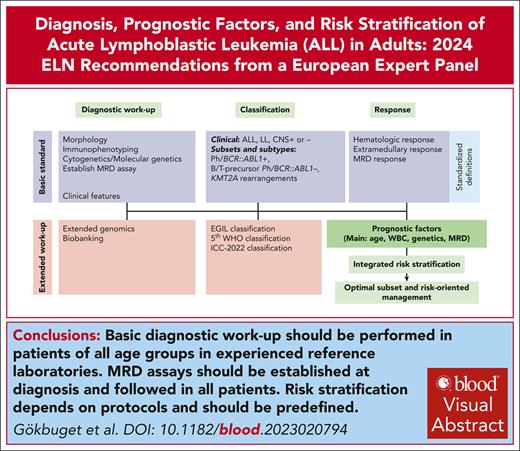
Visual Abstract
Working groups of the European LeukemiaNet have published several important consensus guidelines. Acute lymphoblastic leukemia (ALL) has many different clinical and biological subgroups and the knowledge on disease biology and therapeutic options is increasing exponentially. The European Working Group for Adult ALL has therefore summarized the current state of the art and provided comprehensive consensus recommendations for diagnostic approaches, biologic and clinical characterization, prognostic factors, and risk stratification as well as definitions of endpoints and outcomes. Aspects of treatment, management of subgroups and specific situations, aftercare, and supportive care are covered in a separate publication. The present recommendation intends to provide guidance for the initial management of adult patients with ALL and to define principles as a basis for future collaborative research.
Introduction
Scientific progress in acute lymphoblastic leukemia (ALL) has been driven by the activities of national academic multicenter study groups in Europe and US in the past decades. The European national study groups have founded a collaboration within the European LeukemiaNet named European Working Group for Adult ALL. The group has published a consensus recommendation for the management of adult ALL,1 which was broadly distributed among the participating centers.
ALL is rare compared to other cancers of adulthood. It has many different clinical and biological subgroups and the knowledge on disease biology and therapeutic options is increasing exponentially. There is an unmet need for guidance for clinicians, regulatory agencies, and health care systems. Available guidelines from the National Comprehensive Cancer Center2 or European Society of Medical Oncology3 are relevant but need to be complemented by a broad and comprehensive expert consensus considering the different regulatory and socio-economic framework for ALL management in Europe. Therefore, the group decided to develop European LeukemiaNet Recommendations as published for other entities.4,5 The focus of this article is on diagnostic approaches, biological and clinical characterization, risk stratification, and definitions of endpoints and outcomes. Aspects of treatment, management of subgroups and specific situations, aftercare, and supportive care are covered in a separate publication.6
Methods
The panel includes 17 members representing national study groups. Members met in person and defined topics, tables, and responsibilities of coauthors (supplemental Table 1, available on the Blood website). Coauthors performed literature searches of PubMed database and considered relevant abstracts. The manuscript was reviewed by all coauthors. Formal corrections were performed by the corresponding author. Disagreements were summarized and discussed in the whole group. The whole group agreed on the final version of the manuscript. Due to rapid innovation and availability of new data together with a lack of randomized trials for many essential questions, most of the statements have an evidence level of “expert recommendation” for clinical practice.
Diagnostic procedures and classification
The initial workup should be comprehensive to allow a precise diagnostic characterization and an accurate stratification, as well as to set the basis for minimal residual disease (MRD) monitoring. Morphology, multicolor flow cytometry (MFC), and molecular genetics are obligatory (Table 1). Additional genomic analyses are used to better define molecular subgroups. Diagnosis is usually based on a bone marrow (BM) aspirate, which should be attempted even in cases with high peripheral blast (PB) counts. A BM biopsy is recommended in patients with a dry aspirate; in these cases, BM aspiration can be repeated after the prephase to obtain further material. Biobanking is recommended. It should include nucleic acid (DNA and RNA), viable cells, and possibly germline material. Further workup includes a lumbar puncture with classification and imaging procedures as needed to define potential extramedullary involvement (supplemental Table 2).
Specific diagnostic workup in adult ALL
| . | Recommendation . | Outputs . |
|---|---|---|
| Morphology | Mandatory | Defines infiltration (>25% required for a diagnosis of ALL vs lymphoblastic lymphoma) |
| Allows a differential diagnosis with AML (MPO– vs + MPO+) | ||
| Recognizes L3 (Burkitt-type) subsets | ||
| Flow cytometry | Mandatory | Allows a differential diagnosis with AML |
| Permits to define the cell of origin | ||
| Permits to define the stage of differentiation | ||
| MPO∗ | Mandatory | Allows a differential diagnosis with AML. |
| B-lineage ALL∗: CD19, cCD79a, c/sCD22 (minimal requirement), TdT, CD10, CD20, cIgM, sIg (kappa or lambda) | Mandatory | Pro-B: CD19/CD79a/cCD22+/CD10– (B-I), NG2 |
| Common: CD10+/cIgM– (B-II) | ||
| Pre-B: cIgM+/sIg– (B-III) | ||
| Mature: sIg+ (B-IV), CD20+ | ||
| T-lineage ALL∗: c/sCD3, CD7 (minimal requirement), TdT, CD1a, CD2, CD5, CD4, CD8, TCR α/β or γ/δ | Mandatory | Pro-T: cCD3/CD7+ (T-I) |
| Pre-T: CD2/CD5+ (T-II) | ||
| Cortical-T: CD1a+ (T-III) | ||
| Mature-T: sCD3+/CD1a– (T-IV) | ||
| Molecular genetics | Mandatory | t(9;22)/Ph+/BCR::ABL1 |
| t(4;11)+/KMT2Ar | ||
| t(1;19)+/TCF3::PBX1 | ||
| Other high-risk cytogenetics | ||
| MRD (molecular or MFC) | Mandatory | Individual MRD assay for further follow-up |
| Extended genomics (GEP, CNA, WES, WGS, NGS) | In clinical trials and for research purposes | Identification of novel subgroups with prognostic/biologic significance |
| Identification of molecular targets for targeted therapies |
| . | Recommendation . | Outputs . |
|---|---|---|
| Morphology | Mandatory | Defines infiltration (>25% required for a diagnosis of ALL vs lymphoblastic lymphoma) |
| Allows a differential diagnosis with AML (MPO– vs + MPO+) | ||
| Recognizes L3 (Burkitt-type) subsets | ||
| Flow cytometry | Mandatory | Allows a differential diagnosis with AML |
| Permits to define the cell of origin | ||
| Permits to define the stage of differentiation | ||
| MPO∗ | Mandatory | Allows a differential diagnosis with AML. |
| B-lineage ALL∗: CD19, cCD79a, c/sCD22 (minimal requirement), TdT, CD10, CD20, cIgM, sIg (kappa or lambda) | Mandatory | Pro-B: CD19/CD79a/cCD22+/CD10– (B-I), NG2 |
| Common: CD10+/cIgM– (B-II) | ||
| Pre-B: cIgM+/sIg– (B-III) | ||
| Mature: sIg+ (B-IV), CD20+ | ||
| T-lineage ALL∗: c/sCD3, CD7 (minimal requirement), TdT, CD1a, CD2, CD5, CD4, CD8, TCR α/β or γ/δ | Mandatory | Pro-T: cCD3/CD7+ (T-I) |
| Pre-T: CD2/CD5+ (T-II) | ||
| Cortical-T: CD1a+ (T-III) | ||
| Mature-T: sCD3+/CD1a– (T-IV) | ||
| Molecular genetics | Mandatory | t(9;22)/Ph+/BCR::ABL1 |
| t(4;11)+/KMT2Ar | ||
| t(1;19)+/TCF3::PBX1 | ||
| Other high-risk cytogenetics | ||
| MRD (molecular or MFC) | Mandatory | Individual MRD assay for further follow-up |
| Extended genomics (GEP, CNA, WES, WGS, NGS) | In clinical trials and for research purposes | Identification of novel subgroups with prognostic/biologic significance |
| Identification of molecular targets for targeted therapies |
c, cytoplasmic; GEP, gene expression profiling; MPO, myeloperoxidase; s, surface; WES, whole exome sequencing; WGS, whole genome sequencing.
MPAL: myeloid myeloperoxidase or at least 2 of the following antigens: nonspecific esterase, CD11c, CD14, CD64, and lysozyme; B-lineage: strong CD19 expression plus at least 1 of the following strongly expressed antigens: CD79a, cyCD22, CD10, or weak CD19, with at least 2 of the following strongly expressed antigens: CD79a, cytCD22, CD10; and T-lineage: cCD3 or rarely sCD3.
Morphology and cytochemistry
BM morphology is necessary to evaluate the degree of infiltration and to differentiate ALL from acute myeloid leukemia (AML) and ALL from lymphoblastic lymphoma. There are no specific cytochemistry reactions to classify ALL. Myeloperoxidase is always negative with the exception of mixed-phenotype acute leukemia (MPAL), which may show low/dim/strong levels of expression particularly in B-myeloid cases in which isolated myeloperoxidase (MPO) expression (isoMPO) may occur.
Immunophenotyping
MFC (at least 8 colors) is central. It allows to (1) make a differential diagnosis with AML, (2) establish lineage affiliation and differentiation, (3) define an aberrant phenotype for MRD monitoring, and (4) detect target antigens for immunotherapy. The EuroFlow Consortium has standardized MFC.7 TdT is expressed by all B- and T-cell progenitors, except for mature B-ALL (Burkitt leukemia), while being negative in AML.
B-lineage ALL (B-LIN) accounts for 75% to 80% of cases. The crucial markers are CD19, CD22, and cytoplasmic (cy) CD79a. The EGIL classification8 defines 4 differentiation stages: pro-B, common, pre-B ALL, and mature B-ALL (Table 1). An aberrant coexpression of myeloid markers can be detected in about 40%9 of cases. Pro-B ALL is CD10-negative and often associated with t(4;11)/KMT2A,10 whereas pre-B ALL often carries t(1;19)/TCF3::PBX1 aberration. The identification of surface markers as potential targets for immunotherapy is essential for therapeutic approaches.6
T-lineage ALL (T-LIN) represents 20% to 25% of adult ALL. Crucial markers are cyCD3 and sCD7. Furthermore, CD1a, CD2, sCD3, CD4, CD5, and CD8, as well as T-cell receptor α/β or γ/δ, may be variably expressed. T-ALL can be classified into 4 subtypes: pro–T-ALL, pre–T-ALL, cortical (thymic) T-ALL, and mature T-ALL.8 Pro–T-ALL cases express only cyCD3 and CD7, cortical T-ALL is CD1a positive, whereas mature T-ALL express CD4 and/or CD8, and usually sCD3. The so-called ETP-ALL (early-T precursor),11 represents an entity within pro-T/pre-T ALL with weak CD5 expression which shows at least one myeloid and/or stem cell marker.11 CD34 and myeloid markers are expressed in a proportion of T-ALL.9 The maturation stage correlates with molecular aberrations such a LMO2/HOXA in immature, TLX1/TLX3 in cortical, and TAL1 in mature T-ALL. Besides EGIL there is no uniform international classification of immunophenotypes. To achieve comparability, it is recommended to report results including this classification.
MPAL represents 1% to 5% of acute leukemias12 and is characterized by blasts coexpressing antigens of more than one lineage on the same cells or that have separate populations of blasts of different lineages. The definition of MPAL has been refined in the World Health Organization (WHO) 200813 and 201614 classifications, the latter discriminating B-myeloid from the other MPAL. The combination of antigens used for MPAL recognition is summarized in Table 1.
Cytogenetics and molecular genetics
Karyotyping, fluorescence in situ hybridization, and reverse transcription polymerase chain reaction techniques are used for the genetic characterization.
B-LIN
t(9;22)/BCR::ABL1 is the most frequent aberration. Its detection in the shortest possible time is essential.15 Other subtypes include KMT2A rearrangements, monosomy 7, hypodiploidy/low hypodiploidy (and the related near-triploid), t(1;19)/TCF3::PBX1, and t(17;19)/TCF3::HLF. t(8;14)/MYC/IGH are rarely detected; also t(12;21)/ETV6::RUNX1 and high hyperdiploidy (51-65 chromosomes) are rare in adults.16
T-LIN
Genetic aberrations often comprise the 14q11 and 7q34 breakpoints, leading to the juxtaposition of the TCR loci to transcription factors, namely TAL1, TAL2, LYL1, LMO1, LMO2, TLX1 (HOX11), TLX3 (HOX11L2), HOXA, MYC, and MYB.16 In addition, cryptic ABL-1 rearrangements with different partner genes for example, NUP214, EML1, and ETV6 have been identified.
MPAL
The list of genomic lesions detected in MPAL is rather large, and is likely to increase, given its heterogeneity. As already mentioned, the most frequent rearrangements are BCR::ABL1 detected in about 15% of cases and KMT2A found in roughly 10% of patients; ZNF384 rearrangements (49% of B/MPAL) and BCL11B-r have also been reported.17,24
Extended genomics
Gene expression profiling, copy number alteration analysis (CNA), genome-wide, and next generation sequencing (NGS) techniques have identified new subgroups. These techniques are not part of the standard workup but may be carried out for a refined classification in research laboratories.
The Philadelphia (Ph)-like subgroup is discussed separately.6 ETP-ALL11 is genetically heterogeneous, with mutations of multiple pathways including hematopoietic and lymphoid development, Ras, cytokine receptor, kinase signaling, and loss-of-function mutations targeting epigenetic regulators.18,19
CNA allows to identify aberrations that affect cell cycle (CDKN2A/B and RB1), as well as lymphoid development (IKZF1, PAX5, VPREB1, and LEF1). Among those, IKZF1 deletion (ΔIKZF1)20,21 can be found in about 80% of Ph+ and Ph-like cases.22 The IKZF1-plus genotype (ie, iKZF1 plus CDKN2A/B and/or PAX5 deletions) is considered in some trials.
Recently, further subtypes have been identified in B-LIN. The DUX4- and ERG-rearrangement leads to the formation of an ERG isoform, which in mice models induces leukemic transformation. MEF2D rearrangements have transforming capability in vitro. Finally, ZNF384 deregulation, which can rearrange with several partners (EP300, CREBBP, TAF15, SYNRG, EWSR1, TCF3, and ARID1B) often displays a weak CD10 and CD13/CD33 expression.16
NGS has identified novel mutations and rearrangements; the most frequent involve the RAS pathway (N/K RAS, FLT3, PTPN11, NF1, and BRAF mutations) and can be detected in more than 40% of cases, are prevalent in the hyperdiploid and KMT2A-rearranged cases and tend to increase at relapse. More rare mutations affect the Janus kinase (JAK)/STAT pathway (JAK1/2, JAK3, IL7R, rarely CRLF2, SH2B3, and IL2RB). RAS and JAK/STAT mutations are detected in both B- and T-LIN.23 In T-ALL, NOTCH1/FBXW7 lesions represent the most frequent event (60%) (Table 2).
Molecular subgroups in adult ALL: incidence, prognosis, and molecular findings
| ALL subset . | Prevalence and prognosis∗ . | Related aberration(s) . |
|---|---|---|
| B-lineage ALL | ||
| BCR::ABL1/t(9;22)(q34;q11.2) (Ph+) | 20%-50%, increasing with age; improved by tyrosine kinase inhibitor therapy | BCR::ABL1 rearrangement |
| Ph-like | 25%-27% Unfavorable/controversial for current regimens | Gene expression profile like BCR::ABL1+ ALL but without BCR::ABL1 rearrangement |
| TCF3::PBX1/t(1;19)(q23;p13) | 10%-15% Favorable with intensive therapy | TCF3::PBX1 rearrangement |
| KMT2A(MLL)::AFF1/t(4;11)(q21;q23.3), KMT2A-rearranged/t(v;11q23.3) | ∼5% Unfavorable | KMT2A::AFF1 or KMT2A-other partner gene rearrangement |
| IGH::MYC/t(8,14)(q24;q32) | 1%-5% Unfavorable in B-precursor ALL | IGH::MYC rearrangement |
| TCF3::HLF/t(17;19)(q22;p13.3) | <1% Unfavorable | TCF3::HLF rearrangement |
| iAMP21 | ∼2% Intermediate/unfavorable | — |
| 14q32 translocations | <5%, higher in adolescents Intermediate/unfavorable | IGH fusion with partner genes CRLF2, ID4, CEBP, BCL2, EPOR, LHX4, and IL-3 |
| 9p13 deletions/translocations | ∼25% No impact on outcome | PAX5 fusion with partner genes ETV6, ELN, POM121, PML, FOXP1, MLLT3, JAK2, C20orf112, AUTS2, CHFR, SOX5, and POM121C |
| 7p12.2 focal deletions/mutations | 50%; 80% in Ph+ and Ph–like Controversial prognosis | Deletions of IKZF1 |
| DUX4-rearranged and ERG-deregulated | 3%-7% Favorable | ERG and IKZF1 deletions |
| MEF2D-rearranged ALL | 3%-4% Poor | |
| ZNF384-rearranged ALL | 6%-7% Intermediate | Partner gene EP300, CREBBP, TAF15, SYNRG, EWSR1, TCF3, and ARID1B |
| CDX2::UBTF | 2.7%-4% Poor | High expression of CDX2 and gain (1q); UBTF::ATXN7L3; CDX2-cis-deregulation |
| T-lineage ALL | ||
| TAL and LMO rearrangements/del(1)(p32), t(1;14)(p32;q11), t(1;7)(p32;q34), t(7;9)(q34;q32), t(11;14)(p15;q1), t(11;14)(p13;q1), t(7;11)(q35;p13) | 30%-40% Favorable and partly depending on additional lesions | SIL-TAL1 rearrangement, TCR rearrangements with TAL1, TAL2, LMO1, and LMO2 |
| HOXA aberrations/inv(7)(p15q34), t(7;7)(p15;q34), t(10;11)(p13;q14), t(v;11q23), del(9)(q34) | 20%-25% Outcome depending on additional lesions | TCR-HOXA rearrangement, MLLT10 and MLL rearrangements with various partners, SET-NUP214 rearrangement |
| TLX1-10q24 rearrangements/t(7;10)(q34;q24), t(10;14)(q24;q11) | 20%-30% No impact on prognosis | TCR-TLX11 rearrangement |
| ETP ALL | 10%-15% Unfavorable/controversial | Deregulation of myeloid transcription factors, of members of RAS pathway and of epigenetic regulators |
| TLX3-5q35 rearrangement/t(5;14)(q35;q32) | 10% No impact on prognosis | TLX3-BCL11B rearrangement |
| t(8;14)(q24;q11) | 1% Unfavorable | MYC involvement |
| ABL1 rearrangements | ∼3% Potentially targetable by tyrosine kinase inhibitors | NUP214, EML1; ETV6 |
| LYL/MEF2C rearrangement and immature cluster/t(7;19)(q34;p13), del(5)(q14) | 3%-17% Unfavorable; improved by intensive treatment | TCR with LYL1 MEF2C rearrangements |
| NKX2-1/NKX2-2 rearrangements/inv(14)(q11.2q13), t(7;14)(q34;q13), inv(14)(q13q32.33), t(14;20)(q11;p11) | 6% No impact on prognosis | TCR/IGH-NKX2- or NKX2-2 rearrangements |
| ALL subset . | Prevalence and prognosis∗ . | Related aberration(s) . |
|---|---|---|
| B-lineage ALL | ||
| BCR::ABL1/t(9;22)(q34;q11.2) (Ph+) | 20%-50%, increasing with age; improved by tyrosine kinase inhibitor therapy | BCR::ABL1 rearrangement |
| Ph-like | 25%-27% Unfavorable/controversial for current regimens | Gene expression profile like BCR::ABL1+ ALL but without BCR::ABL1 rearrangement |
| TCF3::PBX1/t(1;19)(q23;p13) | 10%-15% Favorable with intensive therapy | TCF3::PBX1 rearrangement |
| KMT2A(MLL)::AFF1/t(4;11)(q21;q23.3), KMT2A-rearranged/t(v;11q23.3) | ∼5% Unfavorable | KMT2A::AFF1 or KMT2A-other partner gene rearrangement |
| IGH::MYC/t(8,14)(q24;q32) | 1%-5% Unfavorable in B-precursor ALL | IGH::MYC rearrangement |
| TCF3::HLF/t(17;19)(q22;p13.3) | <1% Unfavorable | TCF3::HLF rearrangement |
| iAMP21 | ∼2% Intermediate/unfavorable | — |
| 14q32 translocations | <5%, higher in adolescents Intermediate/unfavorable | IGH fusion with partner genes CRLF2, ID4, CEBP, BCL2, EPOR, LHX4, and IL-3 |
| 9p13 deletions/translocations | ∼25% No impact on outcome | PAX5 fusion with partner genes ETV6, ELN, POM121, PML, FOXP1, MLLT3, JAK2, C20orf112, AUTS2, CHFR, SOX5, and POM121C |
| 7p12.2 focal deletions/mutations | 50%; 80% in Ph+ and Ph–like Controversial prognosis | Deletions of IKZF1 |
| DUX4-rearranged and ERG-deregulated | 3%-7% Favorable | ERG and IKZF1 deletions |
| MEF2D-rearranged ALL | 3%-4% Poor | |
| ZNF384-rearranged ALL | 6%-7% Intermediate | Partner gene EP300, CREBBP, TAF15, SYNRG, EWSR1, TCF3, and ARID1B |
| CDX2::UBTF | 2.7%-4% Poor | High expression of CDX2 and gain (1q); UBTF::ATXN7L3; CDX2-cis-deregulation |
| T-lineage ALL | ||
| TAL and LMO rearrangements/del(1)(p32), t(1;14)(p32;q11), t(1;7)(p32;q34), t(7;9)(q34;q32), t(11;14)(p15;q1), t(11;14)(p13;q1), t(7;11)(q35;p13) | 30%-40% Favorable and partly depending on additional lesions | SIL-TAL1 rearrangement, TCR rearrangements with TAL1, TAL2, LMO1, and LMO2 |
| HOXA aberrations/inv(7)(p15q34), t(7;7)(p15;q34), t(10;11)(p13;q14), t(v;11q23), del(9)(q34) | 20%-25% Outcome depending on additional lesions | TCR-HOXA rearrangement, MLLT10 and MLL rearrangements with various partners, SET-NUP214 rearrangement |
| TLX1-10q24 rearrangements/t(7;10)(q34;q24), t(10;14)(q24;q11) | 20%-30% No impact on prognosis | TCR-TLX11 rearrangement |
| ETP ALL | 10%-15% Unfavorable/controversial | Deregulation of myeloid transcription factors, of members of RAS pathway and of epigenetic regulators |
| TLX3-5q35 rearrangement/t(5;14)(q35;q32) | 10% No impact on prognosis | TLX3-BCL11B rearrangement |
| t(8;14)(q24;q11) | 1% Unfavorable | MYC involvement |
| ABL1 rearrangements | ∼3% Potentially targetable by tyrosine kinase inhibitors | NUP214, EML1; ETV6 |
| LYL/MEF2C rearrangement and immature cluster/t(7;19)(q34;p13), del(5)(q14) | 3%-17% Unfavorable; improved by intensive treatment | TCR with LYL1 MEF2C rearrangements |
| NKX2-1/NKX2-2 rearrangements/inv(14)(q11.2q13), t(7;14)(q34;q13), inv(14)(q13q32.33), t(14;20)(q11;p11) | 6% No impact on prognosis | TCR/IGH-NKX2- or NKX2-2 rearrangements |
Any prognostic statements should be considered carefully; because they depend on protocol, presence of other prognostic features, and are often based on small patient numbers.
WHO and ICC
The updated classifications are largely overlapping; the International Consensus Classification (ICC) identifies more distinct molecular categories16,24 (supplemental Table 3). A variety of methods would be required to establish the respective categories. This is not standard in most countries and beside the most relevant diagnostic characterizations summarized in Table 1 the new subcategories to date have limited immediate clinical consequences. Nevertheless, their identification is of interest in a research context or for potential use for selection of relapse therapy. Further WHO and ICC classifications with more detailed guidance on recommended and standardized methods for identification of subgroups are warranted.
MRD testing
MRD testing aims at detecting and quantifying residual blasts beyond the sensitivity of cytomorphology, which is around 5%.25,26 It is important to establish the target structures for MRD testing based on primary diagnostic material. The panel strongly advises to perform MRD evaluation in reference laboratories participating in standardization approaches.
The most common methods for MRD monitoring are MFC and real-time quantitative PCR (RQ-PCR). MFC can be applied to most cases (>90%), can reach a sensitivity of 0.1% to 0.01% (10–3 to 10–4) and results are promptly available. Disadvantages are as follows: (1) the samples must be rapidly analyzed, (2) the number of evaluable cells is small due to BM hypocellularity, (3) results could be misleading due to phenotypic shift, (4) the interpretation to some extent, is operator dependent, and (5) suboptimal sensitivity. The EuroFlow Consortium has standardized the procedures.27 Further efforts are ongoing to implement the use of next generation flow mainly for better interoperability and sensitivity.
Molecular MRD monitoring of fusion genes (eg, BCR::ABL1) has a sensitivity around 0.01%, is possible in about 40% of cases, is not patient-specific, relatively easy to perform and inexpensive. However, its accuracy is hampered by the variability in the number of RNA transcripts per leukemic cell. The EuroMRD Consortium has standardized MRD detection based on the BCR::ABL1.28
In most patients, a clonal Immunoglobulin/T-cell receptor (IG/TR) rearrangement can be identified for MRD evaluation. The technology is applicable to over 90% of cases29 and has been extensively standardized within the EuroMRD Consortium. Technical issues are as follows: (1) in the most immature forms, IG/TR gene rearrangements may not be found; (2) the designed probes may not be sufficiently sensitive; (3) clonal evolution may lead to false negative results; and (4) the amount of diagnostic DNA may be too low.
Any RQ-PCR may fail to precisely define the amount of residual disease in cases with a very low burden, that is, “positive-not-quantifiable”; their identification is an unmet need in clinical practice. These cases are being investigated with novel techniques, such as digital droplet PCR and NGS. For NGS neither methodology nor reporting is standardized. In part, similar technical issues apply as for IG/TR PCR and particularly the claimed higher sensitivity depends on the amount of DNA. MRD testing by NGS is so far not incorporated in the clinical practice in many countries.30 The various techniques are summarized in supplemental Table 4 and therapeutic application of MRD is discussed separately.6
PFs
Different risk subsets according to disease- and host-related factors can be identified.31 The individual prognosis must be refined by assessing MRD response24,25 and its integration with other prognostic factors (PFs). Patients without poor PF and/or with a favorable postinduction MRD course may represent 50%-60% of all cases and are defined as standard-risk (SR, 5-year overall survival (OS) >50%-60% and up to 70%-80% in selected good-risk subsets), whereas those with any poor PF and/or poor MRD response are classified as high risk (HR, 5-year OS 40% to 50%) (Table 3). This distinction is crucial to develop an effective risk-oriented treatment strategy. One approach is to select HR patients for an allogeneic stem cell transplantation (SCT) and/or other novel therapies, whereas SR patients are treated with chemotherapy with good to very good results and low risk of therapy-related mortality. Risk models depend on treatment protocol, differ among clinical studies and are periodically updated (Table 4).
Potential prognostic and predictive factors in adult ALL
| . | Risk factors . | Annotations . |
|---|---|---|
| Patient-related | ||
| Age (y) | >30-60 y (continuous variable) | Independent PF, usually not affecting risk model (age-adapted protocols) |
| >55 y (older adults and elderly) | ||
| Performance (ECOG) | >1 | Retrospective data; relevance in older patients |
| Disease-related | ||
| WBC (×109/L) | >30 (B), >100 (T) | Variably considered |
| Immunophenotype | Pro-B, CD20+ (B), pro/pre-T, ETP, and mature-T (T) | Variably considered |
| Cytogenetics and fluorescence in situ hybridization | Ph+, t(4;11), hypodiploidy, and complex∗ | Key prognostic elements; beside Ph+ and KMT2Ar variably considered |
| Genetics | BCR::ABL1+, KMT2Ar | Key prognostic elements |
| Ph-like, mutated CLRF2/TP53/JAK-STAT, adverse CNA profile (B), unmutated NOTCH1/FBWX7, and abnormal RAS/PTEN (T) | Variably considered | |
| Miscellaneous | CNS involvement | Occasionally considered |
| Poor treatment compliance and undue treatment reductions and delay | Retrospective data, of greater concern with pediatric-type protocols | |
| Pharmacogenomics (affecting antimetabolite disposition) | Data in children, not usually assessed in adults | |
| Immune marrow microenvironment | Investigational, for research purposes | |
| Drug response profiling | Investigational, for research purposes | |
| Treatment-response dynamics | ||
| Corticosteroid sensitivity (prephase) | Poor prednisone response (PB count ≥1 × 109/L at the end of prephase) | Historical relevance, occasionally considered |
| Early/incomplete blast cell clearance (BM morphology) | Day 8-15 or end of induction BM blasts ≥5% | Variably considered |
| Time to CR (number of courses) | >1 cycle (late CR) | Variably considered |
| MRD (molecular/flow cytometry) | MRD positivity (from end of induction onwards): ≥0.1%/0.01% after induction ≥0.01%/positive after/during consolidation and pre/post-allogeneic SCT | Key and unifying factor predicting outcome |
| . | Risk factors . | Annotations . |
|---|---|---|
| Patient-related | ||
| Age (y) | >30-60 y (continuous variable) | Independent PF, usually not affecting risk model (age-adapted protocols) |
| >55 y (older adults and elderly) | ||
| Performance (ECOG) | >1 | Retrospective data; relevance in older patients |
| Disease-related | ||
| WBC (×109/L) | >30 (B), >100 (T) | Variably considered |
| Immunophenotype | Pro-B, CD20+ (B), pro/pre-T, ETP, and mature-T (T) | Variably considered |
| Cytogenetics and fluorescence in situ hybridization | Ph+, t(4;11), hypodiploidy, and complex∗ | Key prognostic elements; beside Ph+ and KMT2Ar variably considered |
| Genetics | BCR::ABL1+, KMT2Ar | Key prognostic elements |
| Ph-like, mutated CLRF2/TP53/JAK-STAT, adverse CNA profile (B), unmutated NOTCH1/FBWX7, and abnormal RAS/PTEN (T) | Variably considered | |
| Miscellaneous | CNS involvement | Occasionally considered |
| Poor treatment compliance and undue treatment reductions and delay | Retrospective data, of greater concern with pediatric-type protocols | |
| Pharmacogenomics (affecting antimetabolite disposition) | Data in children, not usually assessed in adults | |
| Immune marrow microenvironment | Investigational, for research purposes | |
| Drug response profiling | Investigational, for research purposes | |
| Treatment-response dynamics | ||
| Corticosteroid sensitivity (prephase) | Poor prednisone response (PB count ≥1 × 109/L at the end of prephase) | Historical relevance, occasionally considered |
| Early/incomplete blast cell clearance (BM morphology) | Day 8-15 or end of induction BM blasts ≥5% | Variably considered |
| Time to CR (number of courses) | >1 cycle (late CR) | Variably considered |
| MRD (molecular/flow cytometry) | MRD positivity (from end of induction onwards): ≥0.1%/0.01% after induction ≥0.01%/positive after/during consolidation and pre/post-allogeneic SCT | Key and unifying factor predicting outcome |
∗Definition of complex karyotype: 5 or more chromosomal abnormalities excluding those patients with an established translocation.38
CNS, central nervous system; ECOG, Eastern Cooperative Oncology Group; and ETP, early thymic precursor.
Risk stratification models for allogeneic SCT in adult Ph/BCR::ABL1-negative (Ph–) ALL (European study groups)
| National Study Group . | Patient age (y) . | Risk stratification criteria∗ . | |||
|---|---|---|---|---|---|
| Postinduction MRD . | Cytogenetics/genetics† . | WBC (×109/L) . | Miscellaneous . | ||
| GMALL (Germany) | <55 | ≥0.01% after consolidation (wk 16 onward) | KMT2A+ | >30 (B) | Late CR, pro-B, early/mature-T |
| GIMEMA (Italy) | ≤65 | ≥0.01% after early consolidation (wk 10-16), any positivity (wk 22) | Adverse, KMT2A+ | >100 | Early/mature-T |
| HOVON (The Netherlands) | <40 | ≥0.01% after consolidation (wk 14-16) | Adverse KMT2A, hypodiploidy, complex karyotype | >30 (B), >100 (T) | Late CR |
| PALG (Poland) | <55 | ≥0.1% after induction ≥0.01% during/after consolidation | KMT2A+ | >30 (B), >100 (T) | CNS+ |
| UK NCRI ALL Group (United Kingdom) | <40 | ≥0.1% after induction and consolidation (mathematical risk model integrating MRD, cytogenetics and WBC) | Adverse | High count | — |
| FALL (Finland) | <45 | ≥0.1% after consolidation block B | Abn11q23, hypodiploidy | >100 | Late CR, d15 BM blasts >25% |
| RALL (Russia) | <55 | Positive during/after consolidation | t(4;11), t(1;19), KMT2A+ | — | Age >30 |
| SVALL (Sweden) | <65 | ≥0.1% after consolidation | Hypodiploidy, KMT2A+ | — | EOI BM blasts >5% |
| PETHEMA (Spain) | <55 (60 fit) | ≥0.1% after induction ≥0.01% during/after consolidation | — | — | — |
| GRAALL (France/Belgium/Switzerland) | <60 | ≥0.1% after induction at wk 6 or ≥0.01% after consolidation at wk 12 | — | — | — |
| CELL (Czech Republic) | <65 | ≥0.1% after induction ≥0.01% after consolidation | KMT2A+ | >30 (B) | Early/mature-T |
| National Study Group . | Patient age (y) . | Risk stratification criteria∗ . | |||
|---|---|---|---|---|---|
| Postinduction MRD . | Cytogenetics/genetics† . | WBC (×109/L) . | Miscellaneous . | ||
| GMALL (Germany) | <55 | ≥0.01% after consolidation (wk 16 onward) | KMT2A+ | >30 (B) | Late CR, pro-B, early/mature-T |
| GIMEMA (Italy) | ≤65 | ≥0.01% after early consolidation (wk 10-16), any positivity (wk 22) | Adverse, KMT2A+ | >100 | Early/mature-T |
| HOVON (The Netherlands) | <40 | ≥0.01% after consolidation (wk 14-16) | Adverse KMT2A, hypodiploidy, complex karyotype | >30 (B), >100 (T) | Late CR |
| PALG (Poland) | <55 | ≥0.1% after induction ≥0.01% during/after consolidation | KMT2A+ | >30 (B), >100 (T) | CNS+ |
| UK NCRI ALL Group (United Kingdom) | <40 | ≥0.1% after induction and consolidation (mathematical risk model integrating MRD, cytogenetics and WBC) | Adverse | High count | — |
| FALL (Finland) | <45 | ≥0.1% after consolidation block B | Abn11q23, hypodiploidy | >100 | Late CR, d15 BM blasts >25% |
| RALL (Russia) | <55 | Positive during/after consolidation | t(4;11), t(1;19), KMT2A+ | — | Age >30 |
| SVALL (Sweden) | <65 | ≥0.1% after consolidation | Hypodiploidy, KMT2A+ | — | EOI BM blasts >5% |
| PETHEMA (Spain) | <55 (60 fit) | ≥0.1% after induction ≥0.01% during/after consolidation | — | — | — |
| GRAALL (France/Belgium/Switzerland) | <60 | ≥0.1% after induction at wk 6 or ≥0.01% after consolidation at wk 12 | — | — | — |
| CELL (Czech Republic) | <65 | ≥0.1% after induction ≥0.01% after consolidation | KMT2A+ | >30 (B) | Early/mature-T |
CNS, central nervous system; EOI, end of induction.
Independent risk factors adopted in clinical trials for the definition of HR ALL and the consequent allocation to SCT; adapted from Giebel et al.63
Adverse cytogenetics/genetics (details to be found in single study protocols).
Clinical risk factors: age, WBC, and immunophenotype
Older age and high white blood cell counts (WBCs) are universally recognized as predicting poorer outcome.31 WBC is still a relevant and easily assessable PF. Cutoffs for HR B-ALL are often set >30 × 109/L. A WBC based stratification in T-ALL (>100 × 109/L) is not generally adopted. Other clinical PF are poor performance score (Eastern Cooperative Oncology Group [ECOG] >1), central nervous system–positive ALL in some trials, and undue reductions or delay of postinduction therapy.32,33 Pro–B-ALL is sometimes defined as HR. Some trials report a poorer prognosis in CD20-positive cases,34 unless associated with good MRD response35 or treated with rituximab. In T-ALL the prognosis is worse for the pro/pre-T (or ETP as subentity of immature T-ALL) and mature T subsets, whereas the cortical phenotype has a good prognosis.36 Outcome of HR T-ALL may be improved using pediatric-based chemotherapy in conjunction with SCT.37 In newly diagnosed ALL, the BM blast count differentiating ALL from lymphoblastic lymphoma is not considered as a prognostic feature but as a feature identifying a clinically relevant subtype which has an impact on practical management as outlined below. In relapsed ALL BM, blast count can be predictive for response rates.6
Cytogenetics
Recent cytogenetic classifications suffer from lack of information in up to 50% of the patients, low incidence of some aberrations, and interactions with other PFs.38-40 Good risk karyotypes such as t(12;21) are rare in adults. Most adults fall within an intermediate-risk (IMR) group. According to one of the published classifications, IMR includes the normal diploid subset, hyperdiploidy, and some other abnormalities. Monosomy 7, low hypodiploidy/near triploidy, and complex karyotype with 3-5 simultaneous abnormalities are allocated to a HR cytogenetic category as the t(4;11)/KMT2A rearranged ALL by several groups.40,41 For complex karyotype, a uniform definition is recommended (Table 3). There is no generally accepted cytogenetic classification for adult ALL and it remains open whether the prognostic impact of rare aberrations as outlined above is still valid in modern protocols.
Molecular genetics and genomics
Further genetic aberrations can exert significant prognostic effects (Table 2). Cases with iAMP21, Ph-like ALL,22,42,43 and an altered CNA profile can be associated with a higher relapse risk in both Ph-ALL44,45 and Ph/BCR::ABL1-positive (Ph+) ALL.46,47 An 8-gene CNA profile was validated in pediatric B-ALL.48 The good-risk CNA classifier consisted of no deletion of IKZF1, CDKN2A/B, PAR1, EBF1, and RB1; by an isolated deletion of ETV6, PAX5, and BTG1; or by a deletion of ETV6 with single additional deletion of either BTG1, PAX5, or CDK2A/B. Any other CNA combination exerted a negative prognostic effect. Adult studies identified KMT2A-AFF1, Ph-like, low hypodiploid/near haploid, BCL-2/MYC-rearranged, PAX5alt, ZNF384r- or ZNF384-like, and MEF2D-rearranged as HR or IMR-HR genotypes.49,50 CDX2 deregulated ALL with UBTF::ATXN7L3 has been described as a new unfavorable subtype,51-53 which may benefit from targeted therapies.54
In T-ALL, overexpression of HOX11L2 and ERG, lack of NOTCH1 and FBWX7 mutations, and presence of RAS or PTEN abnormalities yielded unfavorable outcomes.55,56 Mutations/alterations of TP53, JAK/STAT, BCL-2, and MYC are unrestricted to cell lineage and may confer poor prognosis. As for cytogenetics, it is important to note that prognostic annotations are often based on small patient numbers and not always based on outcomes with current protocols.
Early response dynamics
Patients with poor blast cell clearance and late responders requiring more than one chemotherapy course to complete remission (CR) have an inferior outcome. The most useful prognostic information for relapse risk and OS and individual risk stratification comes from MRD testing.2,25,57-59 As a general rule, with RQ-PCR (sensitivity of 0.01%), good prognosis patients reach MRD levels <0.1% to <0.01% (better if negative) at end of induction (weeks 4-6) and <0.01% (better if negative) following 1 to 3 early consolidation courses around weeks 10-16. Therapeutic consequences are discussed separately.6
Integrated risk models
In children, an independent genotype-specific effect on MRD-related outcomes improved the risk stratification.60 In adults, one study demonstrated the usefulness of a mixed MRD and genetic risk classification, showing the independent prognostic effect of a 4-gene classifier (favorable: lack of KMT2A rearrangements and ΔIKZF1 in B-ALL; mutated NOTCH1/FWBX7 without RAS/PTEN abnormalities in T-ALL55). In addition, a comprehensive risk model combined WBC, cytogenetics, and MRD results.61 Both models have not been adopted uniformly.
Recommendations for analysis of PFs and risk stratification
MRD testing is recommended in all cases to optimize risk stratification.59,62 In a European survey, MRD was the only PF shared by 11 national study groups to define HR ALL and to make a decision about SCT indication63 (Table 4). No other PF achieved the same level of consensus, although adverse genetics rank high in HR definitions (8/11 groups). Altogether, it is strongly recommended to perform MRD analysis in all patients with ALL.62 New evidence may prove the independent prognostic power of several genetic abnormalities that in different combinations concur to define novel HR subsets. Appropriate diagnostic identification and clinical data in relevant patient numbers with modern protocols are a prerequisite. New combined risk models are therefore recommended for research purposes to improve the diagnostic and prognostic platforms.
Response criteria and definition of outcomes
Cytologic response in BM
Although criteria for CR are uniformly used, CR with incomplete recovery (CRi) can be an endpoint in clinical trials. Failure or partial remission (PR) (Table 5) can be differentiated or combined. For PR cases a correlation with MRD testing is recommended. If technical criteria for MRD testing are met, PR or CRi with negative MRD status can be considered as CR. On the other hand, patients with MRD of 1% or higher can be considered as failure.66 For future clinical trials, a standardized terminology is requested (Table 5). Publications should also define time point (TP) of response assessment and avoid reporting of cumulative response rates.
Response criteria for ALL
| Category . | Definition . |
|---|---|
| Hematologic response criteria | |
| CR∗ | BM blasts <5% |
| Absence of extramedullary disease | |
| Absolute neutrophil count >1 × 109/L | |
| Platelet count >100 × 109/L (independence of red cell transfusions) | |
| If available: MRD <1%66 | |
| CRi† | All CR criteria except for residual thrombocytopenia |
| <100 × 109/L or neutropenia <1 × 109/L | |
| If available: MRD <1%66 | |
| Morphologic leukemia-free state‡ | BM blasts <5% |
| Absence of extramedullary disease | |
| If available: MRD <1%66 | |
| PR§ | Relevant in the setting of phase 1 and 2 clinical trials only; all hematologic criteria of CR; decrease of BM-blast percentage from 5% to 25%; and decrease of pretreatment BM-blast percentage by at least 50% |
| If available: CR if MRD <1%66 | |
| Failure | None of the above If available: MRD ≥1%66 |
| MRD response criteria (variant 1)|| | |
| Complete MRD response | No detectable MRD¶ |
| MRD failure | MRD above 0.01% (ie, 10–4) |
| MRD other | |
| Negative | MRD negative with insufficient sensitivity |
| Positive/intermediate | MRD positive below 0.01%, quantifiable |
| MRD positive below 0.01%, nonquantifiable | |
| MRD positive, nonquantifiable | |
| MRD response criteria (variant 2)|| | |
| MRD complete response | No detectable MRD¶ |
| MRD persistence | Any quantifiable MRD |
| Criteria for extramedullary response assessment | Published criteria for NHL65 PET in case of CRu/PR according to published criteria for NHL69 |
| Category . | Definition . |
|---|---|
| Hematologic response criteria | |
| CR∗ | BM blasts <5% |
| Absence of extramedullary disease | |
| Absolute neutrophil count >1 × 109/L | |
| Platelet count >100 × 109/L (independence of red cell transfusions) | |
| If available: MRD <1%66 | |
| CRi† | All CR criteria except for residual thrombocytopenia |
| <100 × 109/L or neutropenia <1 × 109/L | |
| If available: MRD <1%66 | |
| Morphologic leukemia-free state‡ | BM blasts <5% |
| Absence of extramedullary disease | |
| If available: MRD <1%66 | |
| PR§ | Relevant in the setting of phase 1 and 2 clinical trials only; all hematologic criteria of CR; decrease of BM-blast percentage from 5% to 25%; and decrease of pretreatment BM-blast percentage by at least 50% |
| If available: CR if MRD <1%66 | |
| Failure | None of the above If available: MRD ≥1%66 |
| MRD response criteria (variant 1)|| | |
| Complete MRD response | No detectable MRD¶ |
| MRD failure | MRD above 0.01% (ie, 10–4) |
| MRD other | |
| Negative | MRD negative with insufficient sensitivity |
| Positive/intermediate | MRD positive below 0.01%, quantifiable |
| MRD positive below 0.01%, nonquantifiable | |
| MRD positive, nonquantifiable | |
| MRD response criteria (variant 2)|| | |
| MRD complete response | No detectable MRD¶ |
| MRD persistence | Any quantifiable MRD |
| Criteria for extramedullary response assessment | Published criteria for NHL65 PET in case of CRu/PR according to published criteria for NHL69 |
This table is modified from Döhner et al.71
All criteria need to be fulfilled; marrow evaluation should be based on a count of 200 nucleated cells in an aspirate with spicules; if ambiguous, consider repeat exam after 5 to 7 days; a marrow biopsy should be performed in cases of dry tap, or if no spicules are obtained; no minimum duration of response required.
CRi is of value in protocols using intensified induction or double induction strategies, in which hematologic recovery is not awaited, but intensive therapy will be continued. In such protocols, CR may even not be achieved during the entire treatment plan. In these instances, the overall remission rate should include CR and CRi patients.
This category may be useful in the clinical development of novel agents within phase 1 clinical trials, in which a transient morphologic leukemia-free state may be achieved at the time of early response assessment.
Marrow should not merely be aplastic; at least 200 cells should be enumerated, or cellularity should be at least 10%.71 Any PR should be confirmed or falsified by parallel MRD assessment.
Confirmation of any MRD response requires the application of standardized methods with minimum technical requirements29 in reference laboratories.
Confirmation of negative MRD requires that technical requirements for establishment of sensitivity (usual: 0.01%) of each individual TP are fulfilled.
MRD response
Response criteria are redefined by integrating MRD assessment. Usually, MRD assessment is only considered in patients with hematologic CR or CRi. In any case the reference population should be clearly stated and evaluable and nonevaluable MRD results should be named. Complete MRD response is defined as no detection of MRD, with a minimum sensitivity of 0.01% at the respective TP.29 MRD persistence is defined as quantifiable MRD, usually 0.01% or greater. Some groups consider any MRD positivity as prognostically relevant; others consider thresholds like 0.1% or 0.01% for different TP or different treatment consequences. Beyond these categories additional MRD results may be reported, including nonquantifiable MRD at different levels of sensitivity or negative MRD with insufficient sensitivity.67,68 For TP with nonquantifiable MRD, additional digital droplet PCR or NGS testing may be used to separate true negative from false positive results.68 Of note, any negative MRD result without the corresponding sensitivity level at a distinct TP does not fulfill technical requirements. The latter are different for MFC, PCR of IG/TR,29 PCR of fusion genes,28 and often not reported for NGS based methods. MRD results should include information on the material tested and specifics: PB vs BM, level, and status of MRD that is, positive vs negative or nonevaluable. Two variants of MRD based response assessment are given in Table 5. The panel is in favor or variant 1 because the exact MRD level (not considered in variant 2) is relevant for any treatment decisions and for comparability of reported results.
For Ph+ ALL MRD, response criteria are often derived from trials for chronic myeloid leukemia. Whether these categories are meaningful for ALL remains open to discussion. Approaches with an ALL-specific definition are considered. Furthermore, discrepancies between BCR::ABL1 based MRD results and other methods may occur if 2 methods are applied in parallel. BCR::ABL1-based MRD may remain positive whereas IG/TR MR is negative. This is probably due to multilineage involvement with BCR::ABL1 also in myeloid precursors. Clinical consequences are not clear so far, but the ICC has now integrated 2 subgroups with BCR::ABL1 (lymphoid and multilineage).16 For MRD–based treatment decisions it is recommended to use in addition to BCR::ABL1–based MRD assessment one additional method that is MFC or IG/TR.
Extramedullary response
Between 20% to 70% of patients with ALL show extramedullary involvement at diagnosis depending on subtype. Type and extent of involvement should be documented. These localizations should have regressed in size at the TP of CR confirmation. Standard criteria for NHL are recommended for classification.65,69 In case of persistent extramedullary involvement after induction and early consolidation, Positron Emission Tomography (PET) analysis can be considered. This approach raises challenges in dose dense treatment protocols for ALL due to the recommended treatment-free interval. For the purposes of clinical trials, patients with CRi or PR, PET negative may be considered as CR. Therapeutic consequences from positive PET remain to be defined.
TP for response assessment
The TPs depend on protocol and treatment consequences. Usually, a CR should be achieved after the initial 1 to 3 phases of standard therapy depending on the protocol. Cases not achieving a CR after this period are considered as primary failures. This definition is important for the purpose of clinical trials. During the follow-up, BM response may be assessed every 2 to 3 months until end of maintenance. Further controls may rely on PB. More frequent assessments are recommended in patients with low level MRD of any level or PET positivity at any TP. Any treatment change based on MRD should be followed by MRD assessment of response. Further essential outcome parameters and major approaches for clinical trials are described in the supplement (supplemental Table 5).
Summary and outlook
The management of ALL is often organized by national study groups and international consortia.70 Their reference laboratories and/or associated biobanks together with clinical data bases are essential for research on disease biology and prognostication and are sources of reliable real-world data. Reference laboratories are crucial to establish and maintain high standards for diagnostic procedures and biologic characterization as basis for risk stratification and optimal management. Cytomorphology, immunophenotyping, and the identification of prognostically relevant molecular markers together with measurement of MRD are the basis for diagnosis and risk adapted therapy.
Further genetic characterization of samples from biobanks has identified many new subtypes of ALL. The prognostic and therapeutic impact, however, often remains open for current treatment regimens and diagnostic identification is not part of standard of care in many countries. It will be essential to define reliable and cost-effective diagnostic procedures to improve classification of ALL. On this basis, future risk stratification may integrate molecular markers, potential treatment targets, risks for relapse, and toxicities as well as the dynamics of MRD. Furthermore, improved understanding of disease biology can contribute to the development of new and targeted precision medicine approaches.
For comparability of data on an international level it will be essential to use uniform and standardized criteria for reporting of results. This applies particularly for the use of MRD as endpoint of clinical trials and for any MRD-based treatment decisions.
Although there is no standard risk model for adult ALL, all experts agree that depending on treatment protocols and clinical subgroup risk-adapted approaches for management are standard.6 Future risk models may also integrate pharmacogenomics to predict and potentially avoid toxicities and in vitro sensitivity testing for identification of potentially active drugs for later-line approaches in relapsed/refractory ALL.
Authorship
Contribution: All authors reviewed literature, contributed to writing parts of the manuscript, reviewed the manuscript, and approved the final manuscript for submission.
Conflict-of-interest disclosure: N.G. has received institutional research funding from Amgen, Clinigen, Incyte, Jazz, Novartis, Pfizer, and Servier and received speaker honoraria or fees for advisory board participation from Amgen, AstraZeneca, Autolus, Celgene, Clinigen, Gilead, Incyte Jazz, Novartis, Pfizer, and Servier. N.B. received honoraria from Amgen, Incyte, Jazz Pharma, Novartis, Pfizer, and Servier and research grants from Amgen, Novartis, Incyte, Jazz Pharma, and Sanofi. S.C. served on the advisory boards of Incyte, Pfizer, and AbbVie and as a consultant for Gilead and Amgen. H.D. received research funding from Amgen, Astellas, Celgene, Incyte, Jazz, Pfizer, and Servier and honoraria from Daiichi Sankyo, Incyte, Jazz, and Servier. M.D. received research support from AstraZeneca. R.F. received speaker honoraria from Amgen, Novartis, and AstraZeneca and served on the advisory board of Merck Sharp & Dohme. S.G. received speaker honoraria from or joined the advisory boards of Amgen, Novartis, Pfizer, and Gilead; received speaker honoraria from Angelini and travel grants from Gilead; and served on advisory boards of Zentiva. M.H. received research support from Novartis, travel support from AbbVie and BeiGene and speaker honoraria from Servier; joined advisory boards of Servier; and was part of advisory boards of Amgen and Clinigen. D.I.M. provided consultancy for Pfizer, Novartis, and Kite. O.O. received speaker honoraria from, joined advisory boards of, and received research grants from Incyte; received research support from Amgen and Celgene; and received honoraria for advisory boards from Autolus. A.R. received research support from Servier. P.R. received research grants from Pfizer and Incyte. J.R. received speaker honoraria from and joined advisory boards of Amgen, Pfizer, Shire, Ariad, and Incyte; joined advisory boards of Takeda; and received research support from Amgen. R.B. joined advisory boards of Novartis and Kite/Gilead and received honoraria or travel support from Amgen, Incyte, Servier, Jazz, and Pfizer. The remaining authors declare no competing financial interests.
Correspondence: Nicola Gökbuget, Department of Medicine II, University Hospital, Theodor Stern Kai 7, 60590 Frankfurt, Germany; email: goekbuget@em.uni-frankfurt.de.
References
Author notes
The online version of this article contains a data supplement.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal