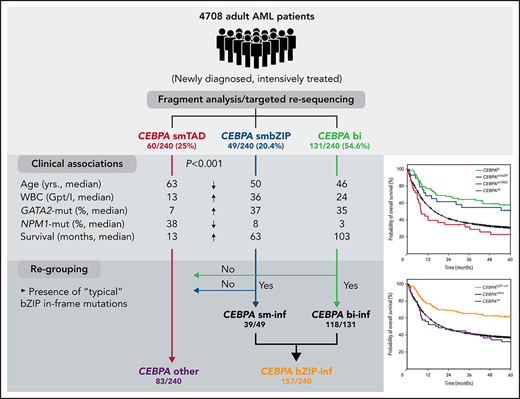
Key Points
CEBPAsmbZIP- and CEBPAbi-mutant AML share clinical and mutational characteristics and are distinct from CEBPAsmTAD-mutant AML.
Only in-frame mutations in CEBPA-bZIP are associated with favorable clinical response in monoallelic and biallelic constellations.
Abstract
Biallelic mutations of the CEBPA gene (CEBPAbi) define a distinct entity associated with favorable prognosis; however, the role of monoallelic mutations (CEBPAsm) is poorly understood. We retrospectively analyzed 4708 adults with acute myeloid leukemia (AML) who had been recruited into the Study Alliance Leukemia trials, to investigate the prognostic impact of CEBPAsm. CEBPA mutations were identified in 240 patients (5.1%): 131 CEBPAbi and 109 CEBPAsm (60 affecting the N-terminal transactivation domains [CEBPAsmTAD] and 49 the C-terminal DNA-binding or basic leucine zipper region [CEBPAsmbZIP]). Interestingly, patients carrying CEBPAbi or CEBPAsmbZIP shared several clinical factors: they were significantly younger (median, 46 and 50 years, respectively) and had higher white blood cell (WBC) counts at diagnosis (median, 23.7 × 109/L and 35.7 × 109/L) than patients with CEBPAsmTAD (median age, 63 years, median WBC 13.1 × 109/L; P < .001). Co-mutations were similar in both groups: GATA2 mutations (35.1% CEBPAbi; 36.7% CEBPAsmbZIP vs 6.7% CEBPAsmTAD; P < .001) or NPM1 mutations (3.1% CEBPAbi; 8.2% CEBPAsmbZIP vs 38.3% CEBPAsmTAD; P < .001). CEBPAbi and CEBPAsmbZIP, but not CEBPAsmTAD were associated with significantly improved overall (OS; median 103 and 63 vs 13 months) and event-free survival (EFS; median, 20.7 and 17.1 months vs 5.7 months), in univariate and multivariable analyses. Additional analyses revealed that the clinical and molecular features as well as the favorable survival were confined to patients with in-frame mutations in bZIP (CEBPAbZIP-inf). When patients were classified according to CEBPAbZIP-inf and CEBPAother (including CEBPAsmTAD and non-CEBPAbZIP-inf), only patients bearing CEBPAbZIP-inf showed superior complete remission rates and the longest median OS and EFS, arguing for a previously undefined prognostic role of this type of mutation.
Introduction
Patients with normal karyotype represent the largest subgroup of patients with acute myeloid leukemia (AML).1 Research performed during the past 20 years has revealed several novel abnormalities that are particularly common in this subgroup of patients (eg, activating mutations of the FLT3 receptor tyrosine kinase2-5 and mutations of the NPM1 gene encoding the nucleophosmin protein.6-8) In addition, mutations of the gene encoding CCAAT/enhancer-binding protein-α (CEBPA) have been described in patients with AML and normal karyotype.9-12 The CEBPA gene encodes a transcription factor that serves as a master regulator of granulopoiesis.13 Targeted disruption of the gene in mice is associated with a block of granulocyte development and downregulation of target genes such as granulocyte colony-stimulating factor receptor.13 The intronless CEBPA gene on chromosome 19q13.1 encodes 2 major protein isoforms, the 42-kDa full-length protein (p42), which has the full transcriptional activity, and a shorter, 30-kDa isoform (p30), produced from a second, alternative start codon and shown to have complex functions, including an inhibitory effect on9 and increased degradation14 of the p42 full-length protein.
Since the initial report,9 several groups have investigated CEBPA mutations in AML.11,12,15-18 Besides the reproducible association with certain morphological and clinical features (eg, FAB M1 and M2 morphology, high CD34 expression on blasts, and predominance in normal karyotype), nearly all studies have shown that patients with CEBPA mutations have a more favorable outcome. However, more recent results have indicated that a good prognosis is confined to patients with biallelic or double mutations (CEBPAbi),19-21 whereas patients with monoallelic CEBPA mutations (CEBPAsm) did not differ in their response to treatment from patients with wild-type CEBPA (CEBPAwt) and had a less favorable outcome.19-21 This finding led to the inclusion of biallelic CEBPA mutations as an independent entity in the most recent World Health Organization classification,22 as well as a favorable prognostic group in the ELN2017 recommendations.23 However, the impact of monoallelic CEBPA mutations has been investigated in more detail in only a few studies, especially in light of the different biological effects of N- and C-terminal mutations.21,24,25
To investigate the prevalence and prognostic role of CEBPA mutations, in particular CEBPAsm mutations, in adult patients with AML, we studied 4708 patients with newly diagnosed AML. In contrast to many previous studies, we included patients of all age groups, as well as secondary AML (sAML) after prior myelodysplasia (MDS) or therapy-related AML (tAML). Because most CEBPA mutations reported so far consist of insertions or deletions,26-29 we used high-resolution fragment analysis for mutation screening and found CEBPA mutations in 5.1% of patients. The results of targeted next-generation sequencing (NGS) indicated profound differences in the co-mutational spectrum of individual CEBPA mutations. Our results point to a differential effect of CEBPAsmbZIP mutations that appear to be associated with similar clinical parameters, co-mutations, and outcome compared with CEBPAbi mutations. If confirmed, these results, generated in one of the largest cohorts of patients with AML analyzed so far, could build the basis of a refined clinical classification of CEBPA mutations.
Patients, materials, and methods
Patients
We screened 4708 adult patients with newly diagnosed AML (n = 3729 with de novo AML; n = 644 with AML and a history of MDS; and n = 335 with tAML) for the presence of mutations in the CEBPA gene. Most individuals (n = 3104; 67.45%) were treated in prospective studies, including the AML96 (n = 1457), AML2003 (n = 1081), AML60+ (n = 359), and SORAML (n = 207) protocols of the Study Alliance Leukemia (SAL). The remaining patients (n = 1604) were recruited to the SAL registry and biorepository. Detailed treatment protocols have been published30-33 and are summarized in the supplemental Data (available on the Blood Web site), including the number of patients treated in each protocol. All studies involved risk-stratified consolidation therapy according to cytogenetic risk groups (CEBPA mutations were not used for risk stratification in any of those studies). Patients <60 years of age received standard cytosine arabinoside (Ara-C)/anthracycline (DA 3 + 7)–based, double-induction chemotherapy followed by consolidation with high-dose Ara-C in patients who had favorable risk. Patients in the intermediate- and high-risk groups had the option of upfront allogeneic transplantation and underwent autologous transplantation or chemotherapy-based consolidation in the absence of suitable donors. In older patients treated with curative intention, the chemotherapy regimen was adjusted according to predefined algorithms that integrate performance status and organ function.
This study was approved by the ethics board of the Technical University Dresden. Each patient gave written informed consent to participate in the respective study protocols.
Patient samples
All materials investigated were obtained at the time of diagnosis. Bone marrow was used whenever available; in all other cases, peripheral blood samples were examined. Genomic DNA was extracted from mononuclear cells by using standard procedures (DNA blood minikit; Qiagen, Hilden, Germany).
PCR for CEBPA
All polymerase chain reaction (PCR) analyses were performed on genomic DNA. Details of the PCR primers and cycling conditions are given in supplemental Tables 1 and 2 and supplemental Figure 1.
PCR-amplified mutant samples were purified and sequenced on an ABI3130xl instrument. Sequences were compared with the WT-CEBPA messenger RNA (mRNA) sequence (U34070).
NGS-based characterization of co-mutations in patients with CEBPA mutations
Profiling of mutations was achieved by targeted NGS-based resequencing with the TruSight Myeloid assay (Illumina, Chesterford, United Kingdom) covering 54 genes frequently mutated in AML, as described recently34 (details in the supplemental Data). Data alignment of demultiplexed FastQ files, variant calling, and filtering were performed with the Sequence Pilot software package (JSI Medical Systems GmbH, Ettenheim, Germany), with default settings and a 5% variant allele frequency cutoff. FLT3-internal tandem duplication (ITD) and NPM1 mutations were evaluated as reported previously.5,8
RNA sequencing
RNA sequencing (RNA-Seq) was performed on total RNA isolated at diagnosis from 20 patients with a CEBPA mutation (5 CEBPAbi-inf, 5 CEBPAsm-inf, 5 CEBPAsm-other, and 5 CEBPAbi-other), by using strand-specific RNA-Seq library preparation (Ultra II Directional RNA Library Prep; New England Biolabs) and sequenced on an Illumina NovaSeq 6000 instrument. The complete workflow and the bioinformatic analyses are detailed in the supplemental Data.
Statistical analysis
Clinical variables across groups were compared by using the χ2 test or 2-sided Fisher’s exact test for categorial variables. The nonparametric Mann-Whitney U test was applied for continuous variables. P < .05 indicated a significant difference. Numerical variables are expressed as the median with interquartile range (IQR). Univariate analyses for the influence of the CEBPA mutational status on complete response (CR) rates were performed by using the χ2 test. The log-rank test was used to evaluate OS and EFS. For multivariable analysis of prognostic factors, Cox proportional hazards regression models were used. Allogeneic hematopoietic stem cell transplantation (allo-HSCT) was modeled as a time-dependent covariate. P-values of association analyses of CEBPA mutations with clinical variables and other molecular abnormalities were adjusted for multiplicity by using the Bonferroni-Holm correction. All statistical analyses were performed with the SPSS software package, version 26 (SPSS, Chicago, IL) and R version 3.5.3 (https://www.R-project.org/).
Results
A total of 371 individual CEBPA mutations were identified in 240 of the 4708 patients (5.1%) by the screening procedure. In all patients with a mutation identified in any part of CEBPA, Sanger sequencing of the entire gene was performed to assess the presence of additional mutations, including single nucleotide variants, and to confirm the alterations identified using fragment analysis. Patients showing the previously described 6-bp polymorphism in TAD226 were included in the WT-CEBPA group.
As illustrated in supplemental Figure 2 and supplemental Table 3 and reported in previous studies, mutations largely clustered in the N-terminal first transactivation domain (TAD1) and the DNA-binding and basic leucine zipper region (bZIP) in the C-terminal part of CEBPA. In our cohort, 131 of 240 patients (54.6%) presented with 2 CEBPA mutations, mostly consisting of combined mutations in bZIP and TAD, denoted as CEBPAbi. CEBPAsm mutations were found in 109 of 240 patients (45.4%), of which 60 had N-terminal (CEBPAsmTAD) and 49 C-terminal (CEBPAsmbZIP) mutations. As reported before, most of the mutations in the TAD domains caused a frameshift, whereas mutations in the bZIP-region were predominantly in-frame insertions and duplications. Eight patients had monoallelic mutations with high variant allele frequency (4 CEBPAsmTAD and 4 CEBPAsmbZIP), indicating a homozygous state.
CEBPA mutations and clinical characteristics
The association of clinical parameters according to the localization of the CEBPA mutation (ie, patients with single N- or C-terminal mutations and patients with biallelic mutations) is summarized in Table 1. Compared with patients bearing CEBPAwt, those with CEBPAbi were significantly younger (median age, 46 years; IQR, 38-59) at diagnosis, similar to those with CEBPAsmbZIP (median age, 50 years; IQR, 39-57), whereas patients with CEBPAsmTAD were significantly older (median age, 63 years; IQR, 55-69.3) and were more comparable to the CEBPAwt group (median, 57 years; IQR, 46-67; P < .001). When patients were categorized in 10-year age intervals (Figure 1), a continuous decrease in CEBPAbi and CEBPAsmbZIP mutations was seen with increasing age, whereas CEBPAsmTAD alterations increased. In line with this finding, only a single patient with a CEBPAsmTAD mutation was observed in the group of patients <30 years of age.
Clinical and cytogenetic characteristics CEBPAwt compared with CEBPAbi, CEBPAsmbZIP and CEBPAsmTAD mutant patients
| . | CEBPAwt n = 4468 . | CEBPAsmTAD n = 60 . | CEBPAsmbZIP n = 49 . | CEBPAbi n = 131 . | P (adj.) . |
|---|---|---|---|---|---|
| Age, y, median (IQR) | 57 (46-67) | 63 (55-69) | 50 (39-57) | 46 (38-59) | <.001 |
| Sex, n (%) | |||||
| Female | 2170 (49) | 32 (53) | 16 (33) | 70 (53) | .26 |
| Male | 2298 (51) | 28 (47) | 33 (67) | 61 (47) | |
| AML type, n (%) | |||||
| de novo | 3510 (78) | 50 (84) | 47 (96) | 122 (93) | .001 |
| sAML | 630 (14) | 8 (13) | 2(4) | 4 (3) | |
| tAML | 328 (7) | 2 (3) | — | 5 (4) | |
| Laboratory, median (IQR) | |||||
| BM blasts, % | 60 (38-80) | 60 (43.3-72.8) | 65.3 (43.9-80) | 64 (50-87) | .823 |
| CD34-positivity, % | 16 (2-47) | 19.2 (3.8-65) | 45 (19.5-68) | 55.6 (32.9-77) | <.001 |
| WBC, ×109/L | 11.7 (3-45.7) | 13.2 (3.3-51.2) | 35.7 (11.5-93) | 23.7 (9.3-64.7) | <.001 |
| LDH, U/L | 412.9 (272-727) | 412 (291-569) | 491 (344-941) | 445 (292-679) | .99 |
| FAB, n (%) | |||||
| M0 | 223 (5) | — | 1 (2) | — | <.001 |
| M1/M2 | 2040 (46) | 40 (67) | 39 (80) | 101 (77) | |
| M4-M7 | 1535 (34) | 15 (25) | 8 (16) | 19 (15) | |
| Unknown | 670 (15) | 5 (8) | 1 (2) | 10 (8) | |
| Cytogenetics, n (%) | |||||
| Normal karyotype | 1923 (43) | 37 (62) | 34 (72) | 102 (78) | <.001 |
| Aberrant karyotype | 2359 (53) | 23 (38) | 15 (28) | 29 (22) | |
| Unknown | 186 (4) | — | — | — | |
| Favorable risk (MRC) | 353 (8) | 1 (2) | 1 (2) | — | <.001 |
| Intermediate risk (MRC) | 2928 (68) | 54 (90) | 43 (92) | 119 (98) | |
| Adverse risk (MRC) | 1001 (23) | 5 (8) | 3 (6) | 2 (2) | |
| Treatment, n (%) | |||||
| Primary allo-HCT | 747 (17) | 3 (5) | 10 (20) | 36 (27) | <.001 |
| Salvage allo-HCT | 915 (20) | 13 (22) | 8 (16) | 22 (17) | .189 |
| . | CEBPAwt n = 4468 . | CEBPAsmTAD n = 60 . | CEBPAsmbZIP n = 49 . | CEBPAbi n = 131 . | P (adj.) . |
|---|---|---|---|---|---|
| Age, y, median (IQR) | 57 (46-67) | 63 (55-69) | 50 (39-57) | 46 (38-59) | <.001 |
| Sex, n (%) | |||||
| Female | 2170 (49) | 32 (53) | 16 (33) | 70 (53) | .26 |
| Male | 2298 (51) | 28 (47) | 33 (67) | 61 (47) | |
| AML type, n (%) | |||||
| de novo | 3510 (78) | 50 (84) | 47 (96) | 122 (93) | .001 |
| sAML | 630 (14) | 8 (13) | 2(4) | 4 (3) | |
| tAML | 328 (7) | 2 (3) | — | 5 (4) | |
| Laboratory, median (IQR) | |||||
| BM blasts, % | 60 (38-80) | 60 (43.3-72.8) | 65.3 (43.9-80) | 64 (50-87) | .823 |
| CD34-positivity, % | 16 (2-47) | 19.2 (3.8-65) | 45 (19.5-68) | 55.6 (32.9-77) | <.001 |
| WBC, ×109/L | 11.7 (3-45.7) | 13.2 (3.3-51.2) | 35.7 (11.5-93) | 23.7 (9.3-64.7) | <.001 |
| LDH, U/L | 412.9 (272-727) | 412 (291-569) | 491 (344-941) | 445 (292-679) | .99 |
| FAB, n (%) | |||||
| M0 | 223 (5) | — | 1 (2) | — | <.001 |
| M1/M2 | 2040 (46) | 40 (67) | 39 (80) | 101 (77) | |
| M4-M7 | 1535 (34) | 15 (25) | 8 (16) | 19 (15) | |
| Unknown | 670 (15) | 5 (8) | 1 (2) | 10 (8) | |
| Cytogenetics, n (%) | |||||
| Normal karyotype | 1923 (43) | 37 (62) | 34 (72) | 102 (78) | <.001 |
| Aberrant karyotype | 2359 (53) | 23 (38) | 15 (28) | 29 (22) | |
| Unknown | 186 (4) | — | — | — | |
| Favorable risk (MRC) | 353 (8) | 1 (2) | 1 (2) | — | <.001 |
| Intermediate risk (MRC) | 2928 (68) | 54 (90) | 43 (92) | 119 (98) | |
| Adverse risk (MRC) | 1001 (23) | 5 (8) | 3 (6) | 2 (2) | |
| Treatment, n (%) | |||||
| Primary allo-HCT | 747 (17) | 3 (5) | 10 (20) | 36 (27) | <.001 |
| Salvage allo-HCT | 915 (20) | 13 (22) | 8 (16) | 22 (17) | .189 |
Bold P-values indicate statistically significant results.
BM, bone marrow; FAB, French-American-British; LDH, lactate dehydrogenase; MRC, medical research council.
Age distribution of the 240 CEBPA mutations (CEBPAbi, CEBPAsmTAD, CEBPAsmbZIP) identified in this study.
Age distribution of the 240 CEBPA mutations (CEBPAbi, CEBPAsmTAD, CEBPAsmbZIP) identified in this study.
As outlined in Table 1, patients with CEBPAsmTAD also had significantly lower WBC counts and significantly lower rates of CD34 positivity, compared with the CEBPAbi and CEBPAsmbZIP groups, and more commonly had prior myelodysplasia syndrome (MDS) or tAML that evolved as a secondary disease.
Association of CEBPA mutations with other molecular abnormalities
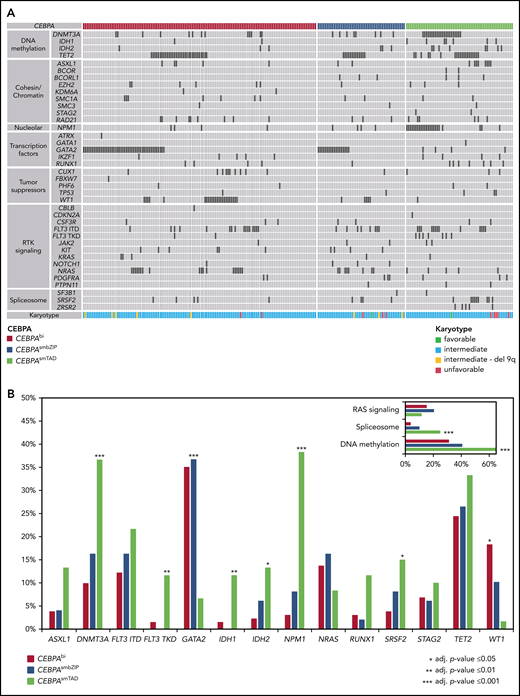
Based on the targeted NGS approach, additional mutations were identified in 208 of 240 patients with a CEBPA mutation (86.7%). The most frequently mutated genes were TET2 (70 of 240; 29.2%), GATA2 (68 of 240; 28.3%), DNMT3A (45 of 240; 18.8%), FLT3-ITD (39 of 240; 16.3%), NPM1 and NRAS (31 of 240; 12.9% each), and WT1 (30 of 240; 12.5%).
Figure 2 illustrates the distribution of co-mutations in the 3 CEBPA subgroups. Significant differences were observed for several genes, the most striking being GATA2, which was mutated in 35.1% of patents with CEBPAbi, 36.7% of those with CEBPAsmbZIP, but in only 6.7% of those with CEBPAsmTAD (P = .001), and NPM1, which was mutated in 3.1% of patients with CEBPAbi and 8.2% of those with CEBPAsmbZIP, but in 38.3% of patients with CEBPAsmTAD (P < .001). Significant differences were also found for mutations in DNTM3A, FLT3-TKD, IDH1, and IDH2, as well as in SRSF2 and WT1 (Figure 2B). In general, the spectrum of mutations of CEBPAsmbZIP was more comparable to that of patients with CEBPAbi and differed markedly from the CEBPAsmTAD group, the latter being more similar to patients with CEBPAwt, who frequently carried mutations in genes associated with AML after prior MDS, such as spliceosome mutations (ie, SRSF2, SF3B1, or U2AF1) and alterations associated with DNA methylation (ie, DNMT3A, ASXL1, TET2, IDH1, and IDH2). Interestingly, only 7 patients in this analysis had mutations in CSF3R, with no significant differences found between the groups (3.1% CEBPAbi, 2% CEBPAsmbZIP, and 3.3% CEBPAsmTAD).
Alignment of additional gene mutations for 240 patients with CEBPAmut. (A) Co-mutations organized by categories of related genes, as labeled on the left. Patients are shown in order by CEBPA subgroup. The heat map includes all mutated genes in patients with CEBPAbi, CEBPAsmbZIP, or CEBPAsmTAD. Each column represents one of the 240 analyzed samples. Mutations in the investigated genes are shown by black bars, light gray bars indicate WT status. (B) Frequency distribution of additional gene mutations identified in patients with CEBPAbi, CEBPAsmbZIP, or CEBPAsmTAD mutations (frequency of at least 10% in 1 subgroup). RAS signaling including KRAS, NRAS, PTPN11, and CBLB; spliceosome, including SF3B1, SRSF2, and ZRSR2; and methylation, including DNMT3A, IDH1, IDH2, and TET2.
Alignment of additional gene mutations for 240 patients with CEBPAmut. (A) Co-mutations organized by categories of related genes, as labeled on the left. Patients are shown in order by CEBPA subgroup. The heat map includes all mutated genes in patients with CEBPAbi, CEBPAsmbZIP, or CEBPAsmTAD. Each column represents one of the 240 analyzed samples. Mutations in the investigated genes are shown by black bars, light gray bars indicate WT status. (B) Frequency distribution of additional gene mutations identified in patients with CEBPAbi, CEBPAsmbZIP, or CEBPAsmTAD mutations (frequency of at least 10% in 1 subgroup). RAS signaling including KRAS, NRAS, PTPN11, and CBLB; spliceosome, including SF3B1, SRSF2, and ZRSR2; and methylation, including DNMT3A, IDH1, IDH2, and TET2.
CEBPA mutations and outcome
The prognostic relevance of CEBPA mutations was analyzed in 4461 intensively treated patients, with a median follow-up time of patients remaining alive of 61 months (IQR, 36-96 months). When analyzed according to the current recommendations (ie, separating only CEBPAbi and CEBPAsm), only patients with CEBPAbi had a significantly higher rate of complete remission (CR; CEBPAbi, 95% vs CEBPAsm, 75% and CEBPAwt, 73%; P < .001), as well as a higher median OS (CEBPAbi, 103.2 months; 95% confidence interval [CI], 70.7-inf vs CEBPAsm, 21.9 months; 95% CI, 12.7-54, and CEBPAwt, 19.3 months; 95% CI, 17.9-21; P < .001) and EFS (CEBPAbi, 20.7 months; 95% CI, 13.1-101.1 vs CEBPAsm, 9.4 months; 95% CI, 5.7-15.3, and CEBPAwt, 7.0 months; 95% CI, 6.5-7.6; P < .001; Figure 3), which was confirmed in multivariable analyses (Table 2).
Survival analysis according to CEBPA mutation status (CEBPAwt, CEBPAbi, and CEBPAsm) within the cohort of 4461 patients receiving intensive treatment. Kaplan-Meier plots showing OS (A) and EFS (B). Numbers below the figures denote the patients at risk. The tables provide the results of pairwise univariate analyses.
Survival analysis according to CEBPA mutation status (CEBPAwt, CEBPAbi, and CEBPAsm) within the cohort of 4461 patients receiving intensive treatment. Kaplan-Meier plots showing OS (A) and EFS (B). Numbers below the figures denote the patients at risk. The tables provide the results of pairwise univariate analyses.
Results of the multivariable analysis for CEBPAbi vs CEBPAsm
| . | OS (HR) . | P . | 95% CI . | EFS (HR) . | P . | 95% CI . | CR1 (OR) . | P . | 95% CI . |
|---|---|---|---|---|---|---|---|---|---|
| Intermediate karyotype | 1 | 1 | 1 | ||||||
| Favorable karyotype | 0.427 | <.001 | (0.35-0.52) | 0.400 | <.001 | (0.34-0.47) | 3.063 | <.001 | (2.07-4.54) |
| Adverse karyotype* | 1.858 | <.001 | (1.68-2.05) | 1.633 | <.001 | (1.49-1.79) | 0.477 | <.001 | (0.40-0.57) |
| Age | 1.032 | <.001 | (1.03-1.04) | 1.023 | <.001 | (1.02-1.03) | 0.948 | <.001 | (0.94-0.95) |
| Log10 WBC | 1.155 | <.001 | (1.08-1.24) | 1.228 | <.001 | (1.15-1.31) | 0.754 | <.001 | (0.66-0.87) |
| De novo AML | 1 | 1 | 1 | ||||||
| sAML | 1.086 | .131 | (0.98-1.21) | 1.069 | .196 | (0.97-1.18) | 0.733 | .002 | (0.60-0.90) |
| tAML | 1.386 | <.001 | (1.20-1.60) | 1.098 | .186 | (0.96-1.26) | 0.691 | .009 | (0.52-0.91) |
| No FLT3-ITDmut | 1 | 1 | 1 | ||||||
| FLT3-ITDmut | 1.206 | <.001 | (1.09-1.33) | 1.242 | <.001 | (1.14-1.36) | 1.017 | .870 | (0.83-1.25) |
| No NPM1mut | 1 | 1 | 1 | ||||||
| NPM1mut | 0.629 | <.001 | (0.57-0.69) | 0.535 | <.001 | (0.49-0.59) | 2.127 | <.001 | (1.75-2.59) |
| CEBPAwt | 1 | 1 | 1 | ||||||
| CEBPAbi | 0.510 | <.001 | (0.39-0.67) | 0.534 | <.001 | (0.42-0.68) | 6.328 | <.001 | (2.73-14.67) |
| CEBPAsm | 0.825 | .122 | (0.65-1.05) | 0.746 | .011 | (0.56-0.94) | 1.117 | .644 | (0.70-1.78) |
| No allo-HSCT in CR1 | 1 | 1 | 1 | ||||||
| Allo-HSCT in CR1 | 0.773 | <.001 | (0.69-0.87) | 0.630 | <.001 | (0.56-0.79) | — | — | — |
| . | OS (HR) . | P . | 95% CI . | EFS (HR) . | P . | 95% CI . | CR1 (OR) . | P . | 95% CI . |
|---|---|---|---|---|---|---|---|---|---|
| Intermediate karyotype | 1 | 1 | 1 | ||||||
| Favorable karyotype | 0.427 | <.001 | (0.35-0.52) | 0.400 | <.001 | (0.34-0.47) | 3.063 | <.001 | (2.07-4.54) |
| Adverse karyotype* | 1.858 | <.001 | (1.68-2.05) | 1.633 | <.001 | (1.49-1.79) | 0.477 | <.001 | (0.40-0.57) |
| Age | 1.032 | <.001 | (1.03-1.04) | 1.023 | <.001 | (1.02-1.03) | 0.948 | <.001 | (0.94-0.95) |
| Log10 WBC | 1.155 | <.001 | (1.08-1.24) | 1.228 | <.001 | (1.15-1.31) | 0.754 | <.001 | (0.66-0.87) |
| De novo AML | 1 | 1 | 1 | ||||||
| sAML | 1.086 | .131 | (0.98-1.21) | 1.069 | .196 | (0.97-1.18) | 0.733 | .002 | (0.60-0.90) |
| tAML | 1.386 | <.001 | (1.20-1.60) | 1.098 | .186 | (0.96-1.26) | 0.691 | .009 | (0.52-0.91) |
| No FLT3-ITDmut | 1 | 1 | 1 | ||||||
| FLT3-ITDmut | 1.206 | <.001 | (1.09-1.33) | 1.242 | <.001 | (1.14-1.36) | 1.017 | .870 | (0.83-1.25) |
| No NPM1mut | 1 | 1 | 1 | ||||||
| NPM1mut | 0.629 | <.001 | (0.57-0.69) | 0.535 | <.001 | (0.49-0.59) | 2.127 | <.001 | (1.75-2.59) |
| CEBPAwt | 1 | 1 | 1 | ||||||
| CEBPAbi | 0.510 | <.001 | (0.39-0.67) | 0.534 | <.001 | (0.42-0.68) | 6.328 | <.001 | (2.73-14.67) |
| CEBPAsm | 0.825 | .122 | (0.65-1.05) | 0.746 | .011 | (0.56-0.94) | 1.117 | .644 | (0.70-1.78) |
| No allo-HSCT in CR1 | 1 | 1 | 1 | ||||||
| Allo-HSCT in CR1 | 0.773 | <.001 | (0.69-0.87) | 0.630 | <.001 | (0.56-0.79) | — | — | — |
The multivariable analysis performed includes the different study regimens as strata.
Adverse karyotype according to ELN 2017.
Given the marked differences in the clinical and molecular alterations associated with the different localization of single-mutant CEBPA, we also looked for the impact of the localization of CEBPAsm on outcome. Patients carrying CEBPAsmbZIP showed a significantly higher CR rate (CEBPAbi 95% vs CEBPAsmbZIP 86% vs CEBPAsmTAD 66% vs CEBPAwt 73%) and a significantly longer OS (CEBPAbi 103.2 months; 95% CI, 70.7-inf, vs CEBPAsmbZIP, 63.3 months; 95% CI, 20.5-inf, vs CEBPAsmTAD, 12.7 months; 95% CI, 7.5-36.5, and CEBPAwt, 17.9 months; 95% CI, 17.9-21) and EFS (CEBPAbi, 20.7 months; 95% CI, 13.1-101.1 vs CEBPAsmbZIP, 17.1 months; 95% CI, 8.3-inf vs CEBPAsmTAD, 5.7 months; 95% CI, 2.6-10, and CEBPAwt, 7.0 months; 95% CI, 6.5-7.6; Figure 4A-B). Multivariable analysis confirmed that CEBPAsmbZIP mutations represented an independent favorable risk for outcome (Table 3).
Survival analysis according to CEBPA mutation status (CEBPAbi, CEBPAsmTAD, CEBPAsmbZIP, and CEBPAwt) within the cohort of 4461 patients receiving intensive treatment. Kaplan-Meier plots showing OS (A) and EFS (B). Numbers of patients at risk and results of log-rank tests from pairwise comparisons are provided below the x-axis.
Survival analysis according to CEBPA mutation status (CEBPAbi, CEBPAsmTAD, CEBPAsmbZIP, and CEBPAwt) within the cohort of 4461 patients receiving intensive treatment. Kaplan-Meier plots showing OS (A) and EFS (B). Numbers of patients at risk and results of log-rank tests from pairwise comparisons are provided below the x-axis.
Results of the multivariable analysis for CEBPAbi vs CEBPAsmbZIP vs CEBPAsmTAD
| . | OS (HR) . | P . | 95% CI . | EFS (HR) . | P . | 95% CI . | CR1 (OR) . | P . | 95% CI . |
|---|---|---|---|---|---|---|---|---|---|
| Intermediate karyotype | 1 | 1 | 1 | ||||||
| Favorable karyotype | 0.426 | <.001 | (0.35-0.51) | 0.400 | <.001 | (0.34-0.47) | 3.076 | <.001 | (2.08-4.56) |
| Adverse karyotype* | 1.853 | <.001 | (1.68-2.04) | 1.625 | <.001 | (1.49-1.78) | 0.476 | <.001 | (0.40-0.58) |
| Age | 1.032 | <.001 | (1.03-1.04) | 1.023 | <.001 | (1.02-1.03) | 0.948 | <.001 | (0.94-0.95) |
| Log10 WBC | 1.158 | <.001 | (1.08-1.24) | 1.231 | <.001 | (1.16-1.31) | 0.751 | <.001 | (0.66-0.86) |
| De novo AML | 1 | 1 | 1 | ||||||
| sAML | 1.088 | .127 | (0.98-1.21) | 1.069 | .189 | (0.97-1.18) | 0.735 | .003 | (0.60-0.90) |
| tAML | 1.380 | <.001 | (1.19-1.60) | 1.098 | .186 | (0.96-1.26) | 0.697 | .009 | (0.53-0.91) |
| No FLT3-ITDmut | 1 | 1 | 1 | ||||||
| FLT3-ITDmut | 1.204 | <.001 | (1.09-1.33) | 1.238 | <.001 | (1.13-1.35) | 1.018 | .866 | (0.83-1.25) |
| No NPM1mut | 1 | 1 | 1 | ||||||
| NPM1mut | 0.625 | <.001 | (0.57-0.69) | 0.531 | <.001 | (0.49-0.58) | 2.151 | <.001 | (1.77-2.62) |
| CEBPAwt | 1 | 1 | 1 | ||||||
| CEBPAbi | 0.507 | <.001 | (0.39-0.67) | 0.530 | <.001 | (0.42-0.67) | 6.367 | <.001 | (2.75-14.77) |
| CEBPAsmbZIP | 0.620 | .019 | (0.42-0.92) | 0.537 | .001 | (0.37-0.77) | 1.876 | .139 | (0.81-4.32) |
| CEBPAsmTAD | 1.024 | .878 | (0.75-1.39) | 0.981 | .891 | (0.74-1.31) | 0.825 | .515 | (0.46-1.48) |
| No allo-HSCT in CR1 | 1 | 1 | 1 | ||||||
| Allo-HSCT in CR1 | 0.776 | <.001 | (0.69-0.87) | 0.632 | <.001 | (0.56-0.71) | — | — | — |
| . | OS (HR) . | P . | 95% CI . | EFS (HR) . | P . | 95% CI . | CR1 (OR) . | P . | 95% CI . |
|---|---|---|---|---|---|---|---|---|---|
| Intermediate karyotype | 1 | 1 | 1 | ||||||
| Favorable karyotype | 0.426 | <.001 | (0.35-0.51) | 0.400 | <.001 | (0.34-0.47) | 3.076 | <.001 | (2.08-4.56) |
| Adverse karyotype* | 1.853 | <.001 | (1.68-2.04) | 1.625 | <.001 | (1.49-1.78) | 0.476 | <.001 | (0.40-0.58) |
| Age | 1.032 | <.001 | (1.03-1.04) | 1.023 | <.001 | (1.02-1.03) | 0.948 | <.001 | (0.94-0.95) |
| Log10 WBC | 1.158 | <.001 | (1.08-1.24) | 1.231 | <.001 | (1.16-1.31) | 0.751 | <.001 | (0.66-0.86) |
| De novo AML | 1 | 1 | 1 | ||||||
| sAML | 1.088 | .127 | (0.98-1.21) | 1.069 | .189 | (0.97-1.18) | 0.735 | .003 | (0.60-0.90) |
| tAML | 1.380 | <.001 | (1.19-1.60) | 1.098 | .186 | (0.96-1.26) | 0.697 | .009 | (0.53-0.91) |
| No FLT3-ITDmut | 1 | 1 | 1 | ||||||
| FLT3-ITDmut | 1.204 | <.001 | (1.09-1.33) | 1.238 | <.001 | (1.13-1.35) | 1.018 | .866 | (0.83-1.25) |
| No NPM1mut | 1 | 1 | 1 | ||||||
| NPM1mut | 0.625 | <.001 | (0.57-0.69) | 0.531 | <.001 | (0.49-0.58) | 2.151 | <.001 | (1.77-2.62) |
| CEBPAwt | 1 | 1 | 1 | ||||||
| CEBPAbi | 0.507 | <.001 | (0.39-0.67) | 0.530 | <.001 | (0.42-0.67) | 6.367 | <.001 | (2.75-14.77) |
| CEBPAsmbZIP | 0.620 | .019 | (0.42-0.92) | 0.537 | .001 | (0.37-0.77) | 1.876 | .139 | (0.81-4.32) |
| CEBPAsmTAD | 1.024 | .878 | (0.75-1.39) | 0.981 | .891 | (0.74-1.31) | 0.825 | .515 | (0.46-1.48) |
| No allo-HSCT in CR1 | 1 | 1 | 1 | ||||||
| Allo-HSCT in CR1 | 0.776 | <.001 | (0.69-0.87) | 0.632 | <.001 | (0.56-0.71) | — | — | — |
The multivariable analysis performed includes the different study regimens as strata.
Adverse karyotype according to ELN 2017.
Effect of co-mutations on outcome
We compared the effect of co-mutations on the survival of the patients according to the mutational status of the 3 most common mutations previously associated with outcome (ie, GATA2, TET2, and WT1 in the 3 CEBPA mutation subgroups (CEBPAbi, CEBPAsmbZIP, and CEBPAsmTAD). Although the presence of GATA2 mutations was associated with improved OS and EFS in the CEBPAbi and CEBPAsmbZIP groups (Figure 5A-B); however, these differences were not significant in pairwise comparisons (all P-values were adjusted for multiple testing with the Bonferroni-Holm procedure). However, for TET2mut (Figure 5B-C) a significant difference was found between CEBPAbi/TET2wt and TET2mut (median OS: CEBPAbi/TET2wt, not reached; 95% CI, 101.1-inf, vs CEBPAbi/TET2mut, 22.5 months; 95% CI, 13.9-inf; P = .012; median EFS: CEBPAbi/TET2wt, 34.7 months; 95% CI, 13.5-inf vs CEBPAbi/TET2mut, 9.5 months; 95% CI, 5.23-50.2; P = .02) and a trend for a different EFS in patients with CEBPAsmbZIP/TET2wt or CEBPAsmbZIP/TET2mut (median EFS CEBPAbZIP/TET2wt, not reached; 95% CI, 9.59-inf vs CEBPAbZIP/TET2mut, 9.2 months; 95% CI, 2.40-66.2; P = .06). WT1 mutations were associated with lower probability of survival (supplemental Figure 4); however, the comparison between the groups did not reach statistical significance.
Survival analysis according to CEBPA and GATA2 mutational status within the cohort of 4461 patients receiving intensive treatment. Kaplan-Meier plots showing OS (A) and EFS (B) of CEBPAwt, compared with CEBPAbi/GATA2wt (n = 82), CEBPAbi/GATA2mut (n = 46), CEBPAsmbZIP/GATA2wt(n = 31), CEBPAsmbZIP/GATA2mut (n = 18) mutant cases. Survival analysis according to CEBPA mutation status and TET2 status within a cohort of 4461 patients. Kaplan-Meier plots showing OS (C) and EFS (D) of CEBPAwt, compared with CEBPAbi/TET2wt (n = 98), CEBPAbi/TET2mut (n = 30), CEBPAsmbZIP/TET2wt (n = 33), CEBPAsmbZIP/TET2mut (n = 16) mutant cases. Numbers of patients at risk and results of log-rank tests from pairwise comparisons are provided below the x-axis.
Survival analysis according to CEBPA and GATA2 mutational status within the cohort of 4461 patients receiving intensive treatment. Kaplan-Meier plots showing OS (A) and EFS (B) of CEBPAwt, compared with CEBPAbi/GATA2wt (n = 82), CEBPAbi/GATA2mut (n = 46), CEBPAsmbZIP/GATA2wt(n = 31), CEBPAsmbZIP/GATA2mut (n = 18) mutant cases. Survival analysis according to CEBPA mutation status and TET2 status within a cohort of 4461 patients. Kaplan-Meier plots showing OS (C) and EFS (D) of CEBPAwt, compared with CEBPAbi/TET2wt (n = 98), CEBPAbi/TET2mut (n = 30), CEBPAsmbZIP/TET2wt (n = 33), CEBPAsmbZIP/TET2mut (n = 16) mutant cases. Numbers of patients at risk and results of log-rank tests from pairwise comparisons are provided below the x-axis.
NPM1 mutations, the most common co-mutation in patients carrying CEBPAsmTAD, were associated with slightly better OS and EFS; however, these differences were also not significant (supplemental Figure 5).
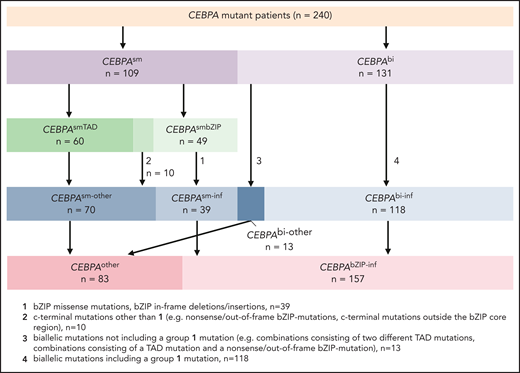
Presence of “typical” bZIP-mutations and outcome
Mutations in the CEBPA-bZIP region are typically in-frame (ie, multiples of 3 bp) and affect the DNA-binding-, fork-, or bZIP-region (for simplicity, summarized as bZIP) between amino acid positions 278 and 345 of the CEBPA protein (supplemental Figure 3), whereas frameshift mutations, the hallmark of mutations affecting the TAD1 and TAD2 domains, are less common in C-terminal mutations. To investigate whether the presence of typical in-frame bZIP mutations (irrespective of the biallelic or monoallelic status) actually represents the decisive molecular factor for the favorable outcome observed, we regrouped the 240 patients with CEBPAmut according to the presence or absence of these mutations. Typical bZIP mutations according to this classification were found in 118 of 131 who had CEBPAbi and 39 of 49 of those with CEBPAsmbZIP (denoted CEBPAbi-inf and CEBPAsm-inf). Thirteen of 131 CEBPAbi had nontypical biallelic mutations (eg, consisting of 2 different TAD domain mutations or a combination of TAD mutations and frameshift or nonsense bZIP mutations, denoted CEBPAbi-other). Patients with mutation in TAD as well as patients with a nontypical single-allele bZIP mutation were grouped as “other single-allele mutations” (CEBPAsm-other). We again observed profound differences in the age distribution, with typical bZIP mutations (double and single allele) predominating in younger adults, whereas other mutations, especially the few nontypical biallelic mutations, were predominantly detected in patients >50 years of age (supplemental Table 4; supplemental Figure 6).
To further characterize potential biological similarities between CEBPAbi-inf and CEBPAsm-inf, we performed RNA-seq of 20 samples from patients with CEBPAbi-inf, CEBPAsm-inf, CEBPAbi-other, and CEBPAsm-other (5 samples per group). As illustrated in the volcano-plots in supplemental Figure 7, we found between 34 and 129 differentially expressed genes in pairwise comparisons between all groups, the only exception being CEBPAbi-inf and CEBPAsm-inf, where no significant differences were found at a false discovery rate of 5% (supplemental Figure 7C).
The survival analysis performed also revealed profound differences. Superimposable, favorable outcomes (OS and EFS) were confined to patients in the CEBPAbi-inf and the CEBPAsm-inf groups, whereas patients with other mutational constellations, in particular patients with nontypical biallelic CEBPA mutations, showed an inferior outcome (supplemental Table 5; supplemental Figure 8). Co-mutations retained their prognostic impact in patients carrying CEBPAbi-inf and CEBPAsm-inf (as illustrated for the most common GATA2 mutations in supplemental Figure 9).
Prognostic classification for CEBPA mutations
Because these results strongly support the notion that mutation constellations containing typical in-frame bZIP mutations (ie, CEBPAbi-inf and CEBPAsm-inf) are comparable with respect to most clinical and molecular factors studied, we combined these 2 mutation types (denoted CEBPAbZIP-inf) and compared this new group to those patients without these mutations (CEBPAother; allocation summarized in Figure 6).
As illustrated in Table 4, the clinical associations already observed for the individual groups were even more pronounced in this analysis, especially the highly divergent median age (46 vs 62 years; P < .001), the difference with respect to the association with sAML (4% vs 17%; P < .001), the median CD34 positivity (51% vs 34%; P < .001), and the median WBC counts (25 × 109/L vs 14.7 × 109/L; P < .001). As expected, the mutational spectrum of patients belonging to these 2 subgroups showed highly significant differences. Only 1 of 157 patients with CEBPAbZIP-inf (0.6%) showed an NPM1 mutation, compared with 30 of 83 patients with CEBPAother mutations (36.1%; P < .001; Figure 7C; supplemental Figure 9). Other mutations that were significantly more common in the CEBPAother subgroup were alterations affecting DNA methylation (43 of 157, 27.4% vs 60 of 83, 72.3%; P < .001) and the spliceosome (4 of 157, 2.6% vs 21 of 83, 25.3%; P < .001).
Survival analysis according to CEBPA mutation status (CEBPAwt, CEBPAbZIP-inf, and CEBPAothers) within the cohort of 4461 patients receiving intensive treatment. Kaplan-Meier plots showing OS (A) and EFS (B). (C) Frequency distribution of additional gene mutations identified in patients with CEBPAbZIP-inf and CEBPAothers mutant genes (frequency of at least 5% in one subgroup). RAS signaling, including KRAS, NRAS, PTPN11, CBLB; spliceosome, including SF3B1, SRSF2, and ZRSR2; and methylation, including DNMT3A, IDH1, IDH2, and TET2. Numbers of patients at risk and results of log-rank tests from pairwise comparisons are provided below the x-axis.
Survival analysis according to CEBPA mutation status (CEBPAwt, CEBPAbZIP-inf, and CEBPAothers) within the cohort of 4461 patients receiving intensive treatment. Kaplan-Meier plots showing OS (A) and EFS (B). (C) Frequency distribution of additional gene mutations identified in patients with CEBPAbZIP-inf and CEBPAothers mutant genes (frequency of at least 5% in one subgroup). RAS signaling, including KRAS, NRAS, PTPN11, CBLB; spliceosome, including SF3B1, SRSF2, and ZRSR2; and methylation, including DNMT3A, IDH1, IDH2, and TET2. Numbers of patients at risk and results of log-rank tests from pairwise comparisons are provided below the x-axis.
Clinical variables in patients with CEBPAbZIP-inf and CEBPAother mutations
| . | CEBPAwt n = 4468 . | CEBPAbZIP-inf n = 157 . | CEBPAother n = 83 . | P (adj.) . |
|---|---|---|---|---|
| Age in years, median (IQR) | 57 (46-67) | 46 (36-57) | 62 (55-69) | <.001 |
| Sex, n (%) | ||||
| Female | 2170 (49) | . 76 (48) | 42 (51) | .885 |
| Male | 2298 (51) | 81 (52) | 41 (49) | |
| AML type, n (%) | ||||
| De novo | 3510 (78) | 149 (96) | 69 (83) | <.001 |
| sAML | 630 (14) | 5 (3) | 9 (11) | |
| tAML | 328 (7) | 2 (1) | 5 (6) | |
| FAB subtype, n (%) | ||||
| M0 | 223 (5) | 1 (1) | / | <.001 |
| M1/M2 | 2040 (46) | 127 (81) | 53 (65) | |
| M4-M7 | 1535 (34) | 22 (14) | 20 (24) | |
| Unknown | 670 (15) | 7 (4) | 9 (11) | |
| Laboratory, median (IQR) | ||||
| BM blasts in % | 60 (38-80) | 64 (50-78) | 62 (43.5-78) | .853 |
| CD34-positivity in % | 16 (2-47) | 51 (29-75.2) | 34 (6.6-72.2) | <.001 |
| WBC in 109/l | 11.7 (3-45.7) | 25 (9.2-70) | 14.7 (4.6-53.4) | .001 |
| LDH in U/l | 412.9 (272-727) | 445 (295-758) | 449 (296-601) | .242 |
| Treatment, n (%) | ||||
| Primary allo-HCT | 747 (17) | 52 (33) | 5 (6) | <.001 |
| Salvage allo-HCT | 915 (20) | 21 (13) | 16 (19) | .209 |
| . | CEBPAwt n = 4468 . | CEBPAbZIP-inf n = 157 . | CEBPAother n = 83 . | P (adj.) . |
|---|---|---|---|---|
| Age in years, median (IQR) | 57 (46-67) | 46 (36-57) | 62 (55-69) | <.001 |
| Sex, n (%) | ||||
| Female | 2170 (49) | . 76 (48) | 42 (51) | .885 |
| Male | 2298 (51) | 81 (52) | 41 (49) | |
| AML type, n (%) | ||||
| De novo | 3510 (78) | 149 (96) | 69 (83) | <.001 |
| sAML | 630 (14) | 5 (3) | 9 (11) | |
| tAML | 328 (7) | 2 (1) | 5 (6) | |
| FAB subtype, n (%) | ||||
| M0 | 223 (5) | 1 (1) | / | <.001 |
| M1/M2 | 2040 (46) | 127 (81) | 53 (65) | |
| M4-M7 | 1535 (34) | 22 (14) | 20 (24) | |
| Unknown | 670 (15) | 7 (4) | 9 (11) | |
| Laboratory, median (IQR) | ||||
| BM blasts in % | 60 (38-80) | 64 (50-78) | 62 (43.5-78) | .853 |
| CD34-positivity in % | 16 (2-47) | 51 (29-75.2) | 34 (6.6-72.2) | <.001 |
| WBC in 109/l | 11.7 (3-45.7) | 25 (9.2-70) | 14.7 (4.6-53.4) | .001 |
| LDH in U/l | 412.9 (272-727) | 445 (295-758) | 449 (296-601) | .242 |
| Treatment, n (%) | ||||
| Primary allo-HCT | 747 (17) | 52 (33) | 5 (6) | <.001 |
| Salvage allo-HCT | 915 (20) | 21 (13) | 16 (19) | .209 |
Bold P-values indicate statically significant results.
BM, bone marrow; LDH, lactate dehydrogenase; FAB, French-American-British; MRC, medical research council.
Outcome analyses performed for CEBPAbZIP-inf patients confirmed a strong prognostic impact of this subgroup, whereas CEBPA mutations in the other patients did not show significant differences compared with CEBPAwt (Figure 7A-B). Interestingly, patients with CEBPAbZIP-inf appear not to benefit from HSCT performed in the first complete remission (CR1; supplemental Figure 10). Multivariable analysis performed for this classification confirmed that CEBPAbZIP-inf positivity was the strongest predictor for achievement of CR (OR: 6.06; 95% CI, 2.78-13.23, P < .001) and a strong prognostic factor for OS (hazards ratio [HR], 0.57; 95% CI, 0.46-0.71; P < .001) and EFS (HR, 0.53; 95% CI, 0.43-0.64, P < .001; Table 5). We also looked for the effect of mutant GATA2, TET2, and WT1 on outcome in the novel subgroups. As observed in the previous analyses, mutant GATA2 and the absence of TET2 and WT1 mutations were associated with improved OS and EFS in patients carrying CEBPAbZIP-inf but not in those with CEBPAother, although these differences were not significant (supplemental Figure 11).
Results of the multivariable analysis for CEBPAbZIP-inf vs CEBPAother
| . | OS (HR) . | P . | 95% CI . | EFS (HR) . | P . | 95% CI . | CR1 (OR) . | P . | 95% CI . |
|---|---|---|---|---|---|---|---|---|---|
| Intermediate karyotype | 1 | 1 | 1 | ||||||
| Favorable karyotype | 0.456 | <.001 | (0.38-0.55) | 0.436 | <.001 | (0.38-0.51) | 3.072 | <.001 | (2.07-4.55) |
| Adverse karyotype* | 1.823 | <.001 | (1.67-2.00) | 1.590 | <.001 | (1.46-1.73) | 0.478 | <.001 | (0.40-0.58) |
| Age | 1.033 | <.001 | (1.03-1.04) | 1.025 | <.001 | (1.02-1.03) | 0.948 | <.001 | (0.94-0.95) |
| Log10 WBC | 1.159 | <.001 | (1.09-1.24) | 1.228 | <.001 | (1.16-1.31) | 0.753 | <.001 | (0.66-0.86) |
| De novo AML | 1 | 1 | 1 | ||||||
| sAML | 1.050 | .356 | (0.95-1.16) | 1.020 | .683 | (0.93-1.12) | 0.734 | .003 | (0.60-0.90) |
| tAML | 1.312 | <.001 | (1.15-1.50) | 1.056 | .413 | (0.93-1.20) | 0.704 | .012 | (0.53-0.93) |
| No FLT3-ITDmut | 1 | 1 | 1 | ||||||
| FLT3-ITDmut | 1.179 | <.001 | (1.08-1.29) | 1.193 | <.001 | (1.10-1.30) | 1.032 | .767 | (0.84-1.27) |
| No NPM1mut | 1 | 1 | 1 | ||||||
| NPM1mut | 0.635 | <.001 | (0.58-0.70) | 0.553 | <.001 | (0.51-0.60) | 2.139 | <.001 | (1.76-2.60) |
| CEBPAwt | 1 | 1 | 1 | ||||||
| CEBPAbZIP-inf | 0.570 | <.001 | (0.46-0.71) | 0.525 | <.001 | (0.43-0.64) | 6.061 | <.001 | (2.78-13.23) |
| CEBPAother | 0.897 | .410 | (0.69-1.16) | 0.964 | .762 | (0.76-1.22) | 1.003 | .989 | (0.61-1.65) |
| No allo-HSCT in CR1 | 1 | 1 | 1 | ||||||
| Allo-HSCT in CR1 | 0.671 | <.001 | (0.60-0.75) | 0.446 | <.001 | (0.40-0.50) | — | — | — |
| . | OS (HR) . | P . | 95% CI . | EFS (HR) . | P . | 95% CI . | CR1 (OR) . | P . | 95% CI . |
|---|---|---|---|---|---|---|---|---|---|
| Intermediate karyotype | 1 | 1 | 1 | ||||||
| Favorable karyotype | 0.456 | <.001 | (0.38-0.55) | 0.436 | <.001 | (0.38-0.51) | 3.072 | <.001 | (2.07-4.55) |
| Adverse karyotype* | 1.823 | <.001 | (1.67-2.00) | 1.590 | <.001 | (1.46-1.73) | 0.478 | <.001 | (0.40-0.58) |
| Age | 1.033 | <.001 | (1.03-1.04) | 1.025 | <.001 | (1.02-1.03) | 0.948 | <.001 | (0.94-0.95) |
| Log10 WBC | 1.159 | <.001 | (1.09-1.24) | 1.228 | <.001 | (1.16-1.31) | 0.753 | <.001 | (0.66-0.86) |
| De novo AML | 1 | 1 | 1 | ||||||
| sAML | 1.050 | .356 | (0.95-1.16) | 1.020 | .683 | (0.93-1.12) | 0.734 | .003 | (0.60-0.90) |
| tAML | 1.312 | <.001 | (1.15-1.50) | 1.056 | .413 | (0.93-1.20) | 0.704 | .012 | (0.53-0.93) |
| No FLT3-ITDmut | 1 | 1 | 1 | ||||||
| FLT3-ITDmut | 1.179 | <.001 | (1.08-1.29) | 1.193 | <.001 | (1.10-1.30) | 1.032 | .767 | (0.84-1.27) |
| No NPM1mut | 1 | 1 | 1 | ||||||
| NPM1mut | 0.635 | <.001 | (0.58-0.70) | 0.553 | <.001 | (0.51-0.60) | 2.139 | <.001 | (1.76-2.60) |
| CEBPAwt | 1 | 1 | 1 | ||||||
| CEBPAbZIP-inf | 0.570 | <.001 | (0.46-0.71) | 0.525 | <.001 | (0.43-0.64) | 6.061 | <.001 | (2.78-13.23) |
| CEBPAother | 0.897 | .410 | (0.69-1.16) | 0.964 | .762 | (0.76-1.22) | 1.003 | .989 | (0.61-1.65) |
| No allo-HSCT in CR1 | 1 | 1 | 1 | ||||||
| Allo-HSCT in CR1 | 0.671 | <.001 | (0.60-0.75) | 0.446 | <.001 | (0.40-0.50) | — | — | — |
The multivariable analysis performed includes the different study regimens as strata.
CR1, first complete remission.
Adverse karyotype according to ELN 2017.
Discussion
We analyzed 4708 adult patients with newly diagnosed AML for CEBPA alterations and identified mutations in CEBPA in 5.1%. Our cohort differed from patient cohorts in previous studies investigating the role of CEBPA mutations,9-12,15,21,24,35-42 because we did not select for age, cytogenetic subgroups, or disease status. Within the entire cohort, 49% showed an aberrant karyotype, and the median patient age at diagnosis was 57 years (IQR, 46-67 years), more accurately reflecting the entirety of patients with AML in general. This fact may explain why the prevalence of CEBPA mutations found in our study is at the lower end of the previously reported range of 4% to 20%.9-12,15,21,24,35-42
Our main focus was the role of monoallelic CEBPA mutations; therefore, we performed an extensive evaluation of this subgroup and a detailed analysis of the individual type and localization of the mutation.
A first aspect observed with respect to the different mutational subgroups (CEBPAbi, CEBPAsmbZIP, and CEBPAsmTAD) was the highly significant difference in the age distribution. Whereas CEBPAbi and CEBPAsmbZIP were predominantly found in younger adults and decreased with age, CEBPAsmTAD mutations were rare in patients up to the age of 40 years and were particularly common in older individuals. The lower age of patients with CEBPAbi mutations has already been reported in previous studies.21,24,41 In contrast, the major age difference between CEBPAsmTAD and CEBPAsmbZIP mutations has not been reported before. Interestingly, a very recently published work in pediatric AML found only monoallelic CEBPAbZIP mutations in their analysis, indicating that these mutations are indeed significantly more prevalent in younger individuals.43CEBPAbi and CEBPAsmbZIP also showed overlapping laboratory profiles, with higher rates of CD34 positivity and higher WBC counts.43 We also observed a favorable prognostic impact of CEBPAsmbZIP comparable to biallelic CEBPA mutations. This finding again is in concordance with the data reported by Tarlock et al in pediatric AML, showing that the outcome of patients with CEBPAbi or CEBPAsmbZIP mutations did not differ.43
Our results indicate a similar spectrum of co-mutations for CEBPAbi and CEBPAsmbZIP (ie, significantly higher rates of GATA2 and WT1 mutations and a mere lack of NPM1 alterations). GATA2 mutations in CEBPAbi have been described previously,44 whereas a similar association of CEBPAsmbZIP with GATA2 mutations has not been reported so far. The presence of GATA2, TET2, and WT1 mutations significantly affected the outcome of patients carrying CEBPAbi, whereas CEBPAsmbZIP mutations showed a significant effect on outcome only for mutations in GATA2. The prognostic impact of mutations in GATA2, TET2, and WT1 has been described recently,45,46 but was confined to biallelic CEBPA mutations. In contrast, our data indicate that these prognostic associations also apply to patients with CEBPAsmbZIP, but not to those with CEBPAsmTAD. Clustering of co-mutations according to functional groups further highlighted a differential spectrum of co-occurring mutations, with CEBPAsmTAD mutations showing a significantly higher prevalence of mutations affecting proteins involved in DNA methylation (ie, DNMT3A, TET2, IDH1, and IDH2) as well as RNA splicing (ie, SRSF2, SF3B1, and ZRSR2), which was not seen in patients with CEBPAbi and CEBPAsmbZIP (Figure 4B). Additional analyses looking in more detail for the individual mutations in CEBPA in our patients suggested that the identified clinical and molecular associations as well as the association with outcome were restricted to typical in-frame mutations within the bZIP region.
The reason for this differential behavior is unclear at present, but based on previous in vitro as well as animal experiments, TAD1 and bZIP mutations have clear functional differences (reviewed in Pulikkan et al47). The frameshift mutations typically observed in the TAD1 domain induce the consecutive translation of the shorter CEBPAp30 protein, instead of the CEBPAp42 full-length protein. In contrast, with both CEBPAp42 and CEBPAp30, mutations in the DBD and bZIP domains result in loss of DNA binding as well as dimerization. Recent animal data modeling the disease suggest functional disparity of different bZIP mutations. Lethally irradiated mice undergoing transplant of hematopoietic stem cells homozygous for a point mutation in bZIP (designated BRM2) start to develop a myeloproliferative disease that transforms into overt AML.48 By transplanting transgenic cells carrying the most common mutation in bZIP (K313dup; K allele), alone or in combination with a TAD1 mutation (designated L-allele), Bereshchenko et al49 documented that cells from mice carrying either the K/K or the K/L genotype, showed similarities in their mRNA expression profiles and higher expansion of immature blasts in the BM, which was distinct from the L/L genotype, as well as CEBPA-WT cells. CEBPA-mutated AML is characterized by specific RNA,50 as well as miRNA35,51 expression profiles. The expression of several key miRNAs, such as miR-34, miR-182, and miR-223, which are involved in stem cell self-renewal, cell migration, and granulocytic differentiation, are physiologically regulated by CEBPA (reviewed in Stavast et al52). Interestingly, recent data suggest that bZIP-mutant CEBPA does not downregulate miR-182, leading to a block of granulocytic differentiation.53 This incapability appears to be mainly restricted to typical in-frame bZIP mutations clustering around the core mutated amino acids 312 and 313 of the CEBPA protein.53 In support of this, one of the genes we found most highly deregulated between CEBPAbi-inf/CEBPAsm-inf and CEBPAsm-other, OSTL/RNF217, coding for a highly conserved RING-finger ubiquitin ligase overexpressed in various leukemia entities,54 has been shown to be regulated by the miRNA cluster miR-183-96-182.55
Taken together, the CEBPAbZIP-inf genotype identified in our analysis describes a subgroup of CEBPA mutations predominantly found in younger adults, indicating characteristics of a more immature and proliferative disease that has an overall prevalence of 3.3% in adult patients with AML, but is found in up to 7% of patients ≤40 years, thus representing a relevant subgroup, especially in younger adults. The mere absence of NPM1 mutations (1 of 157 patients; 0.6%) and the high prevalence of GATA2 mutations in these patients, which was found to be associated with an even better prognosis and a long-term survival in up to 80%, further highlights the special biology of these leukemias. The 2016 World Health Organization classification defines the CEBPA mutational class by the presence of a biallelic mutation, regardless of the localization within the gene. Given that 90% of the patients with the biallelic CEBPA mutation in this analysis actually carried typical bZIP CEBPA mutations, there is obviously a high degree of overlap, which may explain why this difference was undetected in most previous analyses, although there was some evidence in another study.25 However, that only the 90% of patients with biallelic CEBPA mutations containing typical bZIP mutations in fact show a better outcome indicates that only those should be assigned to the favorable risk group.
Acknowledgments
The authors thank all centers and participating physicians of the Study Alliance Leukemia who entered their patients into the study (see Appendix). They also thank M. Böhm and M. Hartwig for skillful technical assistance.
This study was supported in part by grants from the Federal Ministry of Education and Research 01GS0872 (C.T.).
Authorship
Contribution: F.T., J.A.G., and C.T. designed the study; performed the research; collected, assembled, analyzed, interpreted the data; and wrote the manuscript; F.T., J.A.G., S. Stasik., A.P., R.M.-L., and P.J.M.V., performed the molecular analyses and analyzed the data; S.H. performed the molecular analyses; M. Kramer and J.S. performed the statistical analyses; J.M.M., C.R., U.K., A.K., S. Scholl, A.H., T.H.B., R. Naumann, B.S., H.E., M.S., A.B., A.N., K.S.-E., C.S., S.W.K., M.H., R. Nopenney., U.K., C.D.B., M. Kaufmann, F.S., K.S., M.v.B., C.M.-T., U.P., W.E.B., H.S., G.E., M.B., and J.S., treated the patients and collected the clinical data; and all authors approved the final version of the manuscript.
Conflict-of-interest disclosure: C.T. is CEO and co-owner of AgenDix GmbH. The remaining authors declare no competing financial interests.
Correspondence: Christian Thiede, Medizinische Klinik und Poliklinik I, Universitätsklinikum Carl Gustav Carus der Technischen Universität, Fetscherstr 74, 01307 Dresden, Germany; e-mail: christian.thiede@uniklinikum-dresden.de.
Original data can be obtained by e-mail request to the corresponding author (christian.thiede@uniklinikum-dresden.de).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Appendix: participating centers of the Study Alliance Leukemia (SAL)
Tim H. Brümmendorf, Universitätsklinikum Aachen der RWTH, Germany; Armin Schulz-Abelius, Klinikum Altenburger Land GmbH, Altenburg, Germany; Martin Trepel, Klinikum Augsburg, Augsburg, Germany; Martina Teichmann, Sozialstiftung Bamberg, Bamberg, Germany; Alexander Kiani, Klinikum Bayreuth GmbH, Bayreuth, Germany; Bertram Glass, Helios Klinikum Berlin-Buch, Berlin, Germany; Martin Görner, Städt Kliniken Bielefeld gGmbH, Bielefeld, Germany; Dirk Behringer, Augusta-Kranken-Anstalt gGmbH, Bochum, Germany; Johannes Kullmer, Ev Diakonie-Krankenhaus gGmbH, Bremen, Germany; Mathias Hänel, Klinikum Chemnitz gGmbH, Chemnitz, Germany; Christof Lamberti, MVZ Coburg, Coburg, Germany; Martin Schmidt-Hieber, Carl-Thiem-Klinikum Cottbus gGmbH, Cottbus, Germany; Christoph Röllig, Universitätsklinikum Dresden, Dresden, Germany; Michael Flasshove, Krankenhaus Düren gGmbH, Düren, Germany; Andreas Mackensen, Universitätsklinikum Erlangen-Nürnberg, Erlangen, Germany; Maher Hanoun, Universitätsklinikum Essen, Essen, Germany; Michael Kiehl, Klinikum Frankfurt (Oder) GmbH, Frankfurt (Oder), Germany; Hubert Serve, Klinikum der J. W. Goethe Universität, Frankfurt (Main), Germany; H.-G. Höffkes, Klinikum Fulda, Fulda, Germany; Lutz Peter Müller, Universitätsklinikum Halle (Saale), Halle (Saale) Germany; Heinz Albert Dürk, St Barbara-Klinik Hamm, Hamm, Germany; Carsten Müller-Tidow, Universitätsklinikum Heidelberg, Heidelberg, Germany; Ulrich Kaiser, St Bernward Krankenhaus, Hildesheim, Germany; Andreas Hochhaus, Universitätsklinikum Jena, Jena, Germany; Gerhard Held, Westpfalz-Klinikum GmbH, Kaiserslautern, Germany; Roland Peter Repp, Städt Krankenhaus Kiel, Kiel, Germany; Claudia Baldus, Universitätsklinikum Schleswig-Holstein, Kiel, Germany; Jens-Marcus Chemnitz, Gemeinschaftsklinikum Mittelrhein gGmbH, Koblenz, Germany; Uwe Platzbecker, Universitätsklinikum Leipzig, Leipzig, Germany; Stefan Klein, Klinikum Mannheim gGmbH, Mannheim, Germany; Andreas Neubauer, Universitätsklinikum Marburg GmbH, Marburg, Germany; Wolfgang E. Berdel, Universitätsklinikum Münster, Münster, Germany; Martin Wilhelm, Klinikum Nürnberg Nord, Nürnberg, Germany; Jörg Schubert, Elblandklinikum Riesa, Riesa, Germany; Achim Meinhardt, Agaplesion Diakonieklinikum Rotenburg gGmbH, Rotenburg (Wümme), Germany; Thomas Geer, Diakonie-Krankenhaus, Schwäbisch-Hall, Germany; Markus Ritter, Klinikum Sindelfingen-Böblingen, Sindelfingen, Germany; Walter Erich Aulitzky, Robert-Bosch-Krankenhaus, Stuttgart, Germany; Natalia Heinz, HSK Wiesbaden, Wiesbaden, Germany; Markus Schaich, Rems-Murr-Klinikum Winnenden, Winnenden, Germany; and Hermann Einsele, Universitätsklinikum Würzburg, Würzburg, Germany.
REFERENCES
Author notes
F.T. and J.A.G. contributed equally to this study.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal