Abstract
Six Epstein-Barr virus (EBV)-related lymphoproliferative disorders were investigated to verify whether the EBV strain harbored by neoplastic cells had the same EBNA-2 and latent membrane protein-1 (LMP-1) DNA sequences of the virus carried by normal lymphocytes of the same patients. Within each case, the analysis of neoplastic lymph nodes, reactive lymphadenopathies, and/or EBV+ spontaneous lymphoblastoid cell lines gave concordant results with respect to type-specific EBNA-2 region and LMP-1 gene. In particular, five cases showed the same deletion in the 3′ end of the LMP-1 gene in both normal and neoplastic cells. We also determined the prevalence of LMP-1 deletions in a large series of normal peripheral blood mononucleated cells (PBMCs) from Italian individuals. The analysis showed that 50% (9 of 18) of PBMCs from human immunodeficiency virus (HIV)-seronegative donors carried a 30-bp deletion in the C-terminal portion of the LMP-1 gene, whereas a nondeleted fragment was amplified in about 44% (8 of 18) of the cases. Only one sample (5.6%) showed the amplification of a full-length LMP-1 band together with a deleted fragment. Similarly, PBMCs from HIV-infected patients showed an almost equivalent prevalence of full-length (17 of 37, 46%) and deleted (16 of 37, 43.2%) LMP-1 fragments, whereas about 11% of samples (4 of 37) showed evidence of double infections. Of note, deletions in the LMP-1 gene were detected with similar prevalence values in EBV+ Hodgkin's disease (HD) (13 of 30, 43.3%) and non-Hodgkin's lymphoma (NHL) (2 of 5, 40%) cases from HIV-seronegative patients and in HIV-related, EBV+ NHLs (4 of 7, 57.1%). Conversely, a 30-bp LMP-1 deletion was found in 10 of 12 HIV-associated HD cases (83%), a prevalence significantly higher than that detected in HIV-unrelated HD (P = .01). These findings indicate that: (1) the same EBV strain carrying LMP-1 deletions is harbored by normal and neoplastic cells of patients with EBV+ disorders, ruling out that these mutations might result from immunoselection phenomena; (2) in the Italian population, the prevalence of LMP-1 deletion mutants is comparable to that of EBV strains with full-length LMP-1; (3) HIV-induced immunosuppression is not associated with an increased prevalence of LMP-1 deletions in PBMCs; and (4) HIV-related HD cases, but not those of HIV-seronegative Italian patients, are closely correlated with the presence of LMP-1 deletions, suggesting that infection with these strains may increase the risk of developing HD in the HIV setting.
RECENT ADVANCES in diagnostic procedures have allowed a better definition of the spectrum of diseases causally associated with Epstein-Barr virus (EBV) infection. This ubiquitous herpes virus, in fact, is now thought to be involved in the pathogenesis of a wide variety of neoplasms including Burkitt's lymphoma,1 nasopharyngeal carcinoma (NPC),2 Hodgkin's disease (HD),3 lymphomas in immunocompromised patients,4,5 and peripheral T-cell lymphomas (PTLs).6 In several of these disorders, namely NPC, HD, and PTLs, the latent membrane protein-1 (LMP-1) is the only transformation-associated EBV-encoded gene that is almost constantly expressed.6-8 EBV recombinant genetic analyses have indicated that LMP-1 is critical for primary B-lymphocyte immortalization.9 In these cells, LMP-1 can also induce many of the phenotypic changes associated with EBV infection, including increased homotypic adhesion and upregulation of adhesion molecules (leukocyte function-associated antigen [LFA]-1, intercellular adhesion molecule [ICAM]-1, LFA-3),10,11 B-cell activation markers (CD23, CD71),11,12 and the antiapoptotic bcl-2 gene.13 In addition, it has been demonstrated that LMP-1 has transforming effects also in nonlymphoid cells. In fact, rodent fibroblasts transfected with LMP-1 have reduced serum requirements, grow in soft agar, lose contact inhibition, and become tumorigenic in nude mice.14-16
The LMP-1 gene product is an integral membrane protein of 386 amino acids, with a short cytoplasmic amino terminus, six membrane-spanning segments, and a long cytoplasmic domain at the carboxy terminus.17 Recently, a 30-bp deletion at the 3′ end of the LMP-1 gene has been identified in clinically and histologically aggressive European HDs (10% to 30% of the cases),18-20 in most of PTLs,19 and in some angioimmunoblastic lymphadenopathies.21,22 This deletion, resulting in the absence of 10 amino acids (343 to 352) from the carboxy terminus of the LMP-1 protein, is identical to that described in Asian NPC.23,24 Of note, transfectants expressing these LMP-1 deletion mutants have higher tumorigenic potential than cells carrying a full-length gene.24,25 Moreover, experiments performed in a murine carcinoma model showed that LMP-1 deletion mutants were less immunogenic than the wild-type protein,26 indicating that these deletions may have a functional relevance. The results of a comparative sequence analysis of the LMP-1 gene performed in different EBV-associated lymphoproliferative disorders have led to the hypothesis that LMP-1 deletion mutants might be the result of a continuous accumulation of mutations at predilected sites.22 In particular, it has been observed that the sequence coding for the carboxy terminal domain of LMP-1 was relatively stable in the absence of possible cytotoxic T-cell responses against LMP-1–derived epitopes.22 In analogy with what was observed for EBNA-4 epitopes,27 these findings suggested that the pressure exerted by EBV-specific immunity may favor the survival of EBV-infected cells in which mutations within critical LMP-1 epitopes have occurred. Nevertheless, at present, it is not known whether, in patients with EBV-related disorders, the virus carrying LMP-1 mutations and/or deletions is harbored only by malignant cells or, alternatively, is responsible for a systemic infection. In addition, too few data are as yet available on the prevalence of LMP-1 deletion in normal individuals to draw definitive conclusions on the biological role of these particular virus variants. Elucidation of these issues may help us understand the origin of LMP-1 deletion mutants and define their possible pathogenetic role.
In the present study, we investigated the 3′ end of the LMP-1 gene harbored by neoplastic cells of a series of EBV-related lymphoproliferative disorders in comparison with that of the virus carried persistently in the normal lymphocytes of the same patients. In addition, we also determined the prevalence of LMP-1 deletion in normal peripheral blood mononucleated cells (PBMCs) from a large series of Italian individuals including healthy donors, patients with EBV-unrelated disorders, and human immunodeficiency virus (HIV)-infected subjects. Finally, these results were compared with those derived from a similar analysis performed in EBV-associated lymphoproliferative disorders arisen in both HIV-seronegative and HIV-infected Italian patients.
MATERIALS AND METHODS
Clinical samples.The prevalence of deletions in the 3′ end region of the LMP-1 gene was investigated in PBMCs from a total of 124 individuals including 42 healthy donors, 43 patients with EBV-unrelated diseases, and 39 HIV-seropositive patients. PBMCs were purified by centrifugation on Ficoll/Hypaque density gradients. HIV infection was demonstrated by multiple enzyme-linked immunosorbent assays for HIV antibodies in serum and confirmed by Western immunoblot. The analysis was also performed on frozen biopsy material obtained from a series of EBV-associated disorders including 35 HD cases and 5 non-Hodgkin's lymphomas (NHLs) (1 angioimmunoblastic lymphoma,28 2 T-cell–rich B-cell lymphomas,29 and 2 anaplastic large-cell lymphomas28 ) from HIV-seronegative patients and 13 HD and 7 NHL cases (3 Burkitt's lymphomas,28 2 diffuse large B-cell lymphomas,28 and 2 anaplastic large-cell lymphomas28 ) arisen in HIV-infected patients. Reactive lymph nodes obtained from 3 patients of our series (1 HIV-seronegative and 2 HIV-infected) were also included in the study (Table 1). Association with EBV was assessed by EBV-encoded RNA (EBER) in situ hybridization and/or Southern blotting as previously described.30 31
EBV Strains With Similar EBNA-2 and LMP-1 Sequences Are Carried by Neoplastic and Normal Cells of Patients With EBV-Associated Lymphoproliferative Disorders
| Case . | Diagnosis . | Tissues and Cell Lines Investigated . | EBNA-2 Subtype . | LMP-1 3′ Deletions . |
|---|---|---|---|---|
| TR-1 | T-cell–rich–B-cell NHL | 1 involved LN | 1 | — |
| 4 spontaneous LCLs from the lesion | 1 | — | ||
| 1 spontaneous LCL from PBMCs | 1 | — | ||
| HD-1 | HIV-unrelated HD | 1 pathologic LN | 2 | + (30 bp) |
| 1 spontaneous LCL from the lesion | 2 | + (30 bp) | ||
| 1 LN with reactive lymphadenopathy | 2 | + (30 bp) | ||
| HD-2 | HIV-unrelated HD | 1 pathologic LN | 1 | + (30 bp) |
| 1 spontaneous LCL from the lesion | 1 | + (30 bp) | ||
| HD-3 | HIV-unrelated HD | 2 involved LNs | 1 | + (63 bp) |
| 2 spontaneous LCLs from the lesion | 1 | + (63 bp) | ||
| 4 spontaneous LCLs from PBMCs | 1 | + (63 bp) | ||
| NH-1 | HIV-associated B-NHL (Burkitt's lymphoma) | 1 pathologic LN | Und. | + (30 bp) |
| 1 LN with LAS (synchronous) | Und. | + (30 bp) | ||
| HD-4 | HIV-associated HD | 1 involved LN | 1 | + (30 bp) |
| 1 LN with LAS (synchronous) | 1 | + (30 bp) |
| Case . | Diagnosis . | Tissues and Cell Lines Investigated . | EBNA-2 Subtype . | LMP-1 3′ Deletions . |
|---|---|---|---|---|
| TR-1 | T-cell–rich–B-cell NHL | 1 involved LN | 1 | — |
| 4 spontaneous LCLs from the lesion | 1 | — | ||
| 1 spontaneous LCL from PBMCs | 1 | — | ||
| HD-1 | HIV-unrelated HD | 1 pathologic LN | 2 | + (30 bp) |
| 1 spontaneous LCL from the lesion | 2 | + (30 bp) | ||
| 1 LN with reactive lymphadenopathy | 2 | + (30 bp) | ||
| HD-2 | HIV-unrelated HD | 1 pathologic LN | 1 | + (30 bp) |
| 1 spontaneous LCL from the lesion | 1 | + (30 bp) | ||
| HD-3 | HIV-unrelated HD | 2 involved LNs | 1 | + (63 bp) |
| 2 spontaneous LCLs from the lesion | 1 | + (63 bp) | ||
| 4 spontaneous LCLs from PBMCs | 1 | + (63 bp) | ||
| NH-1 | HIV-associated B-NHL (Burkitt's lymphoma) | 1 pathologic LN | Und. | + (30 bp) |
| 1 LN with LAS (synchronous) | Und. | + (30 bp) | ||
| HD-4 | HIV-associated HD | 1 involved LN | 1 | + (30 bp) |
| 1 LN with LAS (synchronous) | 1 | + (30 bp) |
Abbreviations: LN, lymph node; Und, undetermined.
Immunohistochemistry.Expression of the LMP-1 protein was investigated on Bouin-fixed or formalin-fixed, paraffin-embedded tissue sections by the alkaline phosphatase-antialkaline phosphatase (APAAP) method using monoclonal antibodies CS.1-4 (Dakopatts, Glostrup, Denmark). Tissue sections were pretreated with trypsin (0.5 mg/mL in phosphate buffered saline) for 10 minutes at 37°C. Positive controls were included in all test runs and consisted of sections of EBV-positive cell lines. Negative controls consisted of consecutive test sections in which primary antibodies were replaced by nonimmune serum of the same immunoglobulin G subclass (Dakopatts).
Establishment of EBV-infected lymphoblastoid cell lines (LCLs) by spontaneous outgrowth.To obtain spontaneous LCLs carrying the endogenous EBV strain from patients with EBV-related lymphoproliferative disorders, finely minced fragments from biopsy material were placed in fetal calf serum (FCS)-coated 24-well plates and cultured in RPMI 1640 medium (GIBCO Laboratories, Grand Island, NY) supplemented with 10% heat-inactivated FCS, 2 mmol/L L-glutamine, 100 IU/mL penicillin, and 100 IU/mL streptomycin (complete medium). Furthermore, PBMCs were obtained from the same patients by Ficoll/Hypaque gradient centrifugation. Purified PBMCs (106/mL) were seeded in 96-flat well plates and cultured in 200 L of complete medium. Cyclosporine-A (0.1 μg/mL; Sigma, St Louis, MO) was added to the medium to inhibit T-cell activation. Moreover, to prevent the release of infectious EBV virions, cells were cultured in the presence of 100 mol/L trisodium phosphonoformate, a concentration that was shown to efficaciously inhibit virus production in vitro.32 Cultures were maintained by regular refeeding for 8 weeks and observed for the appearance of actively growing foci of lymphoblastoid cells.
Southern blot analysis.Genomic DNA was purified from frozen tissues and LCLs according to conventional methods. Analysis of the molecular conformation of the EBV genome was performed as previously described31 on BamHI-digested DNA (20 μg) hybridized with the following probes: the 1.9-kb Xho I fragment corresponding to the EcoRI D fragment adjacent to the EBV right terminal repeats and the 4.1-kb EcoRI fragment corresponding to the EcoRI I region adjacent to the EBV left terminal repeats.33
Amplification of the 3′ end of the LMP-1 gene by polymerase chain reaction (PCR).The analysis was performed on DNA samples, which in preliminary experiments, were positive for PCR amplification of the β-globin gene fragment. Particular care was taken to avoid contamination of PCR samples: DNA from normal and pathologic material was extracted in a laboratory not working with EBV, blank reactions with no or unrelated DNA were interspersed with the samples, and different rooms were used to set up the reactions. PCR was always performed on 1 μg of DNA, corresponding to approximately 1.5 × 105 diploid cells. The 3′ end of the LMP-1 gene was amplified from DNA of EBV-related lymphoproliferative disorders and LCLs by using one-stage PCR with the primer pair 9/11 from Knecht et al,34 which amplifies a 315-bp DNA fragment. PCR reactions were done in a 50-L solution containing 50 mmol/L KCl, 10 mmol/L Tris-HCl (pH 9), 1.5 mmol/L MgCl2 , 200 mol/L deoxyribonucleoside triphosphates (dNTPs), 0.5 mol/L primers and 1.25 U of Taq DNA polymerase. After incubation at 95°C for 5 minutes, the reaction mixture was subjected to 40 cycles of denaturation at 94°C for 1 minute, annealing at 55°C for 1 minute, and elongation at 72°C for 30 seconds. For final extension, samples were kept at 72°C for 10 minutes. Samples with low EBV load (PBMCs and reactive lymphadenopathies) were analyzed with a seminested PCR protocol that amplifies a 283-bp fragment and uses the MS1 primer22 instead of the LMP9 primer. An aliquot (1 L) of one-stage PCR was added to the reaction mixture and subjected to an additional 20 cycles (1 minute at 94°C, 1 minute at 57°C, 30 seconds at 72°C), preceded by 5 minutes of denaturation at 95°C and followed by a 10-minute incubation at 72°C for final extension. PCR products were subsequently analyzed by electrophoresis in a 4% agarose gel stained with ethidium bromide and optically visualized by ultraviolet (UV) transillumination. The specificity of the amplified fragments was confirmed by Southern blot hybridization using the MS7 primer22 as a specific oligonucleotide-radiolabeled probe. After overnight hybridization, the membranes were washed at 55°C in 6× sodium chloride/sodium citrate buffer (SSC) and 0.05% Na pyrophosphate for 15 minutes. The sensitivity of this PCR protocol was assessed on serial dilutions of known amounts of the 315-bp DNA fragment obtained from an EBV+ LCL with the 9/11 primers. After hybridization of Southern blots and overnight exposure of the autoradiograms, the seminested PCR protocol allows the detection of one to five target molecules. As an alternative approach, on some occasions, a radioactive PCR was performed using the MS1 primer (0.1 mol/L) labeled at the 5′ end by T4 polynucleotide kinase (Boehringer, Mannheim, Germany) and 33P-γadenosine triphosphate (ATP; Amersham, Buckinghamshire, UK) in a 10-L reaction mixture. Denaturing polyacrylamide gel (6%) containing 8 mol/L urea was used to resolve amplification products. The PCR assay was performed at least twice in different experiments for each specimen, and the samples were considered positive in the case of concordant results or when a positive result could be confirmed by a subsequent experiment.
Characterization of EBV subtypes.The analysis was accomplished by a PCR amplification of the EBNA-2 region using primers specific for type 1 and type 2 EBV, as previously described.35 Briefly, the amplification was preceded by an initial denaturation step at 95°C for 7 minutes and followed by a primer extension step at 72°C for 10 minutes. The PCR was performed for 10 cycles of 1 minute at 94°C, 2 minutes at 55°C, and 1 minute at 70°C followed by a further 40 cycles of 1 minute at 90°C, 1 minute at 55°C, and 1 minute at 70°C. PCR products were subsequently analyzed by electrophoresis in a 3.5% agarose gel stained with ethidium bromide and optically visualized by UV transillumination. Specificity of the amplified fragments was confirmed by Southern blot hybridization with type-specific 32P-labeled oligonucleotide probes.35
Single-strand conformation polymorphism (SSCP) analysis.Amplification of LMP-1 fragments for SSCP analysis was performed by using an adapted version of the seminested PCR protocol described above. After one-stage PCR, DNA was subjected to an additional 20 cycles of amplification in a final volume of 10 L of reaction mixture containing 100 nmol/L primers. The MS1 primer was labeled at the 5′ end by T4 polynucleotide kinase (Boehringer) and 33P-γATP (Amersham). The reaction mixture was diluted 1:5 in 0.1% SDS/10 mmol/L EDTA and then mixed 1:1 with a sequencing stop solution (95% formamide, 20 mmol/L EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol); 6 L of reaction mixture were heat-denatured (5 minutes at 95°C), loaded onto MDE gel (AT Biochem, Malvern, PA) and run at 20 to 22 W for 6 to 8 hours with fan cooling. Dried gel was then autoradiographed using a β-max film (Amersham).
DNA sequencing.Double-stranded PCR products, obtained with the primer pair LMP9/LMP11, were purified with the Magic PCR Preps DNA Purification System (Promega, Madison, WI) from 4% low-melting agarose gel and directly sequenced with 33P-dATP (Amersham) using a Sequenase kit (US Biochemicals, Cleveland, OH). In all cases, both strands or two separate PCR products were sequenced. Sequencing primers were MS1 and LMP9 for the coding strand and LMP11 for the noncoding strand. Nucleotide numbering of the LMP-1 gene was performed according to Baer et al.36 Sequences were compared with those of EBV strains B95.836 and CAO NPC.23
Statistical evaluation.Statistical analyses were performed using the two-tailed Fisher's exact test.
RESULTS
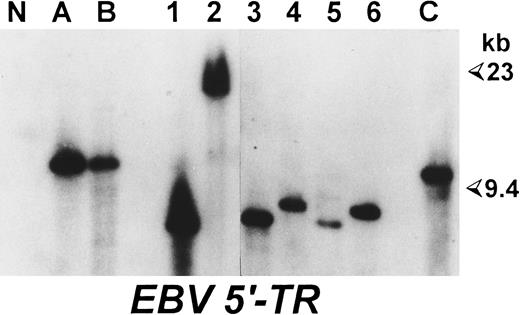
Comparative analysis of LMP-1 and EBNA-2 sequences of EBV strains carried by neoplastic and normal cells of patients with EBV-positive lymphoproliferative disorders.The analysis was performed on six cases of EBV-associated lymphoproliferative disorders (Table 1). As a source of the virus carried persistently by normal lymphocytes, we used EBV+ LCLs obtained by spontaneous outgrowth and/or reactive lymph nodes derived from the same patients. As shown in Table 1, we investigated a total of 13 different spontaneous LCLs, derived from three HIV-unrelated HD cases (HD-1, HD-2, and HD-3) and one HIV-seronegative patient with a T-cell–rich B-cell lymphoma (TR-1). Eight of these LCLs were derived from pathologic lymph nodes, whereas five were obtained from PBMCs (Table 1). All of these cell lines expressed B-lineage–specific surface antigens (CD19, CD20, and CD21) together with several activation markers (CD23, CD25, and HLA-DR) (data not shown). Southern blot analysis confirmed the EBV positivity of these LCLs and demonstrated that each cell line carried a monoclonal episome (Fig 1), consistently with the probable derivation from a single EBV-infected cell. In each case, the size of viral episomes harbored by the different LCLs was different from that of the EBV-infected cellular clone present in the pathologic tissue (Fig 1), ruling out that the LCLs obtained could be derived from the neoplastic EBV-positive cell populations. In addition, we also investigated biopsy material from reactive lymphadenopathies derived from one patient with HIV-unrelated HD (HD-1) and from two HIV-seropositive individuals, one with a concomitant HD (HD-4) and the other carrying a B-NHL (NH-1) (Table 1).
Spontaneous LCLs and neoplastic cells of the same HD patient are derived from independent EBV infections. Southern blot analysis performed on BamHI-digested DNAs hybridized with a probe corresponding to a region flanking the 5′ terminal repeats of EBV genome. (A) and (B) DNA from two pathologic lymph nodes from case HD-3; 1 and 2, DNA from two spontaneous LCLs derived from one lesion of the same HD patient; 3 to 6, DNA from four LCLs obtained by spontaneous outgrowth from the PBMCs of the same patient; N, DNA from a colon carcinoma cell line; (C) DNA from an EBV+ Burkitt's lymphoma as a positive control. Sizes are in kilobases.
Spontaneous LCLs and neoplastic cells of the same HD patient are derived from independent EBV infections. Southern blot analysis performed on BamHI-digested DNAs hybridized with a probe corresponding to a region flanking the 5′ terminal repeats of EBV genome. (A) and (B) DNA from two pathologic lymph nodes from case HD-3; 1 and 2, DNA from two spontaneous LCLs derived from one lesion of the same HD patient; 3 to 6, DNA from four LCLs obtained by spontaneous outgrowth from the PBMCs of the same patient; N, DNA from a colon carcinoma cell line; (C) DNA from an EBV+ Burkitt's lymphoma as a positive control. Sizes are in kilobases.
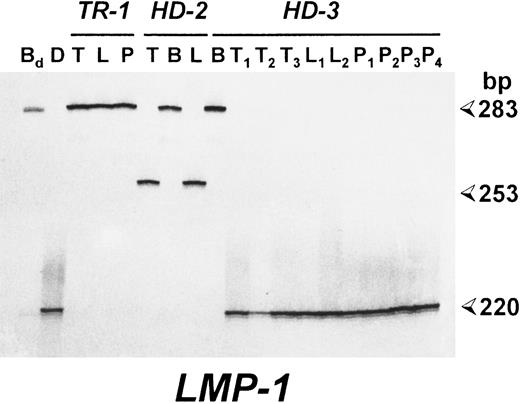
Patients with EBV-associated lymphoproliferative disorders show identical LMP-1 gene deletions in spontaneous LCLs and neoplastic cells. Polyacrylamide gel electrophoresis (PAGE) analysis of radioactive PCR products corresponding to the carboxy terminus of the LMP-1 gene. For each case, the results of the analysis of pathologic biopsy specimens (T) and of spontaneous LCLs derived from the lesion (L) or PBMCs (P) are reported. B95.8-induced LCLs were also investigated in cases HD-2 and HD-3 (B). Bd , DNA from an LCL derived from a healthy donor by transformation with the B95.8 EBV strain; (D) DNA from an EBV+ HD carrying a deleted LMP-1 gene. Sizes are in base pairs.
Patients with EBV-associated lymphoproliferative disorders show identical LMP-1 gene deletions in spontaneous LCLs and neoplastic cells. Polyacrylamide gel electrophoresis (PAGE) analysis of radioactive PCR products corresponding to the carboxy terminus of the LMP-1 gene. For each case, the results of the analysis of pathologic biopsy specimens (T) and of spontaneous LCLs derived from the lesion (L) or PBMCs (P) are reported. B95.8-induced LCLs were also investigated in cases HD-2 and HD-3 (B). Bd , DNA from an LCL derived from a healthy donor by transformation with the B95.8 EBV strain; (D) DNA from an EBV+ HD carrying a deleted LMP-1 gene. Sizes are in base pairs.
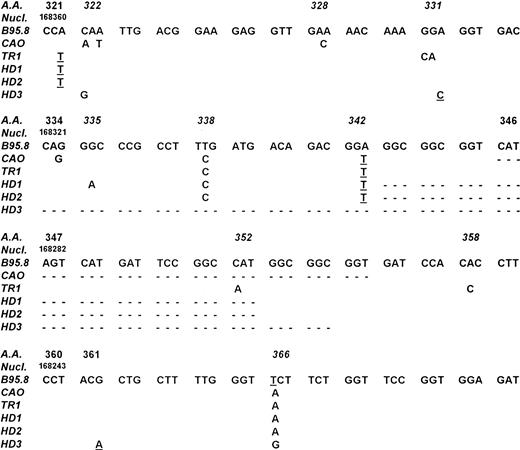
Comparison of LMP-1 DNA sequences from cases TR-1, HD-1, HD-2, HD-3 with those of the EBV strains B95.8 and CAO NPC. Numbering of nucleotides is according to Baer et al.36 Codons containing potential hot spots are reported in italics. Point mutations affecting the amino acid sequence are reported. Silent mutations are underlined and deletions are indicated by dotted lines. Cases HD-1 and HD-2 showed an identical 30-bp deletion starting from codon 343, whereas case HD-3 had a 63-bp deletion starting from codon 334.
Comparison of LMP-1 DNA sequences from cases TR-1, HD-1, HD-2, HD-3 with those of the EBV strains B95.8 and CAO NPC. Numbering of nucleotides is according to Baer et al.36 Codons containing potential hot spots are reported in italics. Point mutations affecting the amino acid sequence are reported. Silent mutations are underlined and deletions are indicated by dotted lines. Cases HD-1 and HD-2 showed an identical 30-bp deletion starting from codon 343, whereas case HD-3 had a 63-bp deletion starting from codon 334.
PCR analysis of the EBNA-2 region showed the presence of the EBV type 1-specific fragment in involved tissues of cases TR-1, HD-2, HD-3, and HD-4, whereas type 2-specific sequences were detected in case HD-1. No amplification with primers for either EBNA-2 subtypes was obtained in case NH-1 (Table 1). In each EBNA-2–positive case, reactive lymphadenopathies and/or spontaneous LCLs carried the same type-specific fragment detected in the corresponding neoplastic tissue (Table 1).
Amplification of the 3′ end of the LMP-1 gene showed the presence of the standard 315-bp LMP-1 fragment of the B95-8 EBV strain in the DNA derived from the biopsy material of patient TR-1 (Table 1). Conversely, a small deletion was detected in the DNA from the pathologic tissue of cases HD-1, HD-2, HD-4, and NH-1, whereas a larger deletion was observed in case HD-3 (Table 1). Similarly to what was observed for the EBNA-2 region, PCR analysis of spontaneous LCLs and/or reactive lymphadenopathies showed in all cases the amplification of LMP-1 fragments comigrating with those obtained from the neoplastic tissue of the corresponding patients (Table 1 and Fig 2). Sequence analysis was then performed in cases TR-1, HD-1, HD-2, and HD-3 to determine the exact location of the deletion and to detect single base mutations. As shown in Fig 3, cases HD-1 and HD-2 showed an identical 30-bp deletion starting from codon 343, whereas a 63-bp deletion starting from codon 334 was detected in case HD-3. A total of 12 mutations affecting the amino acid sequence of LMP-1 were found in the 4 cases investigated (5 in TR-1, 3 in HD-1, and 2 in HD-2 and HD-3) (Fig 3). Eight of these mutations occurred in three codons that were also mutated in the CAO LMP-1 gene. In particular, all 4 cases showed a mutation at codon 366, cases TR-1, HD-1, and HD-2 at codon 338 and case HD-3 at codon 322 (Fig 3). The other 4 missense mutations detected were not present in the sequence of CAO LMP-1 and affected the coding triplet of amino acid 331 (case TR-1), 335 (case HD-1), 352 (case TR-1), and 358 (case TR-1) (Fig 3). These findings are similar to those recently reported by Sandvej et al19 and support the hypothesis that these codons probably constitute mutational hot spots. The analysis also showed the presence of 8 mutations not affecting the amino acid sequence that were located at codons 321 (cases TR-1, HD-1, and HD-2), 331 (case HD-3), 342 (cases TR-1, HD-1, and HD-2), and 361 (case TR-3) (Fig 3).
Prevalence of LMP-1 3′ Deletions in PBMCs
| Source of PBMCs . | LMP-1 DNA Positive Cases (%) . | Full-Length LMP-1* (%) . | LMP-1 3′ Deletion† (%) . | Full-Length + 3′ LMP-1 Deletion‡ (%) . |
|---|---|---|---|---|
| Healthy donors | 9/42 (21.4) | 5/9 (55.6) | 4/9 (44.4) | — |
| Patients with EBV-unrelated disorders | 9/43 (21) | 3/9 (33.3) | 5/9 (55.6)ρ | 1/9 (11.1) |
| Total of HIV-seronegative individuals | 18/85 (21.2) | 8/18 (44.4) | 9/18 (50) | 1/18 (5.6) |
| HIV-seropositive patients | 37/39 (94.9) | 17/37 (46) | 16/37 (43.2) | 4/37 (10.8)ρ |
| Total of PBMC samples analyzed | 55/124 (44.3) | 25/55 (45.4) | 25/55 (45.4) | 5/55 (9.2) |
| Source of PBMCs . | LMP-1 DNA Positive Cases (%) . | Full-Length LMP-1* (%) . | LMP-1 3′ Deletion† (%) . | Full-Length + 3′ LMP-1 Deletion‡ (%) . |
|---|---|---|---|---|
| Healthy donors | 9/42 (21.4) | 5/9 (55.6) | 4/9 (44.4) | — |
| Patients with EBV-unrelated disorders | 9/43 (21) | 3/9 (33.3) | 5/9 (55.6)ρ | 1/9 (11.1) |
| Total of HIV-seronegative individuals | 18/85 (21.2) | 8/18 (44.4) | 9/18 (50) | 1/18 (5.6) |
| HIV-seropositive patients | 37/39 (94.9) | 17/37 (46) | 16/37 (43.2) | 4/37 (10.8)ρ |
| Total of PBMC samples analyzed | 55/124 (44.3) | 25/55 (45.4) | 25/55 (45.4) | 5/55 (9.2) |
Cases with no evidence of 3′ LMP-1 deletions showing the amplification of a single LMP-1 fragment of the same size of that obtained from the DNA of the B95.8 virus strain.
Cases carrying a 30-bp deletion at the 3′ end of the LMP-1 gene.
Cases showing the concomitant amplification of a full-length and a deleted LMP-1 fragment. In 2 cases (1 in d and 1 in e), a 3′ LMP-1 deletion larger than 30 bp was observed.
ρ In one of these cases, a 3′ LMP-1 deletion larger than 30 bp was observed.
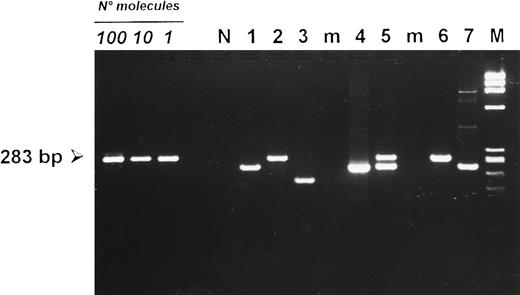
Detection of deletions at the carboxy terminal region of the LMP-1 gene in PBMCs from HIV-seronegative donors (lanes 1 to 3) and HIV-infected patients (lanes 4 to 7 ). Cases 2 and 6 carried a full-length LMP-1 fragment of 283 bp. PBMCs from cases 1, 4, and 7 showed a smaller fragment consistent with a 30-bp deletion. Case 5 showed the concomitant amplification of a full-length and a deleted LMP-1 fragment, whereas case 3 carried a deletion larger than 30 bp. (N) DNA from an EBV-negative squamous cell carcinoma of the head and neck. (M) HaeIII-digested DNA of X-174-RF as a molecular-weight marker. Amplification of 100, 10, and 1 target molecule is also included to show the sensitivity of the seminested PCR protocol used.
Detection of deletions at the carboxy terminal region of the LMP-1 gene in PBMCs from HIV-seronegative donors (lanes 1 to 3) and HIV-infected patients (lanes 4 to 7 ). Cases 2 and 6 carried a full-length LMP-1 fragment of 283 bp. PBMCs from cases 1, 4, and 7 showed a smaller fragment consistent with a 30-bp deletion. Case 5 showed the concomitant amplification of a full-length and a deleted LMP-1 fragment, whereas case 3 carried a deletion larger than 30 bp. (N) DNA from an EBV-negative squamous cell carcinoma of the head and neck. (M) HaeIII-digested DNA of X-174-RF as a molecular-weight marker. Amplification of 100, 10, and 1 target molecule is also included to show the sensitivity of the seminested PCR protocol used.
Prevalence of LMP-1 3′ Deletions in EBV+ Lymphoproliferative Disorders From HIV-Seronegative and HIV-Infected Patients
| . | Hodgkin's Disease . | Non-Hodgkin's Lymphomas . | ||
|---|---|---|---|---|
| . | HIV-Seronegative (%) . | HIV-Seropositive (%) . | HIV-Seronegative (%) . | HIV-Seropositive (%) . |
| Full-length LMP-1 | 18/30 (60) | 2/12 (16.7) | 3/5 (60) | 3/7 (42.9) |
| LMP-1 3′ deletion | 12/303-150 (40) | 10/12 (83.3)3-151 | 2/5 (40) | 4/7 (57.1) |
| . | Hodgkin's Disease . | Non-Hodgkin's Lymphomas . | ||
|---|---|---|---|---|
| . | HIV-Seronegative (%) . | HIV-Seropositive (%) . | HIV-Seronegative (%) . | HIV-Seropositive (%) . |
| Full-length LMP-1 | 18/30 (60) | 2/12 (16.7) | 3/5 (60) | 3/7 (42.9) |
| LMP-1 3′ deletion | 12/303-150 (40) | 10/12 (83.3)3-151 | 2/5 (40) | 4/7 (57.1) |
Two of these cases carried a deletion larger than 30 bp.
The prevalence of LMP-1 deletions in HIV-related HD is significantly higher (P = .01) than that detected in HD from HIV-seropositive patients.
To verify that the EBV strain harbored by neoplastic cells had the same LMP-1 DNA sequence of the virus carried by normal lymphocytes of the same patient, cases TR-1, HD-1, HD-2, and HD-3 were investigated by SSCP analysis. In each case, LMP-1 fragments from normal or neoplastic cells showed an identical migration pattern (not shown), thus excluding the existence of subtle sequence differences.
Prevalence of LMP-1 3′ deletions in PBMCs.The prevalence of deletions in the C-terminal portion of the LMP-1 gene was investigated in 124 PBMC samples. As shown in Table 2, LMP-1 sequences were amplified in 21.2% (18 of 85) of PBMCs from HIV-seronegative individuals. Of note, about 50% of positive samples (9 of 18) carried a 30-bp LMP-1 deletion, whereas a nondeleted, B95.8-like fragment was amplified in about 44% of the cases (8 of 18) (Table 2). When considered separately, PBMCs from HIV-seronegative healthy donors and from patients with EBV-unrelated disorders showed comparable prevalence values of LMP-1 deletions (Table 2). In one PBMC sample, a deletion larger than 30 bp was detected (Fig 4). Only one case (5.6%) showed the amplification of a full-length LMP-1 band together with a deleted LMP-1 fragment (Table 2). A remarkably higher number of PBMCs from HIV-infected patients were positive for LMP-1 DNA sequences (37 of 39, 94.9%) (Table 2). Similarly to what was observed in HIV-seronegative individuals, PBMCs from HIV-infected patients showed an almost equivalent prevalence of full-length (17 of 37, 46%) and deleted (16 of 37, 43.2%) LMP-1 fragments (Table 2). One sample carried a deletion larger than 30 bp, whereas about 11% of PBMCs from HIV-infected individuals (4 of 37) showed the concomitant amplification of a full-length and a deleted LMP-1 fragment (Table 2), consistent with the existence of double infections in these patients.
Prevalence of LMP-1 3′ deletions in EBV-associated lymphoproliferative disorders from HIV-seronegative and HIV-infected patients.Immunohistochemical analysis showed that the LMP-1 protein was expressed by neoplastic cells of all HIV-unrelated HD and NHL cases from our series. Amplification of the 3′ end of the LMP-1 gene was successful in 30 of 35 HD and 5 of 5 NHL cases from HIV-seronegative patients. The prevalence of LMP-1 3′ deletion in EBV+ HD (13 of 30, 43.3%) and NHL (2 of 5, 40%) cases from HIV-seronegative patients was similar to that detected in normal PBMCs samples (Table 3). Two HIV-unrelated HD cases showed the presence of a deletion larger than 30 bp. When HIV-associated lymphoproliferative disorders were investigated, LMP-1 DNA sequences were amplified in 12 of 13 HD and seven of seven NHL samples. A 30-bp LMP-1 deletion was detected in 4 of 7 HIV-related EBV+ NHLs (57.1%), 1 CD30+ anaplastic large-cell lymphoma with high numbers of LMP-1+ cells, and 3 Burkitt's lymphomas negative for LMP-1 expression or showing only rare LMP-1+ elements. All 12 HIV-related HD cases investigated were positive for LMP-1 expression. Of note, a 30-bp LMP-1 deletion was found in 10 of 12 HIV-associated HD cases (83%), a prevalence significantly higher than that detected in HIV-unrelated HD (P = .01) (Table 3). In 1 HIV-related HD case, a deletion smaller than 30 bp was observed. Moreover, LMP-1 deletions were significantly more prevalent in HIV-associated HD cases than in PBMCs from HIV-seropositive individuals (P = .04). Among both HIV-associated and HIV-unrelated HD cases, histology showed that LMP-1 deletions were apparently independent of HD morphologic subtype, the degree of infiltration by Hodgkin and Reed-Sternberg cells, and intensity of staining for the LMP-1 protein (manuscript in preparation).
Relationships between presence of LMP-1 deletions and EBV subtype were investigated in 26 HIV-unrelated and nine HIV-associated EBV+ HD cases (Table 4). LMP-1 deletions were present in 10 of 23 ordinary and 4 of 5 HIV-related HD cases carrying type 1 EBV and in 1 of 2 HIV-unrelated and 3 of 3 HIV-associated HD cases harboring type 2 virus. In one ordinary HD case harboring a deleted LMP-1, no amplification of the EBNA-2 region could be obtained. One HIV-related HD carrying an LMP-1 deletion mutant showed infection with both EBV subtypes (Table 4).
EBNA-2 Typing and Prevalence of LMP-1 3′ Deletions in EBV+ Hodgkin's Disease From HIV-Seronegative and HIV-Infected Patients
| LMP-1 3′ Deletion . | HIV-Seronegative . | HIV-Seropositive . | ||||
|---|---|---|---|---|---|---|
| . | Type 1 . | Type 2 . | Untypable . | Type 1 . | Type 2 . | Types 1 + 2 . |
| + | 10 | 1 | 1 | 4 | 3 | 1 |
| − | 13 | 1 | 0 | 1 | 0 | 0 |
| Total | 23 | 2 | 1 | 5 | 3 | 1 |
| LMP-1 3′ Deletion . | HIV-Seronegative . | HIV-Seropositive . | ||||
|---|---|---|---|---|---|---|
| . | Type 1 . | Type 2 . | Untypable . | Type 1 . | Type 2 . | Types 1 + 2 . |
| + | 10 | 1 | 1 | 4 | 3 | 1 |
| − | 13 | 1 | 0 | 1 | 0 | 0 |
| Total | 23 | 2 | 1 | 5 | 3 | 1 |
DISCUSSION
In the present study, we provide evidence that the same EBV strain is carried by neoplastic cells and normal lymphocytes of patients with EBV-associated lymphoproliferative disorders. Within each case, in fact, EBV+ clonal cells from the lesion and normal EBV-infected lymphocytes not only carried the same type-specyfic EBNA-2 region, but also showed the same DNA sequence encoding for the C-terminal portion of the LMP-1 protein. Of note, 5 of the 6 cases investigated carried LMP-1 deletions identical to those described in Asian NPC23,24 as well as in some European HD and PTL cases.18-20 These results seem to rule out that LMP-1 deletions may arise as a consequence of immunologically mediated selective pressure in patients with EBV-associated lymphoproliferative disorders. This possibility appears unlikely in the light of the recent observation that tumor-infiltrating lymphocytes derived from EBV+ HD lacked EBV-specific cytotoxicity.37 In addition, LMP-1 deletions were also detected in EBV+ lymphomas with no evidence of LMP-1 protein expression, similarly to what was reported by others.38 Moreover, our findings seem to exclude that LMP-1 deletions may result from a progressive accumulation of mutations at the 3′ end of the gene occurring before the onset of the disease and support the hypothesis that EBV strains with LMP-1 deletions are responsible for preexisting persistent infections in these patients. Occasionally, however, LMP-1 deletions may constitute later events, occurring after spreading of the disease, as suggested by the recent finding of two EBV+ HD cases with full-length LMP-1 at diagnosis, but showing a 30-bp LMP-1 deletion at relapse.39
Our PCR analysis performed in normal PBMCs confirmed that EBV strains with deletions in the LMP-1 gene are present in the general population. The large majority of these samples (>95% in HIV-seronegative donors) carried a virus with either full-length or deleted LMP-1, consistent with previous observations indicating that healthy EBV carriers are prevalently infected by a single virus strain.40 Moreover, our results indicate that EBV variants with LMP-1 deletions are relatively frequent in Italy, displaying a prevalence comparable to that of viruses with full-length LMP-1. When compared with HIV-seronegative donors, a significantly higher number of PBMC samples positive for LMP-1 DNA sequences was detected in HIV-infected individuals. This is consistent with the marked increase in the numbers of EBV-infected cells circulating in the blood of HIV-seropositive patients, a phenomenon related to the frequent reactivation of EBV occurring in the context of HIV-associated immunosuppression.41 Of note, as for HIV-seronegative donors, PBMCs from HIV-infected individuals also showed comparable prevalence values of EBV strains with full-length or deleted LMP-1. These findings indicate that the altered immune competence of HIV-seropositive individuals does not increase the prevalence of LMP-1 deletion mutants in these patients, as instead observed for type 2 EBV.42 Moreover, at variance with the data obtained in a murine model,26 these results suggest that the immunogenicity of EBV strains with LMP-1 deletions is not substantially different from that of viruses with full-length LMP-1.
Interestingly, LMP-1 deletions were found as frequently in normal PBMCs as in EBV+ HD and NHL cases from HIV-seronegative Italian patients. Consistently, similar prevalence values of LMP-1 deletions (about 30%) were recently found in tonsils of infectious mononucleosis and HD biopsy specimens from Danish patients.19 These findings suggest that in immunocompetent hosts, EBV strains with deleted or full-length LMP-1 are probably characterized by similar lymphomagenetic potential. On these grounds, infection with EBV variants carrying LMP-1 deletions does not seem to confer an increased risk of developing EBV-associated HD in the general population. The recent finding that Danish (60%) and Malaysian (100%) PTLs showed a high frequency of LMP-1 deletions19 leaves open the possibility that these deletions may be relevant to the pathogenesis of these neoplasms. Nevertheless, studies aimed at defining the prevalence of LMP-1 deletion in the normal population of these countries are needed to elucidate this issue.
Although our results suggest that LMP-1 deletions have no preferential role in the induction of HIV-unrelated HD, there are some data indicating that these mutations can affect the course of the disease. In fact, previous reports have shown that EBV+ HD cases with LMP-1 deletions have clinicopathologic features of more malignant diseases.18,20 This has been related to the fact that deletions in the carboxy terminal region of the LMP-1 gene (between amino acid 334 and 364) prolong the half-life of the protein, leading to increased levels of LMP-1 expression in infected cells.43 Accordingly, immunohistochemical analyses have shown that Hodgkin and Reed-Sternberg cells of HD cases carrying these deletion mutants often show strong signals for LMP-1.18 20 Nevertheless, the number of HD cases investigated so far is limited and prospective studies are still lacking to draw definitive conclusion on the possible prognostic value of LMP-1 deletions in EBV-associated disorders.
Of note, unlike HIV-unrelated cases, HD arising in HIV-seropositive patients was closely correlated with the presence of LMP-1 deletions (83.3%, P = .01). These findings are in keeping with those recently reported by Santón et al,20 who detected a 30-bp LMP-1 deletion in 10 of 10 HIV-related HD cases from Spain. Since the prevalence of LMP-1 deletions in our HIV-associated HD series was significantly higher than that detected in PBMCs from Italian HIV-seropositive patients, infection with LMP-1 deletion mutants seems to confer an increased risk of developing HD in the HIV setting. It is worth considering that HIV-related HDs usually belong to unfavorable histological subtypes, present with advanced disease at diagnosis, and have an aggressive clinical course.44 The possibility that LMP-1 deletions may contribute to the malignant behavior of HIV-associated HD cases constitutes an attractive hypothesis that deserves further investigation.
In our series of HIV-associated and HIV-unrelated HD cases, no correlation was found between the presence of LMP-1 deletions and EBV subtype. The finding that type 2 EBV may also carry LMP-1 deletions confirms that LMP-1 deletion mutants are not a substrain of type 1 EBV, as instead suggested by initial studies.23,45 These results, taken together with the observation that the 3′ end of the LMP-1 gene is a likely mutational hot spot,19,22 suggest that LMP-1 deletions have originated independently in the two EBV strains. Available evidence indicates that type 2 EBV has less efficient transforming properties than type 1 virus.46 Similar to what was recently observed in Danish EBV-associated lymphoproliferative disorders, most of the Italian HD cases with type 2 EBV infection carried LMP-1 deletions.19 These findings support the hypothesis that LMP-1 deletions may be relevant to increase the transforming ability of type 2 EBV.
ACKNOWLEDGMENT
The authors thank S. Rizzo and A. Marzotto for their excellent technical assistance and Dr P. Tonel and P. Pistello for help with the manuscript.
Supported in part by Associazione Italiana per la Ricerca sul Cancro (Milan) and by CNR special project ACRO (95.00504.PF39; Rome, Italy).
Address reprint requests to Mauro Boiocchi, PhD, Division of Experimental Oncology 1, Centro di Riferimento Oncologico, via Pedemontana Occidentale 12, 33081 Aviano (PN) Italy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal