In this issue of Blood, Dickeson et al propose a new pathogenic mechanism for a subset of patients with a swelling disorder called hereditary angioedema (HAE).1
The contact system, which can lead to blood coagulation and inflammation,2 is the neglected stepchild of these pathways. The reason for this neglect is that people with a nonfunctional contact system have no obvious phenotype—they do not bleed or lack inflammatory responses. However, people with an abnormally activated contact system can present with HAE, a disorder stemming from overproduction of bradykinin, a nonapeptide that regulates blood vessel permeability. Excess bradykinin leads to tissue swelling (edema), and depending on where this edema occurs, HAE can be life threatening. For example, tongue swelling can lead to asphyxiation.
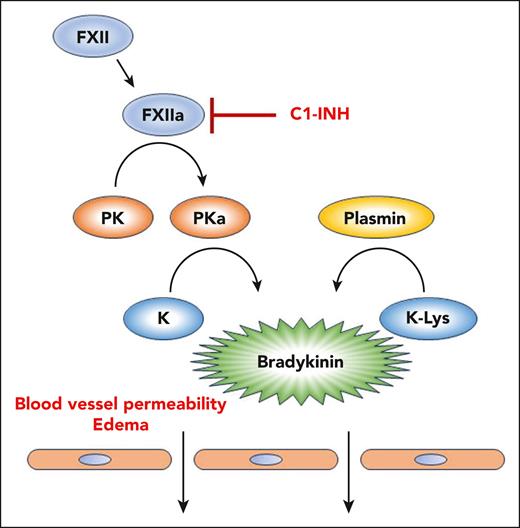
The molecular basis for HAE is most often insufficient or nonfunctional C1 inhibitor (C1-INH). This protein inhibits activated factor XIIa (FXIIa), and reduced C1-INH activity can lead to an unregulated contact system, overproduction of bradykinin, and HAE (see figure). Other genetic causes of HAE include mutations in FXII, heparan sulfate 3-O-sulfotransferase 6, angiopoietin-1, myoferlin, and plasminogen (all classified as HAE normal C1) (reviewed in the study by Dickeson et al).
The kallikrein-kinin system and HAE. Activation of FXII to FXIIa leads to conversion of plasma prekallikrein (PK) to active plasma kallikrein (PKa). PKa can cleave high- and low-molecular-weight kininogens (K) to release bradykinin. Under normal conditions, C1-INH inhibits FXIIa and prevents propagation of the pathway. Many cases of HAE are caused by insufficient or inactive C1-INH. The article by Dickeson et al shows that a methionine-to-lysine missense mutation in kininogen (K-Lys), identified in a family of patients with HAE, renders it susceptible to cleavage by the fibrinolytic protease plasmin. This mutation thus causes excessive bradykinin formation and can lead to HAE.
The kallikrein-kinin system and HAE. Activation of FXII to FXIIa leads to conversion of plasma prekallikrein (PK) to active plasma kallikrein (PKa). PKa can cleave high- and low-molecular-weight kininogens (K) to release bradykinin. Under normal conditions, C1-INH inhibits FXIIa and prevents propagation of the pathway. Many cases of HAE are caused by insufficient or inactive C1-INH. The article by Dickeson et al shows that a methionine-to-lysine missense mutation in kininogen (K-Lys), identified in a family of patients with HAE, renders it susceptible to cleavage by the fibrinolytic protease plasmin. This mutation thus causes excessive bradykinin formation and can lead to HAE.
In the work by Dickeson et al, the authors explore a missense mutation in the kininogen gene kng1 that leads to HAE.3 This mutation substitutes a lysine for a methionine adjacent to the N-terminal position in kininogen (K-Lys) where active plasma kallikrein (PKa) normally cleaves. Possibilities for an HAE mechanism resulting from this mutation included production of an altered and more active form of bradykinin or an increased susceptibility of kininogen to cleavage by PKa. However, the exact mechanism had not been established.
Dickeson et al show that the lysine substitution creates a novel cleavage site for plasmin, a fibrinolytic protease. Cleavage by plasmin results in a form of bradykinin with an additional amino acid at the N-terminus (Lys-bradykinin, kallidin). Lys-bradykinin binds to bradykinin receptors and can also be converted to bradykinin by aminopeptidase B. Plasmin is present in blood and plays a critical role in removing microclots that can disrupt blood flow and promote inflammation.4 Because plasmin is ubiquitously present in blood, having a kininogen that is highly susceptible to plasmin cleavage could promote bradykinin production and initiate an HAE attack.
Complementing these results, it has been found that a mutation in the plasminogen gene (plg), in which a lysine is replaced by a glutamic acid, can also cause HAE.5,6 The plasmin variant stemming from this mutation is more effective in cleaving kininogen and releasing bradykinin.7 This work, coupled with the new results on the K-Lys mutation, shows that plasmin-catalyzed cleavage of kininogen can be a disease-causing mechanism for HAE. Thus, not all patients with HAE will benefit from treatment with FXIIa or PKa inhibitors; if plasmin is involved, bradykinin receptor antagonists or fibrinolytic inhibitors would be more effective.
Edema can occur in contexts other than HAE. For example, treatment of patients with Alzheimer disease with anti–β-amyloid antibodies can lead to amyloid-related imaging abnormalities that are accompanied by edema.8,9 This edema can be serious and therapy limiting in some cases. In addition, increases in the levels of C1-INH have been associated with disease severity in COVID-19.10 Thus, a better understanding of mechanisms by which the kallikrein-kinin pathway is regulated and how edema can occur could have implications beyond HAE. The results also have implications for the use of tissue plasminogen activator (tPA) as a thrombolytic therapy. With tPA treatment, the production of abnormally high plasmin levels could trigger edema even in the absence of mutations in the plg or kng1 gene.
Genetic analysis has revolutionized our understanding of disease. However, in many cases, knowing the genetic mutation that causes a disease does not immediately reveal the mechanism. The article by Dickeson et al is an elegant example of how pursuing the biochemical mechanism based on a genetic mutation can provide insight into pathogenesis. The hope in general, and in this particular case, is that mechanistic understanding can lead to specific and effective therapeutic intervention in genetically defined patients.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal