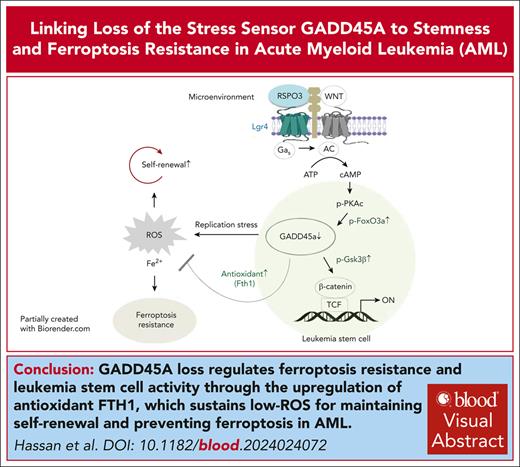
Key Points
GADD45A loss has a dual role in promoting DNA damage while upregulating antioxidants to sustain low ROS essential to safeguard self-renewal.
Deletion of GADD45A enhances leukemia-initiating activity and therapy resistance by suppressing iron and ROS accumulation and ferroptosis.
Visual Abstract
The overall prognosis of acute myeloid leukemia (AML) remains dismal, largely because of the inability of current therapies to kill leukemia stem cells (LSCs) with intrinsic resistance. Loss of the stress sensor growth arrest and DNA damage-inducible 45 alpha (GADD45A) is implicated in poor clinical outcomes, but its role in LSCs and AML pathogenesis is unknown. Here, we define GADD45A as a key downstream target of G protein-coupled receptor (LGR)4 pathway and discover a regulatory role for GADD45A loss in promoting leukemia-initiating activity and oxidative resistance in LGR4/HOXA9-dependent AML, a poor prognosis subset of leukemia. Knockout of GADD45A enhances AML progression in murine and patient-derived xenograft (PDX) mouse models. Deletion of GADD45A induces substantial mutations, increases LSC self-renewal and stemness in vivo, and reduces levels of reactive oxygen species (ROS), accompanied by a decreased response to ROS-associated genotoxic agents (eg, ferroptosis inducer RSL3) and acquisition of an increasingly aggressive phenotype on serial transplantation in mice. Our single-cell cellular indexing of transcriptomes and epitopes by sequencing analysis on patient-derived LSCs in PDX mice and subsequent functional studies in murine LSCs and primary AML patient cells show that loss of GADD45A is associated with resistance to ferroptosis (an iron-dependent oxidative cell death caused by ROS accumulation) through aberrant activation of antioxidant pathways related to iron and ROS detoxification, such as FTH1 and PRDX1, upregulation of which correlates with unfavorable outcomes in patients with AML. These results reveal a therapy resistance mechanism contributing to poor prognosis and support a role for GADD45A loss as a critical step for leukemia-initiating activity and as a target to overcome resistance in aggressive leukemia.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous and malignant clonal disease initiated by self-renewing leukemia stem cells (LSCs) arising from the transformation of hematopoietic stem cells (HSCs) or committed progenitor cells.1 Preleukemic cells transformed from HSCs by oncogenes (e.g., MLL-AF9) can develop a highly aggressive subtype of AML with chemoresistance and expressing genes associated with inferior survival in patients with AML.2,3 Current chemotherapy is insufficient in targeting the intrinsic self-renewal mechanism in LSCs, which are a source of therapy resistance and relapse in AML.4 We have previously shown that the heterogeneity of LSC self-renewal potential can be driven by distinct or partially overlapping regulatory mechanisms, where the interaction of G-protein–coupled receptor LGR4 and its ligand R-spondin 3 (RSPO3) is required for regulation of HSC-derived LSCs through β-catenin activation in AML that depends on HOXA9, a predictor of poor prognosis in a wide variety of human malignancies.5-10 High LSC activity in a heterogeneous LSC pool is associated with poor clinical response to therapy.11,12 It is thus of critical importance to identify key regulators that control leukemia-initiating cell activity essential for acquisition of an aggressive leukemia phenotype and therapeutic resistance.
Here, we found that LGR4 negatively regulates GADD45A, which acts as a crucial stress sensor in response to replication stress-induced DNA damage and genotoxic stress-induced ferroptosis. Inhibition of Lgr4 in HSC-derived leukemic cells induces Gadd45a expression, and deletion of Gadd45a in AML LSCs prevents the reduction of β-catenin induced by Lgr4 depletion, thus impacting a key regulator of self-renewal in hematopoietic development and malignancies. Gadd45a-deleted mice have a normal hematologic phenotype but display genomic instability, impairment in DNA repair, and accumulation of DNA damage and mutations in HSCs.13,14 This is in line with our observation in LSCs, where deletion of Gadd45a causes mutations in DNA repair genes and replication stress while upregulating antioxidants to attenuate stress-induced reactive oxygen species (ROS) and iron production, leading to resistance to ferroptosis (an iron- and ROS-dependent cell death).15 DNA methylation of GADD45A is predictive of poor overall survival in AML and is associated with DNMT3A and IDH1/2 mutations, while downregulation of GADD45A via FLT3-ITD mutation contributes to the myeloid differentiation block.16,17 In contrast, elevated GADD45A expression in normal HSCs induces differentiation via activation of p38 mitogen-activated protein kinase signaling that leads to the removal of damaged HSCs preventing malignant transformation.14,18 GADD45A thus acts in a cell context–specific manner and is differentially required in normal and malignant hematopoiesis. However, the functional consequence of GADD45A loss in AML, especially in LSCs, has been largely unexplored.
Materials and methods
Additional details are provided in the supplemental Methods (available on the Blood website).
Mice
Generation and use of Gadd45a knockout (Gadd45a−/−) and wild-type (Gadd45a+/+) mice have been described previously.19 MLL-AF9 AML mouse models and patient-derived xenografts (PDXs) were established by transplanting leukemic cells into female C57BL/6 or NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (Australian BioResources), as previously described.5,8,10,20 All animal studies were in strict compliance with the Institutional Animal Care and Ethics Committee–approved protocols.
Results
GADD45A is downregulated in the LGR4 pathway acting downstream of LGR4/p-PKAc/p-FOXO
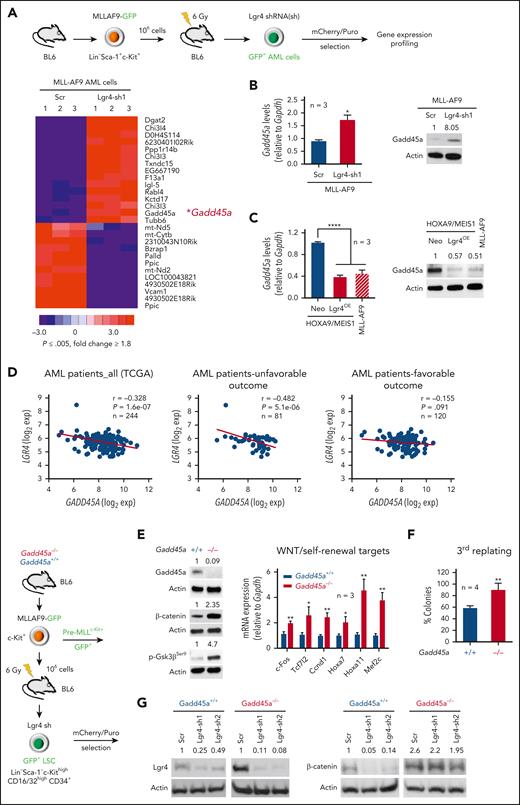
Using a stringent cutoff, our microarray data identified 15 genes upregulated by short hairpin RNA (shRNA)–mediated Lgr4 knockdown (Lgr4-sh1)5 in MLL-AF9–induced AML. Briefly, leukemic cells were generated by transducing HSC-enriched LSK cells (Lin–Sca-1+c-Kit+)1 with MLL-AF9–green fluorescent protein (GFP) and transplanting resultant preleukemic cells into sublethally irradiated C57BL/6 syngeneic mice. After developing AML, GFP+ leukemic cells flow sorted from mouse bone marrow were lentivirally transduced with Lgr4 shRNA and selected for the microarray experiment (Figure 1A; supplemental Figure 1A).
Gadd45a is negatively regulated by Lgr4 and its deletion retains β-catenin activity even in the absence of Lgr4. (A) Heat map analysis of microarray data (n = 3, P ≤ .005, fold change ≥ 1.8) showing differentially expressed genes induced by Lgr4 knockdown in MLL-AF9–induced AML cells. (B) Quantitative polymerase chain reaction (qPCR) (n = 3) and Western blots confirming upregulation of Gadd45a induced by Lgr4 knockdown in MLL-AF9 leukemic cells carrying scrambled control (Scr) vs Lgr4 shRNA1 (sh1). Data are given as mean ± SD. ∗P < .05, unpaired t-test. (C) qPCR (n = 3) and Western blots showing downregulation of Gadd45a induced by Lgr4 overexpression in HOXA9/MEIS1 leukemic cells, compared with endogenous expression of Gadd45a in MLL-AF9 leukemic cells. Data are given as mean ± SD. ∗∗∗∗P < .0001, 1-way analysis of variance (ANOVA). (D) Analysis of TCGA dataset,21,22 revealing the correlation between LGR4 and GADD45A expression in all patients with AML (n = 244, r = −0.328, P = 1.6e-07), patients with AML with unfavorable outcome (n = 81, r = −0.482, P = 5.1e-06), and patients with AML with favorable outcome (n = 120, r = −0.155, P = .091). (E) Western blots confirming efficient Gadd45a knockout with a resultant increase in β-catenin and inactive phospho-Ser9-Gsk3β (p-Gsk3βSer9) in GFP+ pre-MLLc-Kit+ cells. qPCR showing upregulation of Wnt/self-renewal target genes induced by Gadd45a deletion (n = 3). Data are given as mean ± SD. ∗P < .05, ∗∗P < .005, unpaired t-test. (F) Colony formation of Gadd45a−/− vs Gadd45a+/+ pre-MLLc-Kit+ cells. The percentage of colonies (relative to Gadd45a−/−) at the third round of serial replating is shown. n = 4 independent experiments. Data are given as mean ± SD. ∗∗P < .005, unpaired t-test. (G) Western blots confirming efficient Lgr4 knockdown with a resultant change of endogenous β-catenin expression in response to Gadd45a knockout in LSCs (Lin–Sca-1–c-KithighCD16/32highCD34+), flow sorted from the bone marrow (BM) of AML mice following transplantation with pre-MLLc-Kit+ cells.
Gadd45a is negatively regulated by Lgr4 and its deletion retains β-catenin activity even in the absence of Lgr4. (A) Heat map analysis of microarray data (n = 3, P ≤ .005, fold change ≥ 1.8) showing differentially expressed genes induced by Lgr4 knockdown in MLL-AF9–induced AML cells. (B) Quantitative polymerase chain reaction (qPCR) (n = 3) and Western blots confirming upregulation of Gadd45a induced by Lgr4 knockdown in MLL-AF9 leukemic cells carrying scrambled control (Scr) vs Lgr4 shRNA1 (sh1). Data are given as mean ± SD. ∗P < .05, unpaired t-test. (C) qPCR (n = 3) and Western blots showing downregulation of Gadd45a induced by Lgr4 overexpression in HOXA9/MEIS1 leukemic cells, compared with endogenous expression of Gadd45a in MLL-AF9 leukemic cells. Data are given as mean ± SD. ∗∗∗∗P < .0001, 1-way analysis of variance (ANOVA). (D) Analysis of TCGA dataset,21,22 revealing the correlation between LGR4 and GADD45A expression in all patients with AML (n = 244, r = −0.328, P = 1.6e-07), patients with AML with unfavorable outcome (n = 81, r = −0.482, P = 5.1e-06), and patients with AML with favorable outcome (n = 120, r = −0.155, P = .091). (E) Western blots confirming efficient Gadd45a knockout with a resultant increase in β-catenin and inactive phospho-Ser9-Gsk3β (p-Gsk3βSer9) in GFP+ pre-MLLc-Kit+ cells. qPCR showing upregulation of Wnt/self-renewal target genes induced by Gadd45a deletion (n = 3). Data are given as mean ± SD. ∗P < .05, ∗∗P < .005, unpaired t-test. (F) Colony formation of Gadd45a−/− vs Gadd45a+/+ pre-MLLc-Kit+ cells. The percentage of colonies (relative to Gadd45a−/−) at the third round of serial replating is shown. n = 4 independent experiments. Data are given as mean ± SD. ∗∗P < .005, unpaired t-test. (G) Western blots confirming efficient Lgr4 knockdown with a resultant change of endogenous β-catenin expression in response to Gadd45a knockout in LSCs (Lin–Sca-1–c-KithighCD16/32highCD34+), flow sorted from the bone marrow (BM) of AML mice following transplantation with pre-MLLc-Kit+ cells.
Among 15 upregulated genes, Gadd45a has been reported to negatively regulate β-catenin activity in epithelial tumors through promoting Gsk3β activation via dephosphorylation of Ser9.23 We found that Lgr4 depletion increased Gadd45a expression in leukemic cells (Figure 1B). We have recently documented an indispensable role for Lgr4 in regulating self-renewal through cooperation with Hoxa9 in aggressive AML.5 Overexpression of Lgr4 (Lgr4OE) in HOXA9/MEIS1-transformed LSK cells not only produces leukemia with a shortened latency, similar to that in MLL-AF9–induced AML, but also upregulates β-catenin, reaching a level comparable to that in MLL-AF9–transformed LSK cells.5 Consistently, overexpression of Lgr4 downregulated Gadd45a in HOXA9/MEIS1 leukemic cells, reaching a level similar to that in MLL-AF9 leukemic cells (Figure 1C; supplemental Figure 1B). This is in line with a negative correlation between LGR4 and GADD45A in 244 patients with AML from The Cancer Genome Atlas (TCGA) dataset21 that was mainly attributed to patients with unfavorable outcome (Figure 1D). These data indicate LGR4 as a negative regulator of GADD45A in poor-prognosis AML.
We next investigated the mechanism for Lgr4-mediated downregulation of Gadd45a. The TCGA dataset revealed a positive correlation between FOXO3 and GADD45A (Supplemental Figure 1C), which was consistent with a negative correlation between LGR4 and GADD45A in patients with AML with an unfavorable outcome (Figure 1D). This implicates FOXO3-mediated regulation of GADD45A in poor-prognosis AML. Knockdown of Lgr4 decreased the phosphorylation of FoxO3a in MLL-AF9 LSCs (Lin–Sca-1–c-KithighCD16/32highCD34+)1 (supplemental Figure 1D). The phosphorylation of FOXO3A, and its consequent cytoplasmic localization and inactivation, is a poor prognostic factor in AML, particularly in those with FLT3 mutations.24 Consistent with the role of p-PKAc as a downstream effector of Lgr4 pathway,5 the specific inhibitor of PKA, PKI, reduced the phosphorylation of FoxO3a, leading to increased Gadd45a in MLL-AF9 LSCs and in human MLL-AF9 AML (THP-1) cells (supplemental Figure 1E-F). These results support p-PKAc/p-FOXO3A–mediated downregulation of GADD45A.
GADD45A loss increases the WNT/self-renewal activity through phosphorylation of GSK3β
To define a direct role for Gadd45a loss in Lgr4/β-catenin signaling, we tested the effect of Gadd45a deletion on Wnt/β-catenin activity in MLL-AF9-GFP transformed HSC/progenitor-enriched c-Kit+ cells,10 which were isolated from the bone marrow of Gadd45a knockout mice.19 Compared with wild-type Gadd45a (Gadd45a+/+), deletion of Gadd45a (Gadd45a−/−) in GFP+ preleukemic MLL-AF9 cells (pre-MLLc-Kit+) increased endogenous β-catenin and inactivated/phosphorylated Gsk3β (p-Gsk3βSer9), while upregulating key self-renewal signature genes, such as WNT/β-catenin targets (c-Fos, Tcf7l2, and Ccnd1)5 and MLL-fusion targets (Mef2c and HoxA cluster genes Hoxa7 and Hoxa11)1 (Figure 1E). In contrast, knockdown of β-catenin repressed expression of self-renewal signature genes and reduced colony formation in Gadd45a−/− cells (supplemental Figure 1G). Gadd45a−/− cells exhibited enhanced serial-replating capacity, indicative of increased self-renewal (Figure 1F), whereas overexpression of Gadd45a inhibited leukemic cell growth (supplemental Figure 1H). Likewise, knockdown of LGR4 in human AML (THP-1) cells elevated endogenous GADD45A expression, whereas deletion of GADD45A via CRISPR/Cas9 increased inactive p-GSK3βSer9 and nuclear nonphosphorylated (active) β-catenin as well as key WNT/self-renewal target genes (supplemental Figure 2A-D). To further determine the role of Gadd45a loss as a downstream effector of LGR4 signaling, we generated Gadd45a+/+ vs Gadd45a−/− LSCs expressing or depleting Lgr4, by transplanting pre-MLLc-Kit+ cells into syngeneic mice to develop AML followed by lentiviral transduction of LSCs with Lgr4 shRNAs. We observed that knockdown of Lgr4 with 2 independent shRNAs (sh1 and sh2)5 decreased β-catenin expression in Gadd45a+/+ LSCs, but not in Gadd45a−/− LSCs (Figure 1G). These data suggest that loss of GADD45A sustains β-catenin levels and WNT/self-renewal activity through phosphorylation of GSK3β, acting downstream of LGR4 signaling.
Deletion of GADD45A enhances self-renewal and leukemic potential on serial transplantation
We assessed the impact of Gadd45a loss on AML pathogenesis in vivo, and observed a significantly shortened latency for the mice transplanted with Gadd45a−/− compared with Gadd45a+/+ pre-MLLc-Kit+ cells (primary AML; Figure 2A-B; supplemental Figure 3A). This suggests that Gadd45a loss promotes leukemia progression. To test the role of Gadd45a loss in regulating the self-renewal and frequency of LSCs in vivo, we performed serial transplantation and limiting-dilution assays. GFP+ MLL-AF9 leukemic cells flow sorted from mice with primary AML were serially transplanted into syngeneic recipient mice. Gadd45a−/− leukemic cells gradually shortened disease latencies on serial transplantation in mice (secondary/tertiary/quaternary AML; Figure 2C-E) compared with Gadd45a+/+ cells, consistent with a progressively increasing self-renewal potential and an aggressive phenotype similar to that of relapsed human MLL-rearranged AML. In vivo limiting-dilution assay showed that deletion of Gadd45a caused a 10-fold increase in LSC frequency in Gadd45a−/− compared with Gadd45a+/+ mice (1/142 and 1/1650; P = .00114; Figure 2F). Likewise, immunophenotypic analysis revealed a 2-fold increase of Gr-1-/lowc-Kithigh LSC-enriched population in Gadd45a−/− compared with Gadd45a+/+ leukemic cells (Figure 2G; supplemental Figure 3B). We have previously documented that the Gr-1-/lowc-Kithigh population is >100-fold enriched for LSCs compared with the Gr-1+c-Kithigh population in a heterogeneous LSC pool of AML.10 Loss of GADD45A thus results in leukemia-initiating cell enrichment and increased LSC frequency, consistent with a correlation between high LSC frequency at diagnosis and poor prognosis in patients with AML.11
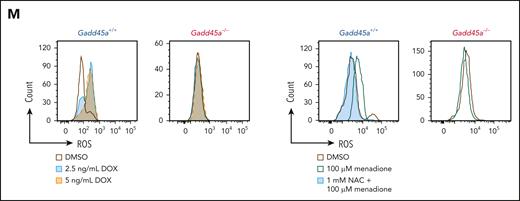
Deletion of Gadd45a enhances oncogenic potential and LSC activity on serial transplantation while inducing mutations. (A) Schematic overview of the experimental procedure. (B-E) Kaplan-Meier mouse survival curves of primary AML (B), secondary AML (C), tertiary AML (D), and quaternary AML (E). Primary AML was generated by transplanting 1 × 106Gadd45a−/− vs Gadd45a+/+ preleukemic cells, induced by MLL-AF9-GFP, into sublethally irradiated C57BL/6 (BL6) recipient mice (n = 6 for each group). Secondary AML, tertiary AML, and quaternary AML were generated by transplanting 1 × 104 GFP+ leukemic bone marrow (BM) cells flow sorted from mice with primary, secondary, and tertiary AML, respectively, into recipient mice (n = 6 for each group). P value was determined by the log-rank test. (F) In vivo limiting dilution transplantation assay showing a 10-fold increase in LSC frequency in Gadd45a−/− vs Gadd45a+/+ mice (1/142 vs 1/1650; P = .00114). LSC frequency was calculated using L-Calc software (StemCell Technologies). Kaplan-Meier survival curves of mice receiving the indicated number of GFP+ leukemic BM cells (n = 6 for each group), and P value was determined by the log-rank test. (G) Scatter plots with a bar graph depicting the percentage of Gr-1-/lowc-Kithigh population in total GFP+ leukemic BM cells from Gadd45a−/− vs Gadd45a+/+ mice (n = 5 for each group). Data are given as mean ± SD. ∗∗∗P < .0005. Unpaired t-test. (H) Stacked bar plots displaying the number of coding mutations identified by whole-genome sequencing (WGS) of Gadd45a−/− vs Gadd45a+/+ LSCs from primary (1st) and tertiary (3rd) AML. Green, yellow, and silver stacked bars represent synonymous, missense, and others (inframe insertion/deletion, frameshit, splice donor variant in coding sequence, start lost, and stop retained/gained), respectively. (I) Distribution of the coding mutations in Gadd45a−/− vs Gadd45a+/+ LSCs from primary AML. (J) Flow cytometry histograms showing decreased intracellular ROS in Gadd45a−/− LSCs compared with Gadd45a+/+ LSCs. (K) The number of colonies following treatment of Gadd45a−/− vs Gadd45a+/+ AML LSCs with dimethyl sulfoxide (DMSO) control or doxorubicin (DOX: 2.5 or 5 ng/mL) for 5 days in methylcellulose (n = 4). Data are given as mean ± SD. ∗P < .05, ∗∗P < .005, ∗∗∗P < .0005; NS, not significant (P > .05). One-way ANOVA. (L) In vivo BrdU proliferation assay with dot plots depicting BrdU+ GFP+ leukemic cells engrafted in the mouse BM at 12 days posttransplantation. Gadd45a−/− or Gadd45a+/+ AML LSCs were pretreated ex vivo with DMSO vs 5 ng/mL DOX for 48 hours, and 1 × 105 treated cells were transplanted into BL6 mice for leukemic cell engraftment and subsequent BrdU proliferation assay. Data are presented as the mean percentage relative to DMSO ± SD (n = 5 mice per group). ∗∗∗∗P < .0001; NS, not significant (P > .05). Unpaired t-test. (M) Flow cytometry histograms showing intracellular ROS levels in Gadd45a−/− vs Gadd45a+/+ AML LSCs treated with DMSO vs DOX (2.5 or 5 ng/mL) for 5 days, or pretreated with 1 mM N-acetylcysteine (NAC) for 1 hour, followed by treatment with 100 μM menadione for an additional 1 hour.
Deletion of Gadd45a enhances oncogenic potential and LSC activity on serial transplantation while inducing mutations. (A) Schematic overview of the experimental procedure. (B-E) Kaplan-Meier mouse survival curves of primary AML (B), secondary AML (C), tertiary AML (D), and quaternary AML (E). Primary AML was generated by transplanting 1 × 106Gadd45a−/− vs Gadd45a+/+ preleukemic cells, induced by MLL-AF9-GFP, into sublethally irradiated C57BL/6 (BL6) recipient mice (n = 6 for each group). Secondary AML, tertiary AML, and quaternary AML were generated by transplanting 1 × 104 GFP+ leukemic bone marrow (BM) cells flow sorted from mice with primary, secondary, and tertiary AML, respectively, into recipient mice (n = 6 for each group). P value was determined by the log-rank test. (F) In vivo limiting dilution transplantation assay showing a 10-fold increase in LSC frequency in Gadd45a−/− vs Gadd45a+/+ mice (1/142 vs 1/1650; P = .00114). LSC frequency was calculated using L-Calc software (StemCell Technologies). Kaplan-Meier survival curves of mice receiving the indicated number of GFP+ leukemic BM cells (n = 6 for each group), and P value was determined by the log-rank test. (G) Scatter plots with a bar graph depicting the percentage of Gr-1-/lowc-Kithigh population in total GFP+ leukemic BM cells from Gadd45a−/− vs Gadd45a+/+ mice (n = 5 for each group). Data are given as mean ± SD. ∗∗∗P < .0005. Unpaired t-test. (H) Stacked bar plots displaying the number of coding mutations identified by whole-genome sequencing (WGS) of Gadd45a−/− vs Gadd45a+/+ LSCs from primary (1st) and tertiary (3rd) AML. Green, yellow, and silver stacked bars represent synonymous, missense, and others (inframe insertion/deletion, frameshit, splice donor variant in coding sequence, start lost, and stop retained/gained), respectively. (I) Distribution of the coding mutations in Gadd45a−/− vs Gadd45a+/+ LSCs from primary AML. (J) Flow cytometry histograms showing decreased intracellular ROS in Gadd45a−/− LSCs compared with Gadd45a+/+ LSCs. (K) The number of colonies following treatment of Gadd45a−/− vs Gadd45a+/+ AML LSCs with dimethyl sulfoxide (DMSO) control or doxorubicin (DOX: 2.5 or 5 ng/mL) for 5 days in methylcellulose (n = 4). Data are given as mean ± SD. ∗P < .05, ∗∗P < .005, ∗∗∗P < .0005; NS, not significant (P > .05). One-way ANOVA. (L) In vivo BrdU proliferation assay with dot plots depicting BrdU+ GFP+ leukemic cells engrafted in the mouse BM at 12 days posttransplantation. Gadd45a−/− or Gadd45a+/+ AML LSCs were pretreated ex vivo with DMSO vs 5 ng/mL DOX for 48 hours, and 1 × 105 treated cells were transplanted into BL6 mice for leukemic cell engraftment and subsequent BrdU proliferation assay. Data are presented as the mean percentage relative to DMSO ± SD (n = 5 mice per group). ∗∗∗∗P < .0001; NS, not significant (P > .05). Unpaired t-test. (M) Flow cytometry histograms showing intracellular ROS levels in Gadd45a−/− vs Gadd45a+/+ AML LSCs treated with DMSO vs DOX (2.5 or 5 ng/mL) for 5 days, or pretreated with 1 mM N-acetylcysteine (NAC) for 1 hour, followed by treatment with 100 μM menadione for an additional 1 hour.
GADD45A loss induces replication stress and mutations in DNA repair and self-renewal genes
As Gadd45a−/− mice exhibit decreased DNA repair and increased mutation frequency,13 we examined whether deletion of Gadd45a promoted AML progression by inducing genomic instability and mutations. Our whole-genome sequencing of LSCs from primary and tertiary AML showed that deletion of Gadd45a induced substantial mutations (Figure 2H-I), particularly missense mutations involved in DNA repair (Ddx11, Rad21),25 self-renewal (HoxA clusters: Hoxa4, Hoxa5, Hoxa9, and Hoxa10),1 and DNA methylation (Dnmt1)26 (supplemental Figure 3C). Sanger sequencing confirmed persistent missense variants of Hoxa5 and Hoxa9 induced by Gadd45a deletion in LSCs from primary and tertiary AML (supplemental Figure 3D), demonstrating the accuracy of whole-genome sequencing data. Notably, Gadd45a deletion induced not only a missense mutation in exon 21 of the Ddx11 gene but also multiple synonymous mutations in exons 19 to 26, leading to decreased expression of DEAD/H-box helicase 11 (Ddx11) (supplemental Figure 3E). Because Ddx11 is a DNA helicase essential for double-stranded break repair during DNA replication,25 loss of Gadd45a likely causes replication stress through DNA repair gene variants, leading to genomic instability and DNA damage accumulation in LSCs. Because no additional mutations were found in key Gadd45a targets over serial transplantation, the enhancement of the leukemic potential in Gadd45a−/− mice was likely because of increased self-renewal, associated with HoxA cluster gene variants, which endowed LSCs with a competitive advantage. This finding aligns with our recent discovery demonstrating a positive cooperation between HOX and LGR4 pathways in promoting self-renewal and leukemic potential in LGR4/HOXA9-dependent AML.5
GADD45A loss fosters stem cell features driving resistance to therapy-associated oxidative stress
We next examined the role of Gadd45a loss as a determinant of therapy resistance in LSCs. Compared with Gadd45a+/+ LSCs, Gadd45a−/− LSCs exhibited a lower level of ROS and a higher proportion of quiescent G0 cells (Figure 2J; supplemental Figure 3F), which are critical features associated with increased resistance to therapy.27 Consistently, treatment with the chemotherapeutic agent doxorubicin (a mainstay of AML therapy, producing high levels of ROS) had little effects on Gadd45a−/− LSCs but reduced the colony-forming capacity of Gadd45a+/+ LSCs (Figure 2K). Subsequent cell proliferation assay at 12 days after injection of mice, following ex vivo pretreatment with doxorubicin, revealed impaired proliferation of Gadd45a+/+ LSCs, but not Gadd45a−/− LSCs (Figure 2L). This is consistent with aberrant ROS production induced by doxorubicin or menadione (ROS inducer) but rescued by N-acetylcysteine (ROS scavenger) in Gadd45a+/+ LSCs, rather than Gadd45a−/− LSCs (Figure 2M), underlining ROS-specific and Gadd45a-dependent effects. These data support a protective role of GADD45A loss against chemotherapy-associated oxidative stress in LSCs, and underscore the importance of GADD45A expression in determining LSC properties essential for leukemia progression and therapy resistance.
Deletion of GADD45A increases tumor burden in AML patient-derived xenograft mice
We have recently demonstrated that RSPO3-LGR4 activation is required for LSC survival in PDX mouse model of relapsed AML harboring mutations, including DNMT3A, RUNX1, and K/NRAS.5 Here, we investigated the impact of GADD45A loss on AML progression in established PDXs expressing firefly luciferase and mCherry. We found that deletion of GADD45A by CRISPR/Cas9 caused increased AML burden, as determined by firefly luciferase–based bioluminescence imaging in PDX mice (Figure 3A-B; supplemental Figure 4A-B). Subsequent secondary transplantation in NSG mice showed that GADD45A deletion enhanced the long-term leukemia-initiating activity of AML cells, which was associated with increased WNT/self-renewal signature (Figure 3C-D). This supports the role for GADD45A loss in promoting in vivo engraftment of human AML cells and leukemia-initiating activity.
Deletion of GADD45A promotes engraftment of human AML PDX cells in NSG mice. (A) Schematic describing in vivo generation of CRISPR/Cas9-mediated knockout (KO) of GADD45A in PDX mice. Also see supplemental Figure 4 for the additional experimental detail. (B) Representative in vivo bioluminescence imaging and total flux (photons/second [p/s]) of GADD45A KO PDX mice (n = 9) vs CRISPR control (Ctr) PDX mice (n = 6) in primary NSG recipients. A total of 5 × 104 mCherry+ GFP+ PDX cells were transplanted into each of recipient mice. Scatter dot plots represent the mean ± SD. ∗∗∗P < .0005, ∗∗∗∗P < .0001, 2-way ANOVA. (C) Representative in vivo bioluminescence imaging and total flux (p/s) of GADD45A KO PDX mice (n = 5) vs CRISPR Ctr PDX mice (n = 6) in secondary NSG recipients. A total of 1 × 105 mCherry+ PDX cells from primary recipient mice were transplanted into each of secondary recipient mice. Data are given as mean ± SD. ∗∗P < .005, ∗∗∗P < .0005, ∗∗∗∗P < .0001, 2-way ANOVA. (D) Quantitative polymerase chain reaction (qPCR) confirming stable knockout of GADD45A and showing relative expression levels of WNT/self-renewal target genes in GADD45A KO vs CRISPR Ctr hCD33+ PDX bone marrow (BM) cells (n = 3). Data are given as mean ± SEM. ∗P < .05, ∗∗P < .005, unpaired t-test.
Deletion of GADD45A promotes engraftment of human AML PDX cells in NSG mice. (A) Schematic describing in vivo generation of CRISPR/Cas9-mediated knockout (KO) of GADD45A in PDX mice. Also see supplemental Figure 4 for the additional experimental detail. (B) Representative in vivo bioluminescence imaging and total flux (photons/second [p/s]) of GADD45A KO PDX mice (n = 9) vs CRISPR control (Ctr) PDX mice (n = 6) in primary NSG recipients. A total of 5 × 104 mCherry+ GFP+ PDX cells were transplanted into each of recipient mice. Scatter dot plots represent the mean ± SD. ∗∗∗P < .0005, ∗∗∗∗P < .0001, 2-way ANOVA. (C) Representative in vivo bioluminescence imaging and total flux (p/s) of GADD45A KO PDX mice (n = 5) vs CRISPR Ctr PDX mice (n = 6) in secondary NSG recipients. A total of 1 × 105 mCherry+ PDX cells from primary recipient mice were transplanted into each of secondary recipient mice. Data are given as mean ± SD. ∗∗P < .005, ∗∗∗P < .0005, ∗∗∗∗P < .0001, 2-way ANOVA. (D) Quantitative polymerase chain reaction (qPCR) confirming stable knockout of GADD45A and showing relative expression levels of WNT/self-renewal target genes in GADD45A KO vs CRISPR Ctr hCD33+ PDX bone marrow (BM) cells (n = 3). Data are given as mean ± SEM. ∗P < .05, ∗∗P < .005, unpaired t-test.
GADD45A loss upregulates pathways involved in self-renewal and antioxidant defense at the single-cell level
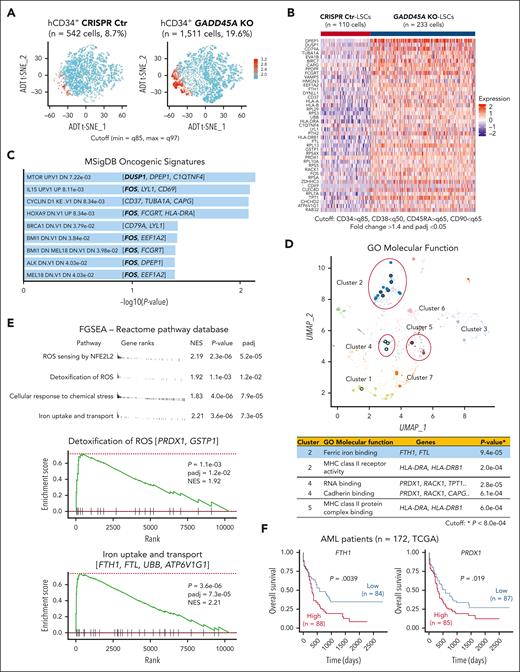
To understand how loss of GADD45A contributes to therapy resistance in aggressive AML, we performed CITE-seq (cellular indexing of transcriptomes and epitopes by sequencing)28 that used DNA-barcoded antibodies to tag cell-surface markers of human LSCs, enabling simultaneous quantification of mRNA and antibody-derived tag on a given single cell. Our results showed that the LSC compartment (CD34+CD38–CD90–CD45RA+)29 in human AML PDX cells was enriched specifically in cluster 2 (supplemental Figure 5A-B). Deletion of GADD45A resulted in a 3-fold increase in the LSC-enriched CD34+ population and a 2-fold increase in the LSC compartment (Figure 4A-B). Single-cell transcriptomic profiling identified 43 differentially expressed genes (DEGs) upregulated in GADD45A-deleted LSCs compared with CRISPR-control LSCs, including genes associated with LSC survival and self-renewal (e.g., DUSP1 and c-FOS)30,31 and iron detoxification and storage (e.g., ferritin heavy/light chain [FTH1/FTL])32,33 (Figure 4B; Supplemental Figure 5C). Pathway enrichment analysis of DEGs using MSigDB uncovered several self-renewal pathways, including the HOXA9 pathway (e.g., c-FOS) (Figure 4C). Strikingly, c-FOS, as a key WNT/β-catenin target that directly interacts with β-catenin for transcriptional activation of downstream components,34 was present in most of the top 9 oncogenic pathways, implicating its crucial role in GADD45A loss-mediated β-catenin self-renewal activation. Subsequent Gene Ontology analysis showed that the DEGs were enriched in molecular function (Figure 4D), particularly in iron ion binding (e.g., FTH1 and FTL). Likewise, our fast preranked gene set enrichment analysis of the Reactome pathway database identified GADD45A loss-induced enrichment of antioxidant-related gene sets, including iron uptake and transport (e.g., FTH1 and FTL) and ROS sensing/detoxification (eg, PRDX1, GSTP1)35,36 (Figure 4E). These observations agree with reduced ROS levels and increased self-renewal in GADD45A-deleted LSCs, underlining a possible involvement of GADD45A loss in antioxidant defense against iron and ROS accumulation (a defining feature of ferroptosis).15
Coupling single cell RNA-sequencing (scRNA-seq) with cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) on AML PDX cells reveals an increased proportion of LSCs and identifies genes/pathways upregulated by GADD45A deletion at a single stem cell level. (A) t-distributed stochastic neighbor embedding (T-SNE) clustering of CITE-seq data showing human CD34+ LSC-enriched subpopulations (in red) with GADD45A KO (n = 1511 cells, 19.6%) vs CRISPR Ctr (n = 542 cells, 8.7%) in the bone marrow (BM) of PDX mice. Cutoff: minimum = q85 and maximum = q97 (q stands for quantile). (B) Heat map of integrated CITE-seq data analysis identifying 43 differentially expressed genes (DEGs) upregulated in GADD45A KO PDX LSCs (n = 233 cells), compared with CRISPR Ctr LSCs (n = 110 cells). The LSC compartment (CD34+CD38–CD45RA+CD90–) was defined by a stringent cutoff of CD34 > q85, CD38 < q50, CD45RA > q65, and CD90 < q65. Wilcoxon rank-sum test, fold change > 1.4, and adjusted P value with Benjamini-Hochberg method < 0.05. (C) Bar chart showing the top 9 enriched cancer-associated pathways from MSigDB oncogenic signatures, along with their corresponding P values and associated DEGs. (D) Scatter plot showing Gene Ontology (GO) function enrichments of the DEGs upregulated in GADD45A KO LSCs, compared with CRISPR Ctr LSCs. Clusters were computed using the Leiden algorithm, and similar gene sets were clustered together. Larger, black-outlined points represent significantly enriched terms. Points are plotted on the first 2 UMAP dimensions. The table lists enriched gene sets with P < 8.0e-04 and associated DEGs. (E) Fast preranked gene set enrichment analysis (FGSEA) of CITE-seq data (cutoff: adjusted P < 0.05 and NES > 1.8) identifying GADD45A loss-induced enrichment of gene sets associated with ROS sensing by NFE2L2, detoxification of ROS, cellular response to chemical stress, and iron uptake and transport, on GADD45A knockout in human AML PDX cells. (F) Kaplan-Meier curves of overall survival for 172 patients with AML, as stratified by expression levels of FTH1 (P = .0039) and PRDX1 (P = .019) in TCGA dataset.
Coupling single cell RNA-sequencing (scRNA-seq) with cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) on AML PDX cells reveals an increased proportion of LSCs and identifies genes/pathways upregulated by GADD45A deletion at a single stem cell level. (A) t-distributed stochastic neighbor embedding (T-SNE) clustering of CITE-seq data showing human CD34+ LSC-enriched subpopulations (in red) with GADD45A KO (n = 1511 cells, 19.6%) vs CRISPR Ctr (n = 542 cells, 8.7%) in the bone marrow (BM) of PDX mice. Cutoff: minimum = q85 and maximum = q97 (q stands for quantile). (B) Heat map of integrated CITE-seq data analysis identifying 43 differentially expressed genes (DEGs) upregulated in GADD45A KO PDX LSCs (n = 233 cells), compared with CRISPR Ctr LSCs (n = 110 cells). The LSC compartment (CD34+CD38–CD45RA+CD90–) was defined by a stringent cutoff of CD34 > q85, CD38 < q50, CD45RA > q65, and CD90 < q65. Wilcoxon rank-sum test, fold change > 1.4, and adjusted P value with Benjamini-Hochberg method < 0.05. (C) Bar chart showing the top 9 enriched cancer-associated pathways from MSigDB oncogenic signatures, along with their corresponding P values and associated DEGs. (D) Scatter plot showing Gene Ontology (GO) function enrichments of the DEGs upregulated in GADD45A KO LSCs, compared with CRISPR Ctr LSCs. Clusters were computed using the Leiden algorithm, and similar gene sets were clustered together. Larger, black-outlined points represent significantly enriched terms. Points are plotted on the first 2 UMAP dimensions. The table lists enriched gene sets with P < 8.0e-04 and associated DEGs. (E) Fast preranked gene set enrichment analysis (FGSEA) of CITE-seq data (cutoff: adjusted P < 0.05 and NES > 1.8) identifying GADD45A loss-induced enrichment of gene sets associated with ROS sensing by NFE2L2, detoxification of ROS, cellular response to chemical stress, and iron uptake and transport, on GADD45A knockout in human AML PDX cells. (F) Kaplan-Meier curves of overall survival for 172 patients with AML, as stratified by expression levels of FTH1 (P = .0039) and PRDX1 (P = .019) in TCGA dataset.
Our analysis of TCGA AML dataset21 revealed a correlation between high expression of antioxidants FTH1 and PRDX1 with unfavorable outcomes in a cohort of 172 patients with AML (Figure 4F). This is in line with a recent report showing that high levels of FTH1 and FTL in a cohort of 525 patients with AML are associated with chemoresistance and enrichment of genes related to oxidative stress and iron pathways.37 Likewise, our functional studies showed that knockdown of GADD45A induced endogenous FTH1 expression (supplemental Figure 6A). Overexpression of FTH1 reduced intracellular ROS and iron, whereas knockdown of FTH1 elevated ROS levels in human AML (THP-1) cells (supplemental Figure 6B-D). These data suggest FTH1 acting downstream of GADD45A loss to provide antioxidant defense against iron and ROS accumulation.
GADD45A loss protects against ferroptosis by upregulating FTH1-mediated antioxidant defense
We next explored the role for GADD45A loss in preventing ferroptosis through upregulation of FTH1. RSL3 (a selective ferroptosis inducer)38 triggered ferroptotic/nonapoptotic cell death and reduced in vitro colony-forming capacity and in vivo cell proliferation in Gadd45a+/+ LSCs, rather than Gadd45a−/− LSCs, from mice with tertiary AML (Figures 2D and 5A-C). The sensitivity of Gadd45a+/+ LSCs to RSL3 treatment could be attributed to the ability of RSL3 to induce Gadd45a expression and reduce Fth1 in Gadd45a+/+ LSCs, but not Gadd45a−/− LSCs (Figure 5D). In support of this notion, knockdown of Fth1 promoted RSL3-induced ferroptosis in both Gadd45a−/− and Gadd45a+/+ LSCs, revealing increased ROS and ferrous iron (Fe2+) accompanied by decreased cell viability (Figure 5E-G). This is in line with the role of Fth1 acting downstream of Gadd45a loss to counteract RSL3-induced ROS and iron and consequently prevent genotoxic stress-induced DNA damage and ferroptosis. The protective effect of Gadd45a deletion against RSL3-induced exogenous DNA damage was demonstrated by the result of RSL3-induced increase in phosphorylated H2AX (γ-H2AX, a biomarker for DNA double-stranded breaks)39 in Gadd45a+/+ LSCs, but not in Gadd45a−/− LSCs, and by a similar result obtained from the alkaline comet assay (genotoxicity testing for DNA-strand breaks)40 (Figure 5H-I). Notably, a higher level of endogenous DNA damage was observed in Gadd45a−/− than Gadd45a+/+ LSCs (Figure 5I), consistent with the role of Gadd45a deletion in increasing replication stress and stress-induced DNA damage (Figure 2H). These data demonstrate GADD45A loss as a key determinant in preventing stress-induced DNA damage and RSL3-induced ferroptosis, primarily by upregulating FTH1-mediated antioxidant defense that scavenges excessively produced ROS and iron.
Deletion of GADD45A prevents RSL3-induced ferroptosis and DNA damage through upregulation of FTH1. (A) Flow cytometry dot plots showing the percentage of nonapoptotic or ferroptotic cell death (annexin V negative, SYTOX blue positive; Q3), following treatment of Gadd45a−/− vs Gadd45a+/+ AML LSCs with dimethyl sulfoxide (DMSO) or 3 μM RSL3 for 4 days in methylcellulose. (B) Percentages of viable cells measured by trypan blue exclusion assay in Gadd45a−/− vs Gadd45a+/+ LSCs, following treatment with DMSO or 3 μM RSL3 for 4 days in methylcellulose (n = 3). Data are given as mean ± SD. ∗∗P < .005; NS, not significant (P > .05). Unpaired t-test. (C) In vivo BrdU proliferation assay with dot plots showing the percentage of BrdU+DAPI− leukemic cells engrafted in the mouse bone marrow (BM) at 21 days posttransplantation. Gadd45a−/− vs Gadd45a+/+ LSCs were pretreated ex vivo with DMSO or 3 μM RSL3 for 4 days in methylcellulose, and 1 × 105 GFP+ treated cells were then transplanted into C57BL/6 (BL6) mice for the engraftment of GFP+ LSCs and subsequent in vivo BrdU cell proliferation assay. Data are presented as the mean percentage relative to DMSO ± SD. ∗∗∗P < .0005; NS, not significant (P > .05). Unpaired t-test. (D) Quantitative polymerase chain reaction (qPCR) (n = 3) showing upregulation of Gadd45a and downregulation of Fth1 induced by RSL3 treatment in Gadd45a+/+ LSCs but not in Gadd45a−/− LSCs. Data are given as mean ± SD. ∗∗P < .005; NS, not significant (P > .05), unpaired t-test. (E) qPCR (n = 3) confirming efficient knockdown of Fth1 by shRNA (Fth1-sh) in Gadd45a−/− LSCs and Gadd45a+/+ LSCs. Data are given as mean ± SD. ∗P < .05, ∗∗∗∗P < .0001, unpaired t-test. (F) Flow cytometry histograms illustrating intracellular iron (Fe2+) and ROS levels in Gadd45a−/− vs Gadd45a+/+ LSCs carrying Scr or Fth1-sh, following treatment with DMSO or 3 μM RSL3 for 4 days in methylcellulose. (G) Percentages of viable cells measured by trypan blue exclusion assay in Gadd45a−/− vs Gadd45a+/+ LSCs carrying Scr or Fth1-sh, following treatment with DMSO or 3 μM RSL3 for 4 days in methylcellulose (n = 3). Data are given as mean ± SD. ∗∗P < .005, ∗∗∗∗P < .0001; NS, not significant (P > .05). One-way ANOVA. (H) Western blots showing increased expression of γH2AX induced by RSL3 treatment in Gadd45a+/+ LSCs but not in Gadd45a−/− LSCs. (I) Alkaline comet assay used to quantify the level of DNA-strand breaks illustrating heightened DNA damage (tail moment) in Gadd45a+/+ LSCs treated in methylcellulose with 3 μM RSL3 compared with DMSO. Scatter plots with a bar graph depicting tail moments (a combined measure of tail length and the amount of migrated DNA) calculated using CometScore. Representative fluorescence images showing DAPI-stained single cells after electrophoresis (×20 magnification). During electrophoresis, damaged DNA migrated out of the nucleus toward the anode, forming a comet tail, whereas undamaged DNA remained in the comet head. The comet tail moment represents the extent of DNA damage in individual cells.
Deletion of GADD45A prevents RSL3-induced ferroptosis and DNA damage through upregulation of FTH1. (A) Flow cytometry dot plots showing the percentage of nonapoptotic or ferroptotic cell death (annexin V negative, SYTOX blue positive; Q3), following treatment of Gadd45a−/− vs Gadd45a+/+ AML LSCs with dimethyl sulfoxide (DMSO) or 3 μM RSL3 for 4 days in methylcellulose. (B) Percentages of viable cells measured by trypan blue exclusion assay in Gadd45a−/− vs Gadd45a+/+ LSCs, following treatment with DMSO or 3 μM RSL3 for 4 days in methylcellulose (n = 3). Data are given as mean ± SD. ∗∗P < .005; NS, not significant (P > .05). Unpaired t-test. (C) In vivo BrdU proliferation assay with dot plots showing the percentage of BrdU+DAPI− leukemic cells engrafted in the mouse bone marrow (BM) at 21 days posttransplantation. Gadd45a−/− vs Gadd45a+/+ LSCs were pretreated ex vivo with DMSO or 3 μM RSL3 for 4 days in methylcellulose, and 1 × 105 GFP+ treated cells were then transplanted into C57BL/6 (BL6) mice for the engraftment of GFP+ LSCs and subsequent in vivo BrdU cell proliferation assay. Data are presented as the mean percentage relative to DMSO ± SD. ∗∗∗P < .0005; NS, not significant (P > .05). Unpaired t-test. (D) Quantitative polymerase chain reaction (qPCR) (n = 3) showing upregulation of Gadd45a and downregulation of Fth1 induced by RSL3 treatment in Gadd45a+/+ LSCs but not in Gadd45a−/− LSCs. Data are given as mean ± SD. ∗∗P < .005; NS, not significant (P > .05), unpaired t-test. (E) qPCR (n = 3) confirming efficient knockdown of Fth1 by shRNA (Fth1-sh) in Gadd45a−/− LSCs and Gadd45a+/+ LSCs. Data are given as mean ± SD. ∗P < .05, ∗∗∗∗P < .0001, unpaired t-test. (F) Flow cytometry histograms illustrating intracellular iron (Fe2+) and ROS levels in Gadd45a−/− vs Gadd45a+/+ LSCs carrying Scr or Fth1-sh, following treatment with DMSO or 3 μM RSL3 for 4 days in methylcellulose. (G) Percentages of viable cells measured by trypan blue exclusion assay in Gadd45a−/− vs Gadd45a+/+ LSCs carrying Scr or Fth1-sh, following treatment with DMSO or 3 μM RSL3 for 4 days in methylcellulose (n = 3). Data are given as mean ± SD. ∗∗P < .005, ∗∗∗∗P < .0001; NS, not significant (P > .05). One-way ANOVA. (H) Western blots showing increased expression of γH2AX induced by RSL3 treatment in Gadd45a+/+ LSCs but not in Gadd45a−/− LSCs. (I) Alkaline comet assay used to quantify the level of DNA-strand breaks illustrating heightened DNA damage (tail moment) in Gadd45a+/+ LSCs treated in methylcellulose with 3 μM RSL3 compared with DMSO. Scatter plots with a bar graph depicting tail moments (a combined measure of tail length and the amount of migrated DNA) calculated using CometScore. Representative fluorescence images showing DAPI-stained single cells after electrophoresis (×20 magnification). During electrophoresis, damaged DNA migrated out of the nucleus toward the anode, forming a comet tail, whereas undamaged DNA remained in the comet head. The comet tail moment represents the extent of DNA damage in individual cells.
Suppression of GADD45A affects ferroptosis sensitivity in primary human AML cells
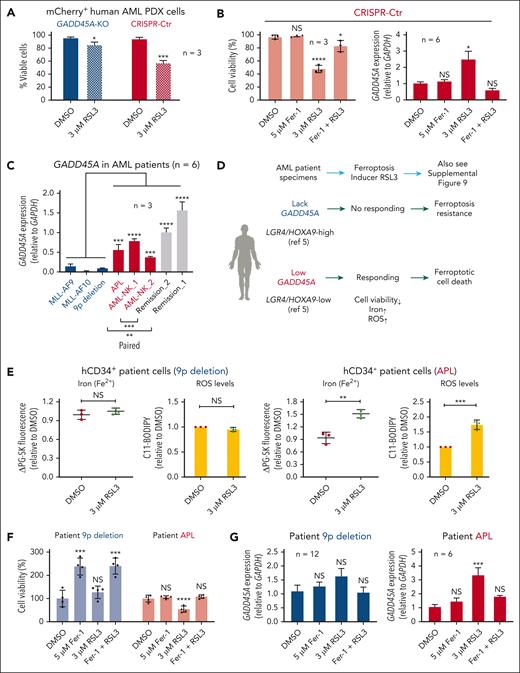
To investigate the role of GADD45A loss in regulating ferroptosis resistance in human AML cells, we treated mCherry+ PDX cells with RSL3. CRISPR/Cas9-mediated deletion of GADD45A prevented RSL3-induced ferroptosis; conversely, CRISPR-control PDX cells that expressed low levels of GADD45A responded to RSL3 treatment, causing ∼40% cell death (Figure 6A; supplemental Figure 7). RSL3-induced ferroptosis could be rescued by pretreatment with a ferroptosis inhibitor, ferrostatin-1 that inhibits ferroptosis by reducing iron levels,15 which suppressed RSL3-induced expression of endogenous GADD45A (Figure 6B). These results support GADD45A as a positive regulator of ferroptosis, and deletion of GADD45A promotes ferroptosis resistance in human AML cells.
Lack of GADD45A influences the response of primary human AML cells to ferroptosis induction. (A) Percentages of viable cells (DAPI negative) in GADD45A-KO vs CRISPR-Ctr mCherry+ human AML PDX cells, following ex vivo treatment with 3 μM RSL3 for 24 hours. n = 3 independent experiments. Data are given as mean ± SD. ∗P < .05, ∗∗∗P < .0005, unpaired t-test. (B) Percentages of viable (DAPI-negative) mCherry+ human AML cells (n = 3, mean ± SD) and quantitative polymerase chain reaction (qPCR) of GADD45A expression (n = 6, mean ± SEM) in CRISPR-Ctr PDX bone marrow (BM) cells, pretreated ex vivo for 18 hours with 5 μM ferrostatin-1 (Fer-1; ferroptosis inhibitor) and subsequently treated with 3 μM RSL3 (ferroptosis inducer) for an additional 24 hours. ∗P < .05, ∗∗∗∗P < .0001; NS, not significant (P > .05). One-way ANOVA. (C) qPCR showing relative expression levels of GADD45A in primary specimens from patients with AML (n = 6), compared with remission samples (n = 2). Data are given as mean ± SD. n = 3 replicates. ∗∗∗P < .0005, ∗∗∗∗P < .0001, one-way ANOVA. Note: two paired diagnostic/remission samples: AML-NK_1/remission_1 and AML-NK_2/remission_2, showing higher levels of GADD45A at remission than paired diagnosis, consistent with higher expression of GADD45A in normal human BM and CD34+ cells than in MLL-rearranged patients with AML (supplemental Figure 8). (D) Schematic overview of AML patient specimens responding to ferroptosis inducer RSL3 ex vivo. (E) Intracellular ferrous iron (Fe2+) was detected using the fluorescent turn-off sensor Phen Green (PG) SK that is quenched on binding iron, while ROS levels were measured using the lipid peroxidation sensor C11-BODIPY (581/591) that shifts its fluorescence from red (∼590 nm) to green (∼530 nm) on oxidation in hCD34+ primary AML patient specimens, following ex vivo treatment with 3 μM RSL3 for 24 hours. ΔPG-SK revealed the reversed value of PG-SK fluorescence quenching, showing an increased intracellular Fe2+ in hCD34+ cells from a patient with AML with acute promyelocytic leukemia (APL) but not 9p deletion. Data are given as mean ± SD. ∗∗P < .005, ∗∗∗P < .0005; NS, not significant (P > .05). Unpaired t-test. Also see supplemental Figure 9 for additional patient samples examined. (F) Percentages of viable cells (n = 4 replicates, mean ± SD) tested using the alamarBlue assay in primary AML patient specimens pretreated ex vivo for 18 hours with 5 μM Fer-1, followed by treatment with 3 μM RSL3 for an additional 24 hours. ∗∗∗P < .0005, ∗∗∗∗P < .0001; NS, not significant (P > .05). One-way ANOVA. (G) qPCR of GADD45A expression (mean ± SEM) in primary AML patient specimens pretreated ex vivo for 18 hours with 5 μM Fer-1, followed by treatment with 3 μM RSL3 for an additional 24 hours. ∗∗∗P < .0005; NS, not significant (P > .05). One-way ANOVA.
Lack of GADD45A influences the response of primary human AML cells to ferroptosis induction. (A) Percentages of viable cells (DAPI negative) in GADD45A-KO vs CRISPR-Ctr mCherry+ human AML PDX cells, following ex vivo treatment with 3 μM RSL3 for 24 hours. n = 3 independent experiments. Data are given as mean ± SD. ∗P < .05, ∗∗∗P < .0005, unpaired t-test. (B) Percentages of viable (DAPI-negative) mCherry+ human AML cells (n = 3, mean ± SD) and quantitative polymerase chain reaction (qPCR) of GADD45A expression (n = 6, mean ± SEM) in CRISPR-Ctr PDX bone marrow (BM) cells, pretreated ex vivo for 18 hours with 5 μM ferrostatin-1 (Fer-1; ferroptosis inhibitor) and subsequently treated with 3 μM RSL3 (ferroptosis inducer) for an additional 24 hours. ∗P < .05, ∗∗∗∗P < .0001; NS, not significant (P > .05). One-way ANOVA. (C) qPCR showing relative expression levels of GADD45A in primary specimens from patients with AML (n = 6), compared with remission samples (n = 2). Data are given as mean ± SD. n = 3 replicates. ∗∗∗P < .0005, ∗∗∗∗P < .0001, one-way ANOVA. Note: two paired diagnostic/remission samples: AML-NK_1/remission_1 and AML-NK_2/remission_2, showing higher levels of GADD45A at remission than paired diagnosis, consistent with higher expression of GADD45A in normal human BM and CD34+ cells than in MLL-rearranged patients with AML (supplemental Figure 8). (D) Schematic overview of AML patient specimens responding to ferroptosis inducer RSL3 ex vivo. (E) Intracellular ferrous iron (Fe2+) was detected using the fluorescent turn-off sensor Phen Green (PG) SK that is quenched on binding iron, while ROS levels were measured using the lipid peroxidation sensor C11-BODIPY (581/591) that shifts its fluorescence from red (∼590 nm) to green (∼530 nm) on oxidation in hCD34+ primary AML patient specimens, following ex vivo treatment with 3 μM RSL3 for 24 hours. ΔPG-SK revealed the reversed value of PG-SK fluorescence quenching, showing an increased intracellular Fe2+ in hCD34+ cells from a patient with AML with acute promyelocytic leukemia (APL) but not 9p deletion. Data are given as mean ± SD. ∗∗P < .005, ∗∗∗P < .0005; NS, not significant (P > .05). Unpaired t-test. Also see supplemental Figure 9 for additional patient samples examined. (F) Percentages of viable cells (n = 4 replicates, mean ± SD) tested using the alamarBlue assay in primary AML patient specimens pretreated ex vivo for 18 hours with 5 μM Fer-1, followed by treatment with 3 μM RSL3 for an additional 24 hours. ∗∗∗P < .0005, ∗∗∗∗P < .0001; NS, not significant (P > .05). One-way ANOVA. (G) qPCR of GADD45A expression (mean ± SEM) in primary AML patient specimens pretreated ex vivo for 18 hours with 5 μM Fer-1, followed by treatment with 3 μM RSL3 for an additional 24 hours. ∗∗∗P < .0005; NS, not significant (P > .05). One-way ANOVA.
We further assessed the association of GADD45A expression with ferroptosis in primary AML patient specimens that express high levels of LGR4/HOXA9 (9p deletion, MLL-AF10, MLL-AF9) vs relatively low levels of LGR4/HOXA9 (acute promyelocytic leukemia, or APL; AML with normal karyotype, or AML-NK), as shown in our recent studies.5 Lower expression of GADD45A was shown in LGR4/HOXA9-high than LGR4/HOXA9-low cells (Figure 6C; supplemental Figure 8A-B). Lack of endogenous GADD45A in AML with 9p deletion or MLL-AF10 rendered LGR4/HOXA9-high cells resistant to RSL3 treatment; conversely, expression of GADD45A in APL or AML-NK, even at much lower levels than paired remission samples, sensitized LGR4/HOXA9-low cells to RSL3 treatment, leading to ferroptosis with increased iron and ROS accompanied by reduced cell viability (Figure 6C-F; supplemental Figure 9A-B). RSL3 induced ferroptosis by upregulating GADD45A that could be suppressed by ferroptosis inhibitor ferrostatin-1 in LGR4/HOXA9-low cells; however, RSL3 had no impact on LGR4/HOXA9-high cells that intrinsically lacked GADD45A (Figure 6C-G). This suggests a threshold level of endogenous GADD45A required for ferroptosis induction. These data underline a proactive role of GADD45A in RSL3-induced ferroptosis; loss of GADD45A provides a shield against this mechanism of cell death in aggressive AML that is dependent on LGR4/HOXA9 expression.
Discussion
LSCs exhibit functional heterogeneity, particularly in HSC-derived AML with adverse/intermediate-risk cytogenetics, characterized by high self-renewal and frequency of leukemia-initiating cells alongside low response to chemotherapy.2,3,5,41 High LSC activity is associated with elevated relapse risk and poor survival in patients with AML.11,12 We have recently demonstrated an indispensable role for RSPO3-LGR4 signaling in the formation and function of HSC-derived LSCs through aberrant activation of p-PKAc/p-CREB/CBP-p300/β-catenin pathway, contributing to aggressive phenotypes in HOXA9-dependent AML.5 Here, we identify GADD45A as a key negative regulator of leukemia-initiating activity and therapy resistance, acting downstream of LGR4/p-PKAc/p-FOXO3A. Loss of GADD45A in a heterogeneous LSC pool increases LSC self-renewal and frequency and decreases chemosensitivity, leading to a more aggressive AML phenotype. Several mechanisms may act in concert to promote GADD45A loss-associated leukemogenic potential: deletion of GADD45A increases WNT/β-catenin self-renewal activity (e.g., c-FOS and HOXA cluster) through GSK3β phosphorylation (inactivation), and induces replication stress and mutations in DNA repair and self-renewal genes contributing to enhanced cooperation of LGR4 and HOX pathways in promoting stemness and self-renewal, while conferring resistance to oxidative stress and ferroptosis via upregulation of FTH1-mediated antioxidant defense that stores iron in a nontoxic form and scavenges excessive ROS production induced by replication stress or therapy-induced genotoxic stress.
ROS is an important determinant of stem cell fate. Low ROS closely correlates with increased self-renewal and quiescence in normal and malignant stem cells,27,42 and a slight increase of intracellular ROS induces LSC differentiation.43 Consistently, deletion of GADD45A in LSCs reduces ROS levels and promotes self-renewal in a mouse model of AML development; however, it also causes replication stress and DNA damage accumulation, which have the potential to increase ROS production.44 This raises a critical question of how low ROS is sustained in LSCs under persistent DNA damage. Our single-cell cellular indexing of transcriptomes and epitopes by sequencing data on patient-derived LSCs and subsequent functional studies directly link GADD45A deletion to aberrant activation of antioxidant defense against ROS and iron accumulation. GADD45A loss serves as a safeguard by upregulating FTH1-mediated antioxidant defense, which attenuates ROS generated by replication stress-induced DNA damage and thereby maintains ROS at a basal nontoxic low level to protect LSC compartment from oxidative stress.
Antioxidant defense upregulated by GADD45A loss also acts as the protective mechanism against genotoxic stress-induced iron and ROS accumulation in response to chemotherapeutic agent doxorubicin and ferroptosis inducer RSL3, leading to therapy resistance. Ferroptosis is an iron (Fe2+)-dependent oxidative cell death caused by ROS generated through Fenton reaction and subsequent lipid peroxidation.15 Ferroptosis resistance remains a significant challenge to therapeutic exploitation of the iron-dependent cell death mechanism. In this study, we identify GADD45A loss as a key regulator of ferroptosis resistance. Deletion of GADD45A in murine LSCs or lack of GADD45A in LGR4/HOXA9-dependent patient cells (indicative of poor outcome) renders cells resistant to ferroptosis induced by RSL3. Our pathway enrichment analysis uncovers abnormal activation of iron detoxification driven by ferritin (FTH1/FTL), an iron storage protein complex.32,33 FTH1 has ferroxidase activity to convert Fe2+ to Fe3+ for iron storage in a nontoxic form via FTL that prevents excess Fe2+ to produce ROS via the Fenton reaction or by forming the doxorubicin-Fe2+ complex.33,45 Upregulation of FTH1 is correlated with unfavorable outcomes in patients with AML, and FTH1 modulates intracellular iron and ROS levels acting downstream of GADD45A in AML LSCs. These findings underline a crucial role of GADD45A loss in therapy resistance by enhancing FTH1-mediated antioxidant defense that limits iron-dependent ROS production, allowing LSCs to survive under persistent genotoxic stress without surpassing a deadly threshold in a subgroup of leukemia with poor prognosis.
In addition to ferritin (FTH1/FTL) as an antioxidant defense system, our single-cell studies also uncover aberrant activation of pathways related to LSC survival (e.g., DUSP1) and antioxidants (e.g., PRDX1 and GSTP1) in GADD45A-deleted human LSCs. Like FTH1, high expression of PRDX1, which can scavenge excess ROS and decreases damage from oxidative stress,35 correlates with unfavorable prognosis in patients with AML. The positive correlation between FTH1 and DUSP1 expression in patients with AML from TCGA (supplemental Figure 10) implies a possible cooperation between FTH1 and DUSP1 pathways in ferroptosis resistance and LSC survival, acting downstream of GADD45A. Thus, loss of GADD45A may play a crucial role in protecting leukemia-initiating cells against oxidative stress and ferroptosis by simultaneous modulation of multiple pathways, suggesting that targeting of individual downstream effectors may not be sufficient to fully restore GADD45A tumor-suppressor function. In this regard, therapeutic approaches to induce GADD45A expression in LSCs likely represent a more robust strategy to compromise LSC function and modulate cell sensitivity to anticancer agents associated with oxidative stress and ferroptosis.
Ferroptosis inducers may be 1 approach to promote GADD45A expression in AML cells expressing this gene even at low levels, but are unable to restore GADD45A in cells lacking its expression. This is because a threshold level of endogenous GADD45A is required for ferroptosis induction. Alternative approaches are needed for AML cells lacking GADD45A. Given our findings that LGR4 pathway inhibition induces GADD45A expression and GADD45A acts downstream of LGR4/p-PKAc/p-FOXO3, restoring GADD45A in LSCs can be achieved by blocking this pathway in a significant group of AML. In particular, we observe the lack of GADD45A in primary patient specimens expressing high LGR4/HOXA9, which critically depend on RSPO3-LGR4 activation for LSC survival and maintenance of myeloid undifferentiated state.5 We have recently demonstrated that a clinical-grade anti-RSPO3 antibody specifically targets LGR4 signaling via disruption of the RSPO3-LGR4 interaction, which compromises in vivo leukemia-initiating activity in primary AML PDX mouse models that harbor high-risk cytogenetics (e.g., MLL-AF9 or 9p deletion)5 and lack GADD45A. Notably, we found that in vivo anti-RSPO3 antibody treatment induced endogenous GADD45A in NSG mice engrafted with primary MLL-AF9 AML patient cells (supplemental Figure 11A-C), thereby offering a potential strategy to overcome resistance to ferroptosis. Nevertheless, we cannot exclude the possibility of other factors, such as DNA methylation, in coordinately regulating GADD45A. We have previously shown that LGR4 decreased intracellular ROS,5 which affects DNA methylation by acting on the activity/expression of DNA methyltransferases (DNMTs).46 The DNMT inhibitor, decitabine, which predominantly inhibits DNMT1 activity,47 induced GADD45A expression in MLL-AF9 LSCs (supplemental Figure 11D). This indicates a coordinated downregulation of GADD45A in LGR4/HOXA9-dependent LSCs by multiple mechanisms, including LGR4/p-PKAc/p-FoxO3a and DNA methylation.
In summary, we have identified a key regulatory role for GADD45A loss in promoting LSC self-renewal and stemness and in driving oxidative resistance, including ferroptosis resistance, in LGR4/HOXA9-dependent AML. Understanding the mechanisms by which GADD45A manipulates iron-dependent ROS accumulation provides additional insights on ferroptosis regulation and unlocks a new arsenal to reverse resistance by restoring the GADD45A activity in combination with ferroptosis induction or chemotherapy for treating this poor prognosis subset of leukemia lacking GADD45A.
Acknowledgments
The authors thank Irmela Jeremias and Karsten Spiekermann for providing acute myeloid leukemia patient-derived xenograft cells; Basit Salik, Karthik B. Polpaya, Gargi Kulkarni, Jayvee Datuin, Sheng X. F. Chen, Jonason Yang, Sayali Gore, and the staff from the Ramaciotti Centre for Genomics at the University of New South Wales for technical assistance; and the Sydney Children’s Tumor Bank Network for providing primary patient samples and related clinical information for this study.
This work was supported by grants from the Leukemia & Lymphoma Society, United States (TRP-23664-23); Cancer Council NSW, Australia (RG 22-06); Anthony Rothe Memorial Trust, Australia (RRE/0700:sz); and Tour de Cure, Australia (RSP-187-2020) (to J.Y.W.).
Authorship
Contribution: N.H. and B.M. performed drug treatment and ferroptosis-related experiments with input from X.D.Z., T.L. and J.W.; H.Y. performed murine model experiments; J.S., A.B., and N.H. performed single-cell experiments with input from D.R.C.; L.G.-B. analyzed cellular indexing of transcriptomes and epitopes by sequencing data with assistance from J.S. and T.C; A.D. performed in vivo bioluminescence imaging; S.E.S., D.A.C., R.J.D., and D.A.L. supplied mouse Gadd45a−/− bone marrow cells and provided input on murine model experiments; R.J.D, D.A.L., S.E.S., D.A.C, J.W., G.M.M., M.N., M.H., M.K., and B.B.C. reviewed the manuscript; and J.Y.W. conceived, designed, and analyzed the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jenny Y. Wang, Cancer and Stem Cell Laboratory, School of Medical Sciences, Faculty of Medicine and Health, University of Sydney, Kolling Institute, Sydney, NSW, Australia; email: jenny.wang@sydney.edu.au.
References
Author notes
N.H. and H.Y. contributed equally to this study.
Microarray data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-4773. The whole-genome sequencing data are available in the US National Library of Medicine under accession number PRJNA994161. Cellular indexing of transcriptomes and epitopes by sequencing data (RNA and antibody-derived tag) have been deposited in the ArrayExpress database at EMBL-European Bioinformatics Institute (EMBL-EBI) (www.ebi.ac.uk/arrayexpress) under the accession number E-MTAB-9803.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.


![Deletion of GADD45A promotes engraftment of human AML PDX cells in NSG mice. (A) Schematic describing in vivo generation of CRISPR/Cas9-mediated knockout (KO) of GADD45A in PDX mice. Also see supplemental Figure 4 for the additional experimental detail. (B) Representative in vivo bioluminescence imaging and total flux (photons/second [p/s]) of GADD45A KO PDX mice (n = 9) vs CRISPR control (Ctr) PDX mice (n = 6) in primary NSG recipients. A total of 5 × 104 mCherry+ GFP+ PDX cells were transplanted into each of recipient mice. Scatter dot plots represent the mean ± SD. ∗∗∗P < .0005, ∗∗∗∗P < .0001, 2-way ANOVA. (C) Representative in vivo bioluminescence imaging and total flux (p/s) of GADD45A KO PDX mice (n = 5) vs CRISPR Ctr PDX mice (n = 6) in secondary NSG recipients. A total of 1 × 105 mCherry+ PDX cells from primary recipient mice were transplanted into each of secondary recipient mice. Data are given as mean ± SD. ∗∗P < .005, ∗∗∗P < .0005, ∗∗∗∗P < .0001, 2-way ANOVA. (D) Quantitative polymerase chain reaction (qPCR) confirming stable knockout of GADD45A and showing relative expression levels of WNT/self-renewal target genes in GADD45A KO vs CRISPR Ctr hCD33+ PDX bone marrow (BM) cells (n = 3). Data are given as mean ± SEM. ∗P < .05, ∗∗P < .005, unpaired t-test.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/144/1/10.1182_blood.2024024072/3/m_blood_bld-2024-024072-gr3.jpeg?Expires=1767750137&Signature=dq2OAaku7IJqn4ecg44xLdlXc~4hm2-l~XZgNcYAs0q0d7tzJi1StrZz2L53ZdsBjMpr9VHi0fGDPJ5LIiD1tbXAHDZFCrsaJP6gd9rhT0F6fyU~2d0Y2-rloQQq40BiEncGAUVe4k7SGJhA5yjrCLniCEwf4~G1hbkUQuzKe5odps-rWxBGITruDZeTtBLnMB4xP8-XiLILsSbQpzxtIhvAV6UWMZ4ZMVhP808vyWUdpfEep8ryIQy6o9nJfWDnT1nW3uOqQZqic61q38a6oBhwj7Q5JsyXhYhKEh~f-dHj4nPLKvUejK~UnZpzWB2LB3YXnSpTObpKtV-hIr7dRg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal