Abstract
Introduction Copy number abnormalities (CNA) and structural variants (SV) are crucial to driving cancer progression and in multiple myeloma (MM). Chr1 CNA are seen in up to 40% of cases and associate with poor prognosis. Variants include deletions, gains, translocations and complex SV events such as chromothripsis (CT), chromoplexy (CP) and templated insertions (TI) which result in aberrant transcriptional patterns. Abnormal expression of genes on chr1 lead to the adverse clinical outcome and studies focussed on 1p12, 1p32.3 and 1q12-21 identified potential causal genes including TENT5C, CDKN2C, CKS1B, PDZK1, BCL9, ANP32E, ILF2, ADAR, MDM2 and MCL1 but none fully explain the clinical behavior. To address this deficiency and to relate chromatin structure to gene deregulation we present a multiomic bioinformatic analysis of SV, CNA, mutation and expression changes in relation to the chromatin structure of chr1.
Methods We analysed data derived from 1,154 CoMMpass trial patients. We analyzed 972 NDMM patients with whole exome for mutations, and 752 whole genomes for copy number, translocations, complex rearrangements such as CP, CT and TI as previously described. Using GISTIC 2.0, we identified hotspots of CNA. This information was then analyzed in conjunction to the RNA-seq data derived from 643 patients to determine the aberrant transcriptional landscape of chr1. Using HiC data derived from U266 MM cell line, we associated these changes with TAD structures, A/B compartments, and histone marks along chr1, to gene expression changes, and recurrent SV. Using the cell line dependency map for CRISPR knockdown of the gene set on chr1 derived from 20 MM cell lines we related cell viability to chr1 copy number status.
Results
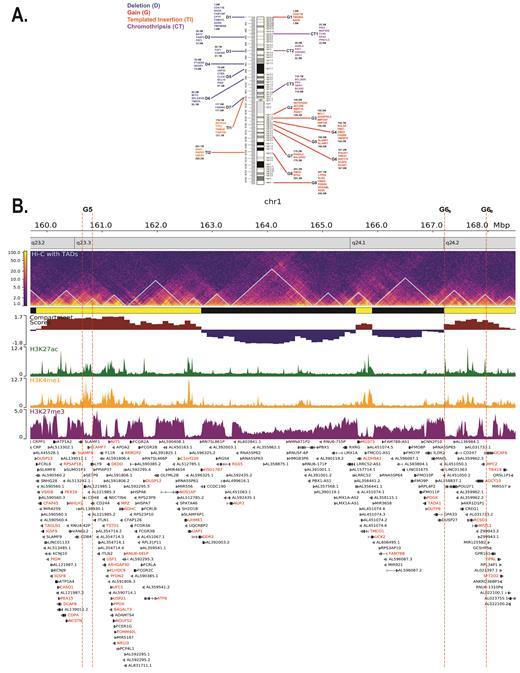
We identified 7 hotspots of deletion, 9 of gain, 3 of CT and 2 of templated-insertion across chr1. We mapped these regions to epigenetic plots and show that gained regions are hypomethylated compared to the rest of chr1 (Wilcoxon, p=0.0002). Overall 69% of gain(1q) and 45% of the non-gained hotspots were in A compartments (χ 2=11, p=0.0009) and had an overall higher compartment score (p=0.01).
The recurrent regions of loss on 1p confirm the clinical relevance of this region. The critical importance of TENT5C, CDKN2C and RPL5 is identified by the impact of deletion, mutation and the rearrangement of superenhancers. Further this convergence of multiple oncogeneic mechanisms to a single locus points to a number of novel candidate drivers including FUB1 and NTRK1.
We provide important new information on 1q21.1-1q25.2 encompassing 145-180Mb a transcriptionally dense region containing 6 GISTIC 2.0 hotspots of gain (G2-G7). The hotspots occur within TAD structures that correlate upregulation of known drivers listed above and also identified novel potential upregulated drivers including POU2F1, a transcription factor, CREG1, an adenovirus E1A protein that both activates and represses gene expression promoting proliferation and inhibiting differentiation (G6) and BTG2 a G1/S transition regulator (G8). These data for copy number gain provides strong evidence for the prognostic relevance of of multiple drivers within deregulated TADs rather than single candidate genes. It also highlights the importance of the chromatin structure of Chr1 in the generation of these events.
Using dependency map CRISPR data we identified 320 essential genes for at least one cell line (>1). A common set of 31 genes were identified including 3 proteasome subunits (PSMA5, PSMB2, PSMB4), three regulators of ubiquitin-protein transferase activity (RPL5, RPL11, CDC20), splicing (SF3B4, SF3A3, SFPQ, RNPC3, SRNPE, PRPF38A, PRPF38B) and DTL. A common dependency for 1q+ or 1p- was not identified but a number of dependencies were identified in more than one cell line including UQCRH, SLCA1, CLSPN in 1p- cell lines and IPO9, PPIAL4G, and MRPS2 in 1q+.
Conclusion We present an elegant anatomic map of chr1 at the genetic and epigenetic levels providing an unprecedented level of resolution for the relationships of structural variants to epigenetic, expression and mutation status. The analysis highlights the importance of active chromatin in gene deregulation by SV and CNA where the importance of multiple gene deregulation within TAD structures is critical to MM pathogenesis. The implications are that we could improve prognostic assignment and identify new targets for therapy by further characterizing these relationships.
Braunstein: Jansen: Membership on an entity's Board of Directors or advisory committees, Research Funding; Celgene: Membership on an entity's Board of Directors or advisory committees, Research Funding; Adaptive: Membership on an entity's Board of Directors or advisory committees; AstraZeneca: Membership on an entity's Board of Directors or advisory committees; Epizyme: Membership on an entity's Board of Directors or advisory committees; Karyopharm: Membership on an entity's Board of Directors or advisory committees; Pfizer: Membership on an entity's Board of Directors or advisory committees; Takeda: Membership on an entity's Board of Directors or advisory committees. Davies: Takeda: Membership on an entity's Board of Directors or advisory committees; Sanofi: Membership on an entity's Board of Directors or advisory committees; Oncopeptides: Membership on an entity's Board of Directors or advisory committees; Constellation: Membership on an entity's Board of Directors or advisory committees; Janssen: Membership on an entity's Board of Directors or advisory committees; Celgene/BMS: Consultancy, Membership on an entity's Board of Directors or advisory committees.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal