against the N′ and C′ termini of GATA1 in: OCI-AML3 cells (left), a GATA1 negative cell line; K562 cells (middle), a cell line expressing both GATA1fl and GATA1s; and CMK (right), a GATA1s+ cell line derived from a ML-DS patient. (E) Schematic representation of a serial dilution of CMK cells. A total of 500 000 CMK cells were serially diluted 11 times into OCI-AML3/K562 cells mix and iFC performed on tubes 1, 3, 5, 7, 8, 9, 10, and 11. Expected frequency of CMK cells is shown below each tube that was tested by iFC. (F) Representative flow cytometry panels of serially diluted GATA1s+ CMK cells (blue gate) in a cell mixture of CMK/OCI-AML3/K562. A total of 500 000 live events were recorded per tube but only a fraction of events is shown for clarity. Each gate contained more than 300 events. Percent of cells in the blue gate from 2 replicates is shown. (G) Top, 3 flow cytometry plots showing the percentage of GATA1WT and GATA1s+ cells in the live CD45lowCD117+ cell compartment in 3 postchemotherapy ML-DS samples in which no GATA1s mutations were identify by NGS. Below, graph of the percentage of GATA1s+ cells live CD45lowCD117+ gate in the isotype controls (4 samples), DS neonates (7 samples), and 3 ML-DS postchemotherapy samples.
Intracellular flow detection of GATA1s+ cells is highly sensitive and can be used to measure MRD in ML-DS. (A) Representative flow cytometry panels of ML-DS samples before (top) and after chemotherapy (bottom) in which additional markers previously described as aberrant (CD235A, CD41, CD7, CD36, and CD34) were examined in GATA1s+ cells (blue gate) and GATA1WT (red gate). (B) Bar graph showing the proportion of GATA1s+ (blue), GATA1WT (red) and GATA1 negative (light green) cells within the CD45lowCD117+ gate in 5 prechemotherapy and 3 postchemotherapy ML-DS samples. (C) Violin plots showing the percent of cells positive for CD235A, CD41, CD36, CD34, and CD7 in GATA1s+ cells before chemotherapy (blue), GATA1WT after chemotherapy (red), and GATA1− cells after chemotherapy (light green). (D) Flow cytometry plots of dual immunostaining with antibodies against the N′ and C′ termini of GATA1 in: OCI-AML3 cells (left), a GATA1 negative cell line; K562 cells (middle), a cell line expressing both GATA1fl and GATA1s; and CMK (right), a GATA1s+ cell line derived from a ML-DS patient. (E) Schematic representation of a serial dilution of CMK cells. A total of 500 000 CMK cells were serially diluted 11 times into OCI-AML3/K562 cells mix and iFC performed on tubes 1, 3, 5, 7, 8, 9, 10, and 11. Expected frequency of CMK cells is shown below each tube that was tested by iFC. (F) Representative flow cytometry panels of serially diluted GATA1s+ CMK cells (blue gate) in a cell mixture of CMK/OCI-AML3/K562. A total of 500 000 live events were recorded per tube but only a fraction of events is shown for clarity. Each gate contained more than 300 events. Percent of cells in the blue gate from 2 replicates is shown. (G) Top, 3 flow cytometry plots showing the percentage of GATA1WT and GATA1s+ cells in the live CD45lowCD117+ cell compartment in 3 postchemotherapy ML-DS samples in which no GATA1s mutations were identify by NGS. Below, graph of the percentage of GATA1s+ cells live CD45lowCD117+ gate in the isotype controls (4 samples), DS neonates (7 samples), and 3 ML-DS postchemotherapy samples.
Finally, receiver operating characteristic analysis determined the sensitivity (100%), specificity (92.31%), and a cutoff value (0.18%) for of the iFC method and showed that it is superior to conventional surface flow cytometry in identifying malignant cells (supplemental Figure 3; supplemental Tables 4 and 5). To confirm this cutoff value, we serially diluted CMK cells, a cell line derived from an ML-DS patient, into a mixture of GATA1-nonexpressing (OCI-AML3) and GATA1-expressing (K562) cells (Figure 2E-F) and measured the percentage of GATA1s+ cells. GATA1s+ cells were detected when present at a frequency greater than ∼0.1%. We then tested 3 postchemotherapy ML-DS samples and did not detect a GATA1s+ population at a sensitivity of 0.1% (Figure 2G). In all 3 samples, GATA1s mutation was not detected by NGS and the patients remain in remission 4 years after treatment.
In summary, iFC is a sensitive, simple, and rapid method to identify and track malignant cells in TAM and ML-DS samples and has a sensitivity of 0.18%. Its value is in the rapid diagnosis of TAM and ML-DS in most hospital laboratories that perform flow cytometry, without need for more expensive and time-consuming NGS technology. Its use as an MRD tool will need to be validated in larger studies with appropriate follow-up. Caution has to be exercised, and further studies are needed, before adopting this method to make a diagnosis of silent TAM.
E-mail the corresponding author for original material, data sets, and protocols.
The online version of this article contains a data supplement.
Acknowledgments
This work was supported by Lady Tata Memorial Trust (D.C.H.). P.V. and I.R. are supported by Bloodwise Specialist Programme grant 13001 and by the NIHR Oxford Biomedical Centre Research Fund. P.V. is supported by program grants from the MRC Molecular Haematology Unit (MC UU 12009/11).
Authorship
Contribution: D.C.H. designed, performed experiments, and analyzed data and wrote the manuscript; M.M. performed and analyzed experiments and data; N.E., A.P.d.G., and B.U. performed experiments; and P.V. and I.R. analyzed the data and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paresh Vyas, John Radcliffe Hospital, University of Oxford, Oxford OX3 9DU, United Kingdom: e-mail: paresh.vyas@imm.ox.ac.uk.
REFERENCES
Author notes
D.C.H. and M.M. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal