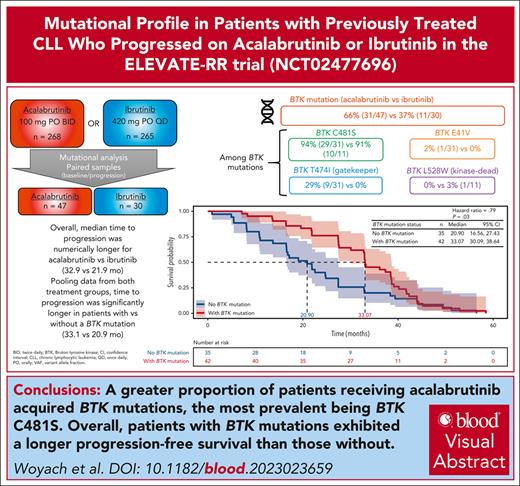
Key Points
BTK C481S was most common in both groups; in the acalabrutinib arm, low-VAF T474I (9/47; 8 co-occurring with C481S) but no L528W was seen.
More patients receiving acalabrutinib acquired BTK mutations, though overall, patients with BTK mutation did not fare worse vs those without.
Visual Abstract
Chronic lymphocytic leukemia (CLL) progression during Bruton tyrosine kinase (BTK) inhibitor treatment is typically characterized by emergent B-cell receptor pathway mutations. Using peripheral blood samples from patients with relapsed/refractory CLL in ELEVATE-RR (NCT02477696; median 2 prior therapies), we report clonal evolution data for patients progressing on acalabrutinib or ibrutinib (median follow-up, 41 months). Paired (baseline and progression) samples were available for 47 (excluding 1 Richter) acalabrutinib-treated and 30 (excluding 6 Richter) ibrutinib-treated patients. At progression, emergent BTK mutations were observed in 31 acalabrutinib-treated (66%) and 11 ibrutinib-treated patients (37%; median variant allele fraction [VAF], 16.1% vs 15.6%, respectively). BTK C481S mutations were most common in both groups; T474I (n = 9; 8 co-occurring with C481) and the novel E41V mutation within the pleckstrin homology domain of BTK (n = 1) occurred with acalabrutinib, whereas neither mutation occurred with ibrutinib. L528W and A428D comutations presented in 1 ibrutinib-treated patient. Preexisting TP53 mutations were present in 25 acalabrutinib-treated (53.2%) and 16 ibrutinib-treated patients (53.3%) at screening. Emergent TP53 mutations occurred with acalabrutinib and ibrutinib (13% vs 7%; median VAF, 6.0% vs 37.3%, respectively). Six acalabrutinib-treated patients and 1 ibrutinib-treated patient had emergent TP53/BTK comutations. Emergent PLCG2 mutations occurred in 3 acalabrutinib-treated (6%) and 6 ibrutinib-treated patients (20%). One acalabrutinib-treated patient and 4 ibrutinib-treated patients had emergent BTK/PLCG2 comutations. Although common BTK C481 mutations were observed with both treatments, patterns of mutation and comutation frequency, mutation VAF, and uncommon BTK variants varied with acalabrutinib (T474I and E41V) and ibrutinib (L528W and A428D) in this patient population. The trial was registered at www.clinicaltrials.gov as #NCT02477696.
Introduction
Covalent Bruton tyrosine kinase (BTK) inhibitors (BTKis) are highly effective in the treatment of chronic lymphocytic leukemia (CLL) and have resulted in a paradigm shift in the management of the disease.1 However, disease progression in patients receiving covalent BTKis eventually occurs in most patients and is often characterized by B-cell receptor pathway mutations at relapse, which commonly occur in the BTK and PLCG2 genes.2-8BTK mutations often occur at the C481 residue and disrupt binding to, and inactivation of, BTK by all covalent BTKis.9-11 C481 mutations preclude irreversible binding of ibrutinib to BTK, resulting in a greatly reduced drug potency; these mutations were subsequently associated with resistance to acalabrutinib and zanubrutinib.10,12,13 This has led to the development of noncovalent BTKis designed to avoid the resistance mechanisms associated with these mutations.14 Mutations in PLCG2, which acts downstream of BTK, also allow for B-cell receptor signaling irrespective of BTK inhibition,10 which also may affect the efficacy of noncovalent BTKis.15
Ibrutinib is a first-generation covalent BTKi first approved in 2013 for relapsed/refractory (R/R) mantle cell lymphoma, and, subsequently, other B-cell malignancies (CLL, Waldenström macroglobulinemia, and marginal zone lymphoma).16,17 Acalabrutinib is a selective next-generation covalent BTKi first approved for R/R mantle cell lymphoma in 2017 and for CLL in 2019.18,19 In the head-to-head ELEVATE-RR trial (NCT02477696), which comprised a population of patients with R/R CLL and higher-risk genetic features (del(17p) and/or del(11q)), acalabrutinib demonstrated noninferior progression-free survival (PFS) with an improved safety and tolerability profile, including fewer cardiovascular adverse events, vs ibrutinib. However, to our knowledge, no data to date have directly compared the mutational profiles of patients who progress on acalabrutinib and ibrutinib. Herein, we report comparative clonal evolution of genes implicated in resistance, including but not limited to BTK and PLCG2, in patients with CLL progression on acalabrutinib or ibrutinib in the ELEVATE-RR clinical trial.
Methods
Study design
The study design and primary results of ELEVATE-RR have been published previously.20 Briefly, in this phase 3, randomized, multicenter, open-label, noninferiority trial, eligible patients were adults with previously treated CLL, an Eastern Cooperative Oncology Group performance status ≤2, and presence of del(17p) and/or del(11q). Cytogenic testing of peripheral blood was performed by a central laboratory using fluorescence in situ hybridization and stimulated karyotyping. Complex karyotype was defined based on the patient having ≥3 chromosomal abnormalities and ≥1 structural abnormalities. The study was conducted in accordance with local laws, the protocol, the Declaration of Helsinki, and International Conference on Harmonization Guidelines for Good Clinical Practices. All patients provided written informed consent. Patients were randomly assigned to receive acalabrutinib 100 mg twice daily or ibrutinib 420 mg once daily until disease progression or unacceptable toxicity.
Mutational analysis
Peripheral blood samples were collected at baseline and at relapse. DNA was extracted from enriched CD19+ cells (RoboSep) and subjected to a 50-gene AmpliSeq next-generation sequencing assay (LifeTech)6 covering the full BTK and PLCG2 coding regions with a mean depth of 2000 to 4000 reads, producing a validated sensitivity cutoff of 0.5% variant allele fraction (VAF) for resistance-associated mutations. Forty-eight other genes associated with CLL were assessed at 1% to 2% VAF (based on call depth/quality), including full coding regions of ASXL1, B2M, BCL2, BCOR, BCORL1, BIRC3, BRAF, CARD11, CXCR4, DDX41, DNMT3A, ELANE, EZH2, ETV6, FBXW7, GATA2, GNA13, KLF2, KRAS, MAP2K1, MEF2B, NOTCH1, NOTCH2, NRAS, PIM1, POT1, PTEN, PTPRD, SAMHD1, SETD2, SF3B1, SH2B3, STAT6, TERC, TERT, TET2, TP53, and ZRSR2 and the recurrently mutated regions (from public variant databases) of CD79B, CREBBP, KIT, MYD88, PIK3CA, PIK3CD, PIK3CG, RPS15, U2AF1, and XPO1. Mutational data were examined in relation to PFS, which was defined as the time from random assignment to disease progression or death from any cause. Data cutoff was the same as the published primary analysis (15 September 2020).
Statistical analysis
Survival analysis using Kaplan-Meier analysis was performed to determine the median time of PFS between acalabrutinib and ibrutinib for patients who developed mutations during the trial. Proportional-hazards Cox regression analysis was used to calculate the hazard ratio and corresponding P value to assess whether a significantly increased risk of developing a mutation in 1 treatment arm vs the other existed. Median VAFs were calculated as the median of the maximum VAF values derived by gene mutation and by patient. For genes with mutations present in both treatments in ≥2 patients, P values were calculated using Wilcoxon rank sum test to determine significant difference at 95% confidence.
The study protocol and informed consent were approved by the appropriate institutional review board/independent ethics committee at each of the study sites before initiation of the study and during the study.
Results
Patients
In total, 268 and 265 patients were randomly assigned to receive acalabrutinib and ibrutinib, respectively. Demographics and baseline characteristics were reported previously.20 At baseline, 45.1% and 45.3% of acalabrutinib- and ibrutinib-treated patients, respectively, had del(17p), 62.3% and 66.0% had del(11q), 37.3% and 42.3% had TP53 mutations, 82.1% and 89.4% had unmutated immunoglobulin heavy chain variable region genes (IGHV), and 46.3% and 47.2% had a complex karyotype.20,21
Mutation analysis
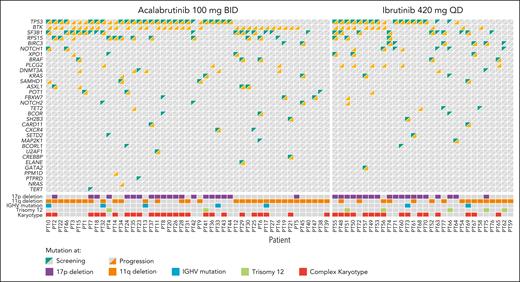
Paired (baseline and progression) samples were available and included in the analysis for 47 of 82 (57.3%) and 30 of 68 patients (44.1%) who experienced disease progression while receiving acalabrutinib and ibrutinib, respectively (supplemental Figure 1, available on the Blood website). One additional acalabrutinib-treated patient and 6 additional ibrutinib-treated patients who had Richter transformation as their mode of progression were excluded from analysis; mutational analysis for these patients can be found in supplemental Figures 2 and 3. Full data (excluding patients with Richter transformation) including mutations at screening and emergent mutations for both treatment arms are presented in Figure 1. The most common mutations at screening were TP53 mutations in both groups (supplemental Figure 4). Baseline cytogenetics for patients included in this analysis are summarized in Table 1. A summary of the change in mutations by the end of treatment is presented in supplemental Figure 5. Among patients with paired samples, the median time to progression was numerically longer for acalabrutinib vs ibrutinib (32.9 months vs 21.9 months, respectively), but the PFS hazard ratio was not significantly different between treatment groups (supplemental Figure 6).
Baseline genetics
| Baseline genetics, n (%) . | Acalabrutinib, 100 mg BID (n = 47) . | Ibrutinib, 420 mg QD (n = 30) . | P value . |
|---|---|---|---|
| 11q deletion | 32 (68.1) | 16 (53.3) | .232 |
| 17p deletion | 20 (42.6) | 19 (63.3) | .103 |
| Unmutated IGHV | 42 (89.4) | 28 (93.3) | .699 |
| Complex karyotype | 25 (53.2) | 16 (53.3) | 1 |
| TP53 mutation | 24 (51.1) | 16 (53.3) | 1 |
| Trisomy 12 positive | 2 (4.3) | 5 (16.7) | .103 |
| Baseline genetics, n (%) . | Acalabrutinib, 100 mg BID (n = 47) . | Ibrutinib, 420 mg QD (n = 30) . | P value . |
|---|---|---|---|
| 11q deletion | 32 (68.1) | 16 (53.3) | .232 |
| 17p deletion | 20 (42.6) | 19 (63.3) | .103 |
| Unmutated IGHV | 42 (89.4) | 28 (93.3) | .699 |
| Complex karyotype | 25 (53.2) | 16 (53.3) | 1 |
| TP53 mutation | 24 (51.1) | 16 (53.3) | 1 |
| Trisomy 12 positive | 2 (4.3) | 5 (16.7) | .103 |
BID, twice daily; QD, once daily.
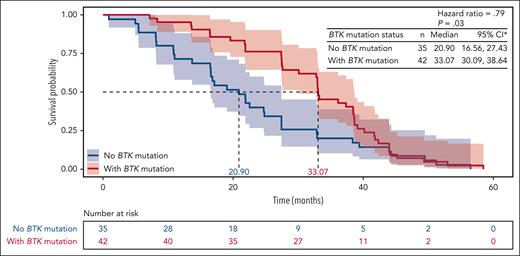
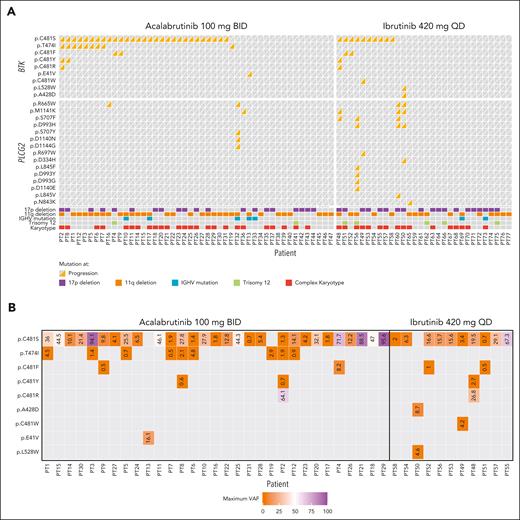
No BTK mutations were observed at screening. Acquired BTK mutations were observed in 31 acalabrutinib-treated patients (66.0%) and 11 ibrutinib-treated patients (36.7%) at time of progression (Table 2). The median VAF for BTK mutations was not significantly different in the acalabrutinib group (16.1%) vs the ibrutinib group (15.6%; supplemental Table 1). When analyzed by BTK mutation status in both treatment arms combined, time to progression was significantly (P = .03) longer in patients with vs without a BTK mutation (Figure 2). Among those with acquired BTK mutations, 29 of 31 acalabrutinib-treated (93.5%) and 10 of 11 ibrutinib-treated patients (90.9%) had C481S mutations; 2 (6.5%) and 2 (18.2%) had C481F mutations; 2 (6.5%) and 1 (9.1%) had C481Y mutations; and 1 (3.2%) and 1 (9.1%) had C481R mutations (Figure 3A). The VAF for C481S mutations ranged from 0.7% to 95.6% with acalabrutinib and from 2.0% to 67.3% with ibrutinib (Figure 3B). Of 31 acalabrutinib-treated patients, 9 (29.0%) had T474I mutations (gatekeeper mutation), only 1 of which did not co-occur with BTK C481 mutations; VAF ranged from 0.5% to 4.5%. A novel E41V mutation within the pleckstrin homology domain of BTK was seen in 1 acalabrutinib-treated patient, with a VAF of 16.1% (Figure 3A-B). Initial preclinical investigation in TMD8 cells suggests this E41V mutation may not independently confer resistance to acalabrutinib (see methodology and results in supplemental Figure 7). In the ibrutinib group, L528W (kinase-dead mutation) and A428D comutations were observed in 1 patient (and did not co-occur with C481S mutation), with VAFs of 4.6% and 8.7%, respectively, and 1 patient had C481W mutation (VAF, 4.2%; Figure 3A-B). No statistical difference was seen in the proportions of acalabrutinib- or ibrutinib-treated patients who acquired BTK mutations among patients with del(17p), del(11q), complex karyotype, unmutated IGHV, or trisomy 12 positivity (Table 2).
Emergent mutations summary
| Gene, n (%) . | Acalabrutinib, 100 mg BID (n = 47) . | Ibrutinib, 420 mg QD (n = 30) . | P value . |
|---|---|---|---|
| BTK | 31 (66.0) | 11 (36.7) | .0185 |
| By baseline cytogenetics∗ | |||
| 11q deletion | 24 (77.4) | 5 (45.5) | .0664 |
| 17p deletion | 12 (38.7) | 7 (63.6) | .18 |
| Unmutated IGHV | 28 (90.3) | 11 (100.0) | .554 |
| Complex karyotype | 18 (58.1) | 8 (72.7) | .485 |
| Trisomy 12 positive | 1 (3.2) | 2 (18.2) | .163 |
| TP53 | 6 (12.8) | 2 (6.7) | .472 |
| DNMT3A | 5 (10.6) | 1 (3.3) | .395 |
| PLCG2 | 3 (6.4) | 6 (20.0) | .142 |
| ASXL1 | 1 (2.1) | 0 | — |
| KRAS | 1 (2.1) | 0 | — |
| NRAS | 1 (2.1) | 0 | — |
| PPM1D | 1 (2.1) | 0 | — |
| RPS15 | 1 (2.1) | 0 | — |
| SAMHD1 | 1 (2.1) | 0 | — |
| TET2 | 1 (2.1) | 1 (3.3) | 1 |
| XPO1 | 1 (2.1) | 0 | — |
| FBXW7 | 0 | 1 (3.3) | — |
| NOTCH2 | 0 | 1 (3.3) | — |
| NOTCH1 | 0 | 2 (6.7) | — |
| POT1 | 0 | 1 (3.3) | — |
| Any emergent non-BTK mutation† | 14 (29.8) | 10 (33.3) | .804 |
| Any emergent mutation‡ | 36 (76.6) | 16 (53.3) | .0464 |
| Gene, n (%) . | Acalabrutinib, 100 mg BID (n = 47) . | Ibrutinib, 420 mg QD (n = 30) . | P value . |
|---|---|---|---|
| BTK | 31 (66.0) | 11 (36.7) | .0185 |
| By baseline cytogenetics∗ | |||
| 11q deletion | 24 (77.4) | 5 (45.5) | .0664 |
| 17p deletion | 12 (38.7) | 7 (63.6) | .18 |
| Unmutated IGHV | 28 (90.3) | 11 (100.0) | .554 |
| Complex karyotype | 18 (58.1) | 8 (72.7) | .485 |
| Trisomy 12 positive | 1 (3.2) | 2 (18.2) | .163 |
| TP53 | 6 (12.8) | 2 (6.7) | .472 |
| DNMT3A | 5 (10.6) | 1 (3.3) | .395 |
| PLCG2 | 3 (6.4) | 6 (20.0) | .142 |
| ASXL1 | 1 (2.1) | 0 | — |
| KRAS | 1 (2.1) | 0 | — |
| NRAS | 1 (2.1) | 0 | — |
| PPM1D | 1 (2.1) | 0 | — |
| RPS15 | 1 (2.1) | 0 | — |
| SAMHD1 | 1 (2.1) | 0 | — |
| TET2 | 1 (2.1) | 1 (3.3) | 1 |
| XPO1 | 1 (2.1) | 0 | — |
| FBXW7 | 0 | 1 (3.3) | — |
| NOTCH2 | 0 | 1 (3.3) | — |
| NOTCH1 | 0 | 2 (6.7) | — |
| POT1 | 0 | 1 (3.3) | — |
| Any emergent non-BTK mutation† | 14 (29.8) | 10 (33.3) | .804 |
| Any emergent mutation‡ | 36 (76.6) | 16 (53.3) | .0464 |
Abbreviations are explained in Table 1.
Percentages based on patients with BTK mutation.
Patients who had any newly emergent mutation during treatment in non-BTK genes (ie, a new mutation that was not present at baseline, excluding patients with BTK mutations).
Patients who had any newly emergent mutation during treatment in any gene including BTK (ie, a new mutation that was not present at baseline).
PFS by BTK mutation status. ∗Kaplan-Meier survival estimate. CI, confidence interval.
PFS by BTK mutation status. ∗Kaplan-Meier survival estimate. CI, confidence interval.
Emergent BTK and PLCG2 variants. (A) BTK and PLCG2 variants; (B) BTK mutation variant allele frequency.
Emergent BTK and PLCG2 variants. (A) BTK and PLCG2 variants; (B) BTK mutation variant allele frequency.
No PLCG2 mutations were observed at screening. Emergent PLCG2 mutations occurred in 3 acalabrutinib-treated (6.4%) and 6 ibrutinib-treated patients (20.0%; P = .142; Table 2), with median VAFs of 1.9% and 9.5%, respectively (supplemental Table 1); only 1 acalabrutinib-treated patient had co-occurrence of BTK and PLCG2 mutations vs 4 ibrutinib-treated patients (Figure 3A). One patient in the acalabrutinib group and 3 in the ibrutinib group had ≥4 co-occurring PLCG2 mutation variants. The most common acquired PLCG2 variants in acalabrutinib- and ibrutinib-treated patients, respectively, were M1141K (1 vs 3 patients), S707F (0 vs 3 patients), D993H (0 vs 3 patients), and R665W (2 vs 2 patients; Figure 3A). All other variants occurred in 1 patient each.
Preexisting TP53 mutations were present in 25 (53.2%) and 16 patients (53.3%) at screening, among whom 1 acalabrutinib-treated patient and 2 ibrutinib-treated patients lost TP53 mutation by the end of treatment (all 3 had del(17p) at baseline; supplemental Figure 5). After BTK, TP53 mutations were the next most frequent emergent mutation in the acalabrutinib arm (n = 6 [12.8%]); 2 patients (6.7%) in the ibrutinib arm had emergent TP53 mutations (Table 2); the median VAF was 6.0% in the acalabrutinib arm and 37.3% in the ibrutinib arm (supplemental Table 1). The VAF for TP53 mutations at screening and the end of treatment for each patient is shown in supplemental Figure 8. Six acalabrutinib-treated patients and 1 ibrutinib-treated patient had TP53 and BTK comutations (the 1 ibrutinib-treated patient had co-occurring TP53, BTK, and PLCG2 mutations; supplemental Figure 9).
Additional emergent mutations observed with both acalabrutinib and ibrutinib were DNMT3A and TET2 (Table 2); among the few patients who had mutations in these genes at screening, these mutations were lost by the end of treatment (supplemental Figures 4 and 5). Data regarding additional mutations and associated VAF at screening and the end of treatment are shown for the SF3B1 gene in supplemental Figure 10 and for the RPS15, BIRC3, and NOTCH1 genes in supplemental Figure 11.
Discussion
This analysis of the ELEVATE-RR study provided an opportunity to further characterize the profile of mutations observed in patients who progress on acalabrutinib and ibrutinib within a well-defined cohort of patients. Overall, the most common emergent mutation with both treatments, BTK C481S, was similar; however, the distribution of other mutations, including their respective VAFs, varied between treatment arms. Not all patients who progressed had a mutation in BTK C481, but in those who did, the VAF was highly variable; therefore, other mechanisms of resistance may exist. Baseline cytogenetics did not appear to result in differences between treatment arms in the proportion of patients with BTK mutations. No particular pattern regarding the proportion of patients with non-BTK mutations was observed in either treatment arm. Emergent BTK mutation was associated with longer time to progression than the absence of BTK mutation.
The rate of emergent BTK mutations reported with acalabrutinib at relapse in our study (66%) was consistent with the rate of BTK mutations at relapse reported previously with acalabrutinib in a single-center study of patients with treatment-naive or R/R CLL (69%).12 However, the proportion of emergent BTK mutations seen in the ibrutinib arm (37%) at relapse in our study was much lower than that previously reported in the literature (49%-67%), including real-world evidence.11,22 A greater number of samples were available for analysis in the acalabrutinib arm vs ibrutinib arm (n = 47 vs n = 30) and a greater number of patients in the ibrutinib arm (n = 6) were also excluded vs the acalabrutinib arm (n = 1) from the analysis due to Richter transformation, resulting in a greater proportion of paired samples being included for acalabrutinib (57% of progressed patients) than ibrutinib (44% of progressed patients), which could have affected the results from our analysis. With covalent BTKis, C481 mutations are typically the most common resistance mutations encountered,11 and most of the BTK mutations (>50%) in both treatment groups of this study were C481 mutations.
Mutations occurring at codon T474 are considered a gatekeeper change because they often interfere with BTKi (both covalent and noncovalent) binding to BTK, allowing for normal B-cell signaling.23 In patients with ibrutinib resistance, the T474I mutation has been previously observed co-occurring with the C481S mutation.7 Co-occurring mutations in BTK have been observed as a potential additional escape mechanism for BTKis based on preclinical data.24 Both of these mutations are considered kinase proficient, still allowing BTK kinase activity in the presence or absence of BTKis.25 T474I mutations were observed in 9 acalabrutinib-treated patients in our study, albeit at low VAF. In all but 1 of these patients, BTK C481S was also present.
The L528W mutation results in a kinase-dead BTK, hindering BTK catalytic activity; however, B-cell signaling is thought to continue via a BTK scaffolding mechanism that recruits other kinases for B-cell signaling.25,26 A recent study of the covalent BTKi zanubrutinib also identified BTK C481 mutations in 5 of 8 patients with zanubrutinib resistance, 1 of whom also harbored an L528W mutation.13 Another study showed that the L528W mutation was more prevalent in patients with CLL who had disease progression while receiving zanubrutinib than those receiving ibrutinib.27 Similarly, mutational analysis of the phase 3 ALPINE study showed L528W mutation in 2 of 5 patients with BTK mutations treated with zanubrutinib and in none of the 3 patients with BTK mutations treated with ibrutinib.28 In this study, L528W mutation was observed in 1 ibrutinib-treated patient and no acalabrutinib-treated patients. We also observed a novel E41V mutation at relapse in 1 acalabrutinib-treated patient but no ibrutinib-treated patients. A previous in vitro study in murine NIH 3T3 cells demonstrated the E41K mutation to be a BTK-activating mutation.29 Mutations at this location in the pleckstrin homology domain of BTK have resulted in higher binding affinity for inositol 1,2,3,4,5,6-hexakisphosphate, which may be involved in hematopoietic cell differentiation by activating the BTK/Tec/ITK family.30 To our knowledge, our study is the first time a BTK mutation at this residue has been observed in a treated population. The clinical relevance of the VAF of the specific mutations discussed above (C481, T474I, L528W, and E41K), however, is not well understood and is an area for further research.
Regarding other gene mutations observed in our study, PLCG2 mutations have been shown previously to confer resistance to ibrutinib by promoting B-cell receptor signaling despite continued inhibition of BTK by ibrutinib.10PLCG2 mutations were the second most frequent emergent mutations observed with ibrutinib after BTK mutations in our study, although the difference between treatment arms was not significant (P = .142). Certain PLCG2 variants have been shown previously to be associated with ibrutinib resistance31 and appeared with both acalabrutinib and ibrutinib in our analysis; PLCG2 mutations R665W and M1141K were reported in both arms, whereas other mutations were seen only in the ibrutinib arm (S707F, D993H, D993Y, L845F, and L845V) or only in the acalabrutinib arm (S707Y and D1140N). TP53 mutations were the second most common emergent mutations in acalabrutinib-treated patients after BTK mutations. Although TP53 mutation typically predisposes patients to relapse, TP53 mutation is not a known cause of disease progression with BTKi therapy, whereas mutated BTK is often associated with relapse.32 There were 3 patients whose preexisting TP53 mutations were no longer detectable at progression (1 treated with acalabrutinib and 2 treated with ibrutinib), and all 3 patients had 17p deletion. Loss of preexisting TP53 mutations has been observed previously in ibrutinib-treated patients.22
Despite shared resistance mutations such as C481S, differing patterns of mutation frequency, mutation VAFs, and uncommon BTK variants were observed with acalabrutinib vs ibrutinib in this R/R CLL population. For example, T474I occurred with acalabrutinib but not with ibrutinib. This analysis established a mutational profile in this population using a unique comparative data set; however, because of the limited sample size, the clinical significance of the mutations data reported herein is not known. In addition, for ∼33% of patients with disease progression, no emergent mutations in BTK were detected, and these patients had shorter PFS, suggesting additional research is needed to better understand mechanisms of resistance that may occur outside the B-cell receptor pathway. It is also not clear whether the higher-risk genomic features of patients in ELEVATE-RR contributed to genetic instability or affected the generalizability of the results. With limited data, the mechanism of resistance to BTKi is becoming increasingly complex. The ability to clinically sequence covalent to noncovalent BTKis may be dependent on the combination of co-occurring mutations that impart resistance to covalent and noncovalent BTKis and the VAF of each mutational clone in the tumor. It will become increasingly important to further understand the patterns of mutations commonly observed with the various BTKis to best optimize and appropriately sequence these important drugs for their maximal clinical benefit.
Acknowledgments
The study was funded by AstraZeneca. Medical writing assistance, funded by AstraZeneca, was provided by Robert J. Schoen of Peloton Advantage, LLC, an OPEN Health company, under the direction of the authors. J.A.W. is a clinical scholar of the Leukemia & Lymphoma Society.
Authorship
Contribution: J.C.B. designed the study; J.A.W., W.J., T.R., Á.I., A.P.K., P.G., J.C.B., and J.F.S. were study investigators; J.A.W., W.J., T.R., Á.I., A.P.K., P.G., J.C.B., J.F.S., and R.L. provided patients or study materials; D.J., W.J., T.R., Á.I., J.C.B., S.L., S.B., T.-H.L., G.d.B., and R.L. participated in collection and assembly of data; D.J., J.C.B., S.L., S.B., N.M., G.D.J., and S.R. participated in data analysis; J.A.W., D.J., Á.I., A.P.K., P.G., J.C.B., J.F.S., S.B., T.-H.L., N.M., G.D.J., and V.M. participated in data interpretation; J.A.W., D.J., W.J., Á.I., P.G., J.C.B., T.-H.L., N.M., and G.D.J. participated in manuscript preparation; and all authors participated in the critical review and revision of the manuscript and provided approval for the manuscript submission.
Conflict-of-interest disclosure: J.A.W. reports research funding from AbbVie, Janssen, Karyopharm Therapeutics, Loxo/Lilly, Pharmacyclics, and Schrodinger; holds a consultant or advisory role for AbbVie, AstraZeneca, BeiGene, Genentech, Janssen, Merck, Loxo/Lilly, Newave, and Pharmacyclics. D.J. reports research funding from AbbVie, Acerta/AstraZeneca, Pharmacyclics, Novartis, and MingSight; and support from The Ohio State University for high sensitivity Bruton tyrosine kinase mutation profiling. W.J. reports research funding from AbbVie, AstraZeneca, BeiGene, Janssen, Lilly, Roche, and Takeda; and consultant or advisory role fees from AbbVie, AstraZeneca, BeiGene, Lilly, Roche, and Takeda. T.R. reports research funding, consultant or advisory role fees, and honoraria from AstraZeneca, BeiGene, and Janssen. Á.I. reports research funding from Takeda and Seattle Genetics; and honoraria from Janssen, Celgene, Novartis, Pfizer, Takeda, and Roche. A.P.K. reports research funding from AstraZeneca, Bristol Myers Squibb (BMS), Roche/Genentech, Janssen, and AbbVie; serves as a consultant or adviser for AstraZeneca, BMS, Roche/Genentech, Janssen, AbbVie, and LAVA; and other fundings from Janssen, LAVA, AbbVie, and AstraZeneca. P.G. reports research funding from AbbVie, AstraZeneca, Janssen, and BMS; and honoraria from AbbVie, AstraZeneca, BeiGene, Janssen, BMS, Merck Sharp & Dohme, Loxo Oncology/Lilly, and Roche. J.C.B. reports research funding from Zencor and Pharmacyclics; holds a consultant or advisory role for Janssen, Novartis, Syndax, Newave, AstraZeneca, Kura, Vincerx Pharma, Trillium, and AbbVie; and stock ownership in Vincerx Pharma. J.F.S. reports research funding from AbbVie, Celgene, Janssen, and Roche; serves as a consultant or adviser for AbbVie, AstraZeneca, Celgene, Genentech, Genor Biopharma, Gilead, Janssen, MorphoSys, Roche, Sunesis, and TG Therapeutics; and other fundings from AbbVie, Celgene, Roche, and TG Therapeutics. G.D.J. reports employment and stock ownership in AstraZeneca. R.L. and S.R. report employment in AstraZeneca. G.d.B. reports employment in Acerta Pharma BV. V.M. reports employment in AstraZeneca; and stock ownership in AstraZeneca and Gilead Sciences. The remaining authors declare no competing financial interests.
Correspondence: Jennifer A. Woyach, Division of Hematology, The Ohio State University Comprehensive Cancer Center, 455A Wiseman Hall 410 W 12th Ave, Columbus, OH 43210; email: jennifer.woyach@osumc.edu.
References
Author notes
Presented in part at the 17th International Conference on Malignant Lymphoma, Lugano, Switzerland, 13 to 17 June 2023.
Data underlying the findings described in this manuscript may be obtained in accordance with AstraZeneca’s data sharing policy described at https://astrazenecagrouptrials.pharmacm.com/ST/Submission/Disclosure. Data for studies directly listed on Vivli can be requested through Vivli at www.vivli.org. Data for studies not listed on Vivli can be requested through Vivli at https://vivli.org/members/enquiries-about-studies-not-listed-on-the-vivli-platform/. AstraZeneca Vivli member page is also available outlining further details at https://vivli.org/ourmember/astrazeneca/.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal