Abstract
In this study, we sought to identify factors responsible for the positive modulation of lactoferrin (LF), a neutrophil-specific, secondary-granule protein gene. Initial reporter gene transfection assays indicated that the first 89 base pairs of the LF promoter are capable of directing myeloid-specific LF gene expression. The presence of a C/EBP site flanked by 2 Sp1 sites within this segment of the LF promoter prompted us to investigate the possible role of these sites in LF expression. Cotransfection studies of LF-89luc plasmid with increasing concentrations of a C/EBP expression vector in myeloid cells resulted in a linear transactivation of luciferase reporter activity. Electrophoretic mobility shift assays found that the C/EBP site is recognized by C/EBP and that both LF Sp1 binding sites bind the Sp1 transcription factor specifically in myeloid cells. Mutation of either Sp1 site markedly reduced activity of the LF-89luc plasmid in myeloid cells, and neither Sp1 mutant plasmid was transactivated by a C/EBP expression plasmid to the same extent as wild-type LF-89luc. We also transfected LF-89luc into Drosophila Schneider cells, which do not express endogenous Sp1, and demonstrated up-regulation of luciferase activity in response to a cotransfected Sp1 expression plasmid, as well as to a C/EBP expression plasmid. Furthermore, cotransfection of LF-89luc plasmid simultaneously with C/EBP and Sp1 expression plasmids resulted in an increase in luciferase activity greater than that induced by either factor alone. Taken together, these observations indicate a functional interaction between C/EBP and Sp1 in mediating LF expression.
During granulopoiesis, the developing neutrophil undergoes a series of commitment steps that limit its proliferative capacity while increasing its morphologic and biochemical maturity.1 The acquisition of primary (azurophilic) and secondary (specific) granules marks progressive stages of neutrophil differentiation. Acute myeloid leukemias are characterized by the lack of secondary granules and their content proteins. Hence, an understanding of the molecular mechanisms underlying neutrophil development is crucial to defining defects associated with leukemia.
Lactoferrin (LF) is one of the secondary-granule protein (SGP) genes, a group of genomically unlinked and functionally diverse genes expressed late in neutrophil maturation. We previously demonstrated that SGP gene expression is coordinately regulated at the transcriptional level during neutrophil maturation.2-5 Additionally, we showed that CCAAT displacement protein (CDP/cut), a highly conserved silencing factor that binds the LF promoter, coordinately represses expression of all SGP genes.6 7 In this study, we sought to identify factors responsible for the up-regulation of LF in order to describe possible shared positive regulators of SGP gene expression. We identified a C/EBP binding site flanked by 2 Sp1 binding sites within the first 89 base pairs (bps) of the LF promoter and investigated the possible importance of these sites in regulating LF expression during granulocytic maturation.
The role of transcription factors in hematopoietic proliferation, differentiation, and survival is becoming increasingly clear.8,9 Maturation of the multipotent progenitor stem cell into specialized blood cells (eg, lymphocytes, erythrocytes, neutrophils, monocytes, eosinophils, and others) is thought to be partly regulated by a well-orchestrated interplay of transcription factors capable of instructing expression of a specific set of lineage-specific genes.9 Current evidence suggests that a combination of transcription factors, both lineage restricted and ubiquitous, is required to achieve differentiation within a given hematopoietic lineage. In the myeloid compartment, several granulocyte-macrophage–specific genes are regulated by combinations of the transcription factors C/EBP, PU.1, c-Myb, AML-1, and Sp1,9 and target genes have been found to have multiplecis-acting elements for these transcription factors in their promoters. Gene-disruption experiments showed that PU.1 and C/EBPα transcription factors are indispensable for normal progression of the myeloid development program.10-12
CCAAT-enhancer binding proteins belong to a family of the basic region–leucine zipper (bZip) class of transcription factors that recognize the consensus DNA-binding sequence 5′ ATTGCGCAAT 3′ in the regulatory regions of target genes. C/EBP family proteins bind as either homodimers or heterodimers. This family of transcription factors currently includes C/EBPα C/EBPβ, C/EBPγ C/EBPδ, C/EBPε, and CHOP-GADD 153, all of which contain highly homologous C-terminal dimerization (leucine-zipper) domains and DNA-binding (basic-region) motifs but differ in their N-terminal transactivation domains (except for CHOP-GADD 153, which lacks this domain).13 With the exception of C/EBPε, which is expressed at high levels mainly in the late stages of granulopoiesis, C/EBP family members are expressed in a wide variety of cells, including liver, adipocyte, lung, intestine, adrenal gland, placenta, and peripheral blood mononuclear cells.13 Profound hematopoietic abnormalities have been reported in mice nullizygous for C/EBPα, C/EBPβ, and C/EBPε. C/EBP family members are known to exert pleiotropic effects in the tissues in which they are expressed. This may be because of their tissue- and stage-specific expression, their ability to dimerize both with members of their own family and with the Fos/Jun and ATF/CREB families of transcription factors, their ability to interact with other transcription factors, such as NF-κB and Sp1, or a combination of these factors.9
Sp1 is a ubiquitous DNA-binding transcriptional activator that recognizes GC-rich sequences in the promoters of several TATA-less genes.14,15 Although Sp1 is considered to be a constitutive activator of gene expression, several studies indicated that Sp1-dependent transactivation is mediated through a variety of signals, including transforming growth factor-β induction on the p15 gene16 and cyclic adenosine monophosphate activation of the CYP11A gene.17 Sp1 is abundantly expressed in myeloid cells,18 and several studies have implicated this transcription factor in mediating myeloid-specific gene expression. For example, in cooperation with the ets factor GABP, Sp1 achieved high levels of myeloid-specific expression of the CD18 promoter.19 Additionally, the C/EBP family of proteins was found to interact functionally with Sp1 to regulate the activity of the CD11c integrin gene promoter.20 The aim of this study was to identify the role of Sp1 and C/EBP in mediating expression of the LF gene in myeloid cells.
Materials and methods
Plasmid construction
5′-end deletions of the LF promoter were obtained from a previously described LF genomic clone containing 950 bps 5′ of the transcription start site.2 Subcloning of the LF649 fragment into a pGL3basic promoter-less vector upstream of the luciferase reporter gene (Promega Biotech, Madison, WI) was described previously.6 This clone was used to prepare shorter LF promoter constructs, which were then subcloned into pGL3basic. The LF649-pGL3basic plasmid was digested with Spe1 (which cuts at position −237 in the LF promoter) and BglII. The resultant 240-bp fragment was subcloned into the pGL3basic vector previously digested with NheI and BglII enzymes to yield the LF237-pGL3basic plasmid. Next, 5 μg of this plasmid was linearized with SpeI and treated with 0.5 U Bal 31(NEB, Beverly, MA) at 37°C for up to 5 minutes to generate a series of 5′-end deletions in the LF promoter. At 1-minute intervals, aliquots of the reaction were removed and added to 20 mmol/L ethyleneglycotetraacetic acid (EGTA) to stop the Bal 31reaction. Bal 31–digested DNA fragments were digested with BglII enzyme and subsequently subcloned intoSmaI/BglII–digested pGL3basic vector. The resulting series of LF 5′-deleted clones were sequenced. Clones LF167 and LF89 were picked for further analysis. A mutant C/EBP site was created in the LF89-pGL3basic clone as follows. Sense and antisense oligomers containing the first 89 bps of the LF promoter with an NheI site at the 5′ end and an XhoI site at the 3′ end were synthesized with a mutation in the C/EBP site: 5′ TTGGGCAAC 3′ (wild type) was changed to 5′ CCTTTAGGC 3′ (mutant C/EBP).
Complementary oligomers were annealed and ligated into the pGL3basic vector, which had been previously digested withNheI/XhoI. Six subclones containing inserts were sequenced to confirm the presence of the mutation in the C/EBP site in the LF promoter. A similar strategy was used to construct mutations in the 2 Sp1 sites in the LF89-pGL3 clone. The distal Sp1 site was mutated from 5′ AGTGGGGA 3′ (wild type) to 5′ AGTAAAAA 3′ (mutant Sp1A), and the pGL3basic plasmid was referred to as mutant LFSp1A. Similarly, the proximal Sp1 site was mutated from 5′ GGGCGGGG 3′ (wild type) to 5′ TTATATAT 3′ (mutant Sp1B), and the pGL3basic plasmid was referred to as mutant LF89Sp1B. Large-scale plasmid preparations (Qiagen, Valencia, CA) of all LF promoter–pGL3basic plasmids were made, divided into aliquots, and stored at −20°C. The mutations introduced into the C/EPB, Sp1A, Sp1B sites did not give rise to or overlap with the binding site for any known transcription factor, as judged by a comparison with transcription factors in 2 data bases.
Tissue culture, transient transfections, and luciferase assay
Mouse erythroleukemic (MEL) cells were obtained from the American Type Culture Collection (ATCC) and were maintained and grown in Dulbecco modified Eagle medium (Gibco, Grand Island, NY) supplemented with 10% heat-inactivated fetal-calf serum (FCS; Gemini Bioproducts, Calabasas, CA), 0.2 mmol/L glutamate, 50 units/mL penicillin, and 50 μg/mL streptomycin. 32Dwt18 cells, a gift from Dr Daniel Link (University of Washington, St Louis, MO), were grown in Iscove modified Dulbecco medium supplemented with 10% FCS and 10% WEHI-conditioned medium, as a source of interleukin 3 (IL-3).21U937-C/EBPα cells, which were described previously,22were a gift from Dr Daniel G. Tenen (Harvard Medical School, Boston, MA). These cells were maintained in RPMI 1640 medium (Gibco) supplemented with 10% FCS (Gemini Bioproducts) and 850 μg/mL G418 (Gibco). All cells were maintained at 37°C in a humidified 5% carbon dioxide (CO2) incubator. U937-C/EBPα cells were induced to express the C/EBPα gene, under the control of the metallothionein promoter, by the addition of 100 μmol/L zinc sulfate. Maturation of all inductions was monitored with use of Wright-Giemsa staining.
For transient transfection experiments, approximately 1 × 107 cells were gently pelleted, washed twice with phosphate-buffered saline (PBS), and resuspended in 180 μLN-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES)–buffered saline in electroporation cuvettes. Then, 10 to 20 μg of each reporter plasmid construct and 2 μg of pCMVβgal (Clontech, Palo Alto, CA), an internal control plasmid used to monitor transfection efficiency, were added to each aliquot of cells. After a 5-minute incubation at room temperature, the DNA-cell samples were electroporated by using a Bio-Rad Gene Pulser (Hercules, CA). MEL and U937-C/EBPα cells were electroporated at 200 mV with 960-μF capacitance. 32Dwt18 cells were electroporated at 400 mV with a capacitance of 250 μF. Transiently transfected cells were incubated in growth medium at 37°C in 5% CO2 for 16 to 20 hours. Luciferase activity was then determined with an assay kit (Promega Biotech), according to the manufacturer's instructions. Luciferase expression levels were normalized to the levels of β-galactosidase expression or as per microgram of total protein, as described previously.6
Schneider cells (ATCC CRL-1963; Drosophila melanogaster embryo line 2) were grown in Shang M3 medium (Difco Labs, Detroit, MI) supplemented with 10% FCS and Bactopeptone (12.5 g/L; Difco Labs) and 5 g/L technetium Yeastolate (Difco Labs) and incubated at 25°C. These cells were transfected by using 30 μL Lipofectamine (Gibco), 10 μg of LF89-pGL3basic plasmids, and 5 μg of effector plasmids pPac-Sp1 (containing full-length Sp1 complementary DNA [cDNA] in the pPAC expression vector [a gift from Robert Tjian, Berkeley, CA]) or pPac-C/EBPα (or both), which was constructed by isolating the full-length rat C/EBPα cDNA from pMSVC/EBPα plasmid (a gift from Dr Alan Friedman, Johns Hopkins University, Baltimore, MD) withBamHI and subcloning it into the BamHI site of the pPac expression vector. The correct orientation of the C/EBPα cDNA was determined by sequence analysis of the resultant clones. Salmon-sperm DNA (Sigma, St Louis, MO) was used to normalize the total amount of DNA in each transfected sample. Luciferase activity was measured 48 hours after transfection.
Preparation of nuclear extracts
Nuclear extracts were prepared essentially as described previously.6 Briefly, 1 × 107 cells were washed twice in ice-cold PBS and once in buffer A containing 10 mmol/L HEPES-potassium hydroxide (KOH) (pH 7.9), 1.5 mmol/L magnesium chloride (MgCl2), 10 mmol/L potassium chloride (KCl), 0.5 mmol/L dithiothreitol (DTT), and 0.5 mmol/L phenylmethylsulfonyl fluoride (PMSF). Cells were lysed after 5 minutes of incubation on ice in buffer A with 0.1% NP-40. Nuclei were recovered by centrifugation at 4°C for 15 minutes. The nuclei were then lysed in high-salt buffer C (20 mmol/L HEPES-KOH [pH 7.9], 10% glycerol, 420 mmol/L sodium chloride, 10 mmol/L KCl, 0.2 mmol/L EDTA, and 0.5 mmol/L DTT), and nuclear extracts were recovered by centrifugation at 4°C for 15 minutes. The salt concentration in the nuclear extracts was adjusted by adding buffer D (20 mmol/L HEPES-KOH [pH 7.9], 20% glycerol, 0.05 mmol/L KCl, 0.2 mmol/L EDTA, 0.5 mmol/L DTT, and 0.5 mmol/L PMSF). All buffers used included the following mixture of protease inhibitors at a final concentration of : pepstatin A (10 μg/mL), leupeptin (10 μg/mL), aprotinin (1 μg/mL), and perfabloc (1 mg/mL) (Boehringer Mannheim, Indianapolis, IN). Total protein concentration in the nuclear-extract preparations was assayed by using a Bio-Rad kit according to the manufacturer's instructions. Most preparations yielded 1.5 to 2 μg/μL protein. Aliquots of nuclear-protein extracts were frozen immediately and stored at −70°C until used.
Oligonucleotides and electrophoretic mobility shift assays (EMSAs)
Complementary oligonucleotides were annealed and labeled at their 5′ ends by using γ-phosphorus 32 adenosine triphosphate (222 × 1012 Bq/mmol; Amersham, Buckinghamshire, United Kingdom) and T4 polynucleotide kinase (NEB). Radiolabeled double-stranded oligonucleotides were separated from unincorporated nucleotide by passage through a Sephadex G-25 spin column (Boehringer Mannheim). Probes were stored at −20°C.
EMSAs were performed by incubating 15-μg nuclear extracts or 1 foot-printing unit of affinity-purified Sp1 protein (Promega Biotech) with 20 000 cpm of double-stranded oligonucleotide in a 20-μL reaction mixture containing 10 mmol/L HEPES-KOH buffer (pH 7.9), 50 mmol/L KCl, 2.5 mmol/L MgCl2, 1 mmol/L DTT, 10% glycerol, 1 μg acetylated bovine serum albumin (NEB), and 0.5 μg poly(dI-dC) at 25°C for 20 minutes. For competition analysis, a 100-fold molar excess of unlabeled oligonucleotides was added to the nuclear extracts before addition of the labeled probe. For the supershift assay, polyclonal C/EBPα and Sp1 antibodies (Santa Cruz Biotech, Santa Cruz, CA) were incubated with nuclear extracts for 15 minutes after the addition of radiolabeled probe. Binding reactions were resolved on a 4% nondenaturing polyacrylamide gel containing 1 × TBE (0.089 mol/L Tris-borate, 0.089 mol/L boric acid, and 0.002 mol/L EDTA) and electrophoresed at 150 V for 3 hours at 4°C. Gels were exposed to x-ray film with an intensifying screen overnight at −80°C.
The oligonucleotide used in the EMSA analysis had the following sequence (the C/EBP and Sp1 sites are underlined): 5′ GGGAGGGAAGGGTGTCTAGG 3′ LF-C/EBP, 5′ GC- GGGAA 3′ LF Sp1A oligomer; 5′ CAAC- AAAGC 3′ LF Sp1B oligomer; and 5′ ATTCGAT- AGC 3′ consensus Sp1 site. The CD18 probe that includes the proximal CD18 Sp1 binding site corresponding to CD18-85/−3719 was 5′ AACCCACCACTTCCTCCAAGGAGGAGCTGAGA- GGAACAGGAAGTGTCAG 3′. The irrelevant (nonspecific) probe that lacks an Sp1 binding site (corresponding to CD18-903/−88319 was 5′ GCGAAGCTTGCAGTGAGCTGAGATCACGGATCCGCG 3′.
Results
Transient transfection of LF promoter plasmids in myeloid and nonmyeloid cells
In an attempt to identify factors responsible for the up-regulation of LF during myeloid maturation, LF promoter fragments (LF89, LF167, LF237, and LF 649) cloned into the promoterless pGL3basic vector harboring the luciferase reporter gene were transiently transfected into myeloid (32Dwt18) and nonmyeloid (MEL) cells. 32Dwt18 is an IL-3–dependent subline of 32Dcl3 cells. Transfected cells were harvested 24 hours after transfection and luciferase reporter gene activity was measured. As shown in Figure1A, all LF promoter plasmids expressed high levels of luciferase activity in 32Dwt18 cells but not in MEL cells. LF89 conferred the highest level of luciferase activity in 32Dwt18 cells (56-fold above that of pGL3basic alone). Similarly high levels of luciferase activity for the LF89 plasmid were detected in other myeloid cell lines, such as NB4 and EPRO (data not shown). LF89-luc activity in the nonmyeloid Cos-7 cell line, on the other hand, was similar to that observed in MEL cells (data not shown). A 4-fold decline in luciferase activity between LF89 and LF167 suggested the presence of a negative regulatory element or elements between these coordinates. Differences in reporter gene activity between LF237 and LF649 suggested the presence of additional regulatory elements within those coordinates of the LF promoter. Notable sequences in this portion of the promoter are an overlapping estrogen receptor and retinoic acid response element, which is thought to mediate LF expression in mammary gland and hematopoietic cells,23 and a mitogen response unit capable of binding AP-1 and CREB in mammary epithelial cells.24
Lactoferrin (LF) promoter transfection and nucleotide sequence.
(A) Transient transfection of LF promoter plasmids in 32Dwt18 (myeloid) and MEL (nonmyeloid) cells. LF promoter fragments (LF89, LF167, LF237, and LF 649) were cloned into the promoterless pGL3B vector and transiently transfected into 32Dwt18 and MEL cells, along with pCMVβgal plasmid. Cells were harvested 24 hours after transfection and luciferase reporter gene activity was measured. The figure represents the mean ± SE value from 3 independent experiments, each performed in duplicate. Normalized luciferase values are represented as a ratio of luciferase activity to pGL3B luciferase activity. (B) Nucleotide sequence of the human LF promoter and its 5′ flanking sequence. The position of the LF89 (−89 base pairs [bps]) is indicated. Putative binding sites for Sp1, Ets, C/EBP, and Myb transcription factors are indicated.
Lactoferrin (LF) promoter transfection and nucleotide sequence.
(A) Transient transfection of LF promoter plasmids in 32Dwt18 (myeloid) and MEL (nonmyeloid) cells. LF promoter fragments (LF89, LF167, LF237, and LF 649) were cloned into the promoterless pGL3B vector and transiently transfected into 32Dwt18 and MEL cells, along with pCMVβgal plasmid. Cells were harvested 24 hours after transfection and luciferase reporter gene activity was measured. The figure represents the mean ± SE value from 3 independent experiments, each performed in duplicate. Normalized luciferase values are represented as a ratio of luciferase activity to pGL3B luciferase activity. (B) Nucleotide sequence of the human LF promoter and its 5′ flanking sequence. The position of the LF89 (−89 base pairs [bps]) is indicated. Putative binding sites for Sp1, Ets, C/EBP, and Myb transcription factors are indicated.
Because the first 89-bp region of the LF promoter is capable of directing myeloid-specific expression, we examined more closely the sequence of this region of the human LF promoter, the sequence of which we described previously.2 Figure 1B illustrates the sequence of the first 170 bps of the LF promoter. Putative binding sites for C/EBP, Sp1, ETS, and c-Myb were recognized within the first 89 bps of the LF promoter. Using supershift EMSA analysis, we demonstrated that the ETS site in the LF promoter does not bind PU.1, Ets-1, Ets-2, or GABP but may bind another unidentified member of the Ets family of transcription factors (data not shown). Additionally, transient cotransfection analysis in 32Dwt18 cells and in NIH-3T3 cells using expression plasmids for PU.1 or c-Myb did not appear to transactivate the LF89-luc plasmid markedly (data not shown). Because preliminary analysis of the ETS and Myb sites in the LF promoter suggested that they did not play an important role in regulating LF gene expression, we did not further investigate these sites in the current study.
Binding of Sp1 and C/EBP to their respective binding sites in the LF89 promoter fragment indicated by EMSA analysis
To demonstrate binding of the Sp1 and C/EBP transcription factors to the putative binding sites within the first 89 bps of the LF promoter, we performed an EMSA using fragments of the LF89 promoter fragment containing the 2 Sp1 sites (Sp1A and Sp1B) and the C/EBP site. Nuclear extracts from the U937 cell line were used as a source of nuclear proteins. U937 is a human myelomonocytic cell line that can be made to undergo partial granulocytic maturation on induction with all-trans retinoic acid (ATRA).22 ATRA-induced U937 cells, however, do not express LF (Khanna-Gupta A, Berliner N, unpublished data). In a study by Radomska et al,22 U937 cells were stably transfected with a plasmid harboring rat C/EBPα cDNA under the influence of a zinc-inducible human metallothionein promoter. Conditional expression of C/EBPα in these U937-C/EBPα cells induced a granulocytic maturation program, complete with the expected morphologic and biochemical changes, including expression of LF at the messenger RNA level.22
Using a probe spanning coordinates −89 bp to −66 bp (Sp1A probe) of the LF promoter, an EMSA using U937 nuclear extracts was performed. As shown in Figure 2A, several DNA-protein complexes were formed as a result of the interaction of probe Sp1A and U937 nuclear extracts (lane 2); however, only one of the complexes appeared to be specifically eliminated competitively by the addition of a 100-fold molar excess of unlabeled Sp1A probe (lane 3). The same complex was eliminated competitively by the addition of a 100-fold molar excess of either a probe containing a consensus Sp1 binding site (Figure 2A, lane 4) or a probe harboring an Sp1 site from the CD18 promoter19 (lane 5) but not by the addition of a nonspecific probe (lane 6). Preincubation of this complex with an anti-Sp1 antibody but not with preimmune serum (data not shown) resulted in the appearance of a supershifted complex (Figure 2A, lane 7, arrow marked “supershift”). Furthermore, the complex in question (Figure 2A, arrow marked “Sp1”) in fact migrated to the same extent in the nondenaturing gel as did a complex formed by using purified Sp1 protein. Taken together, these observations suggest that the Sp1 protein specifically binds to the LF promoter between the −89-bp and −66-bp coordinates.
Electrophoretic mobility shift assays (EMSAs) of fragments within the first 89 bps of the LF promoter.
(A) An EMSA was performed by using double-stranded phosphorus 32–labeled LF −89 bp to −66 bp (SP1A) as a probe with nuclear extracts from U937 cells. Binding of Sp1 (lower arrow) was subjected to competition with a 100-fold molar excess of self cold competitor (CC) (lane 3), Sp1 consensus probe (lane 4), and a CD18 Sp1 cold competitor (lane 5) but not a 100-fold molar excess of a nonspecific (n.s.) probe (lane 6). Preincubation of Sp1A probe and U937 nuclear extracts with antiserum to Sp1 resulted in a band shift (lane 7, upper arrow [supershift]). Purified Sp1 protein also bound to the Sp1A probe (lane 8). (B) Results of EMSA analysis using nuclear extracts from uninduced (U937/U) and zinc-induced (U937/I) U937-C/EBPα cells and LF −53 bp to −35 bp (SP1B) as a probe. In both uninduced and induced extracts, Sp1 was eliminated competitively by the addition of a 100-fold molar excess of Sp1B probe (self CC, lanes 3 and 7) and an Sp1 consensus probe (lanes 4 and 8) but not by a nonspecific probe (n.s., lanes 5 and 9, lower arrow). Preincubation of protein-DNA complexes with anti-Sp1 antiserum resulted in a supershifted band (upper arrow) in both uninduced (lane 9) and induced (lane 10) nuclear extracts. (C) EMSA supershift analysis was performed on the radiolabeled LF-C/EBP binding site (LF −74 bp to −51 bp) by using nuclear extracts from uninduced U937-C/EBPα cells and from U937-C/EBPα cells induced with zinc. C/EBPα bound to the LF-C/EBP site from both uninduced (lane 2) and induced (lane 5) U937-C/EBPα extracts (lower arrow). The protein-DNA complex was supershifted (upper arrow) by anti-C/EBPα antiserum (lanes 3 and 6) but not by anti-C/EBPβ antiserum (lanes 4 and 7).
Electrophoretic mobility shift assays (EMSAs) of fragments within the first 89 bps of the LF promoter.
(A) An EMSA was performed by using double-stranded phosphorus 32–labeled LF −89 bp to −66 bp (SP1A) as a probe with nuclear extracts from U937 cells. Binding of Sp1 (lower arrow) was subjected to competition with a 100-fold molar excess of self cold competitor (CC) (lane 3), Sp1 consensus probe (lane 4), and a CD18 Sp1 cold competitor (lane 5) but not a 100-fold molar excess of a nonspecific (n.s.) probe (lane 6). Preincubation of Sp1A probe and U937 nuclear extracts with antiserum to Sp1 resulted in a band shift (lane 7, upper arrow [supershift]). Purified Sp1 protein also bound to the Sp1A probe (lane 8). (B) Results of EMSA analysis using nuclear extracts from uninduced (U937/U) and zinc-induced (U937/I) U937-C/EBPα cells and LF −53 bp to −35 bp (SP1B) as a probe. In both uninduced and induced extracts, Sp1 was eliminated competitively by the addition of a 100-fold molar excess of Sp1B probe (self CC, lanes 3 and 7) and an Sp1 consensus probe (lanes 4 and 8) but not by a nonspecific probe (n.s., lanes 5 and 9, lower arrow). Preincubation of protein-DNA complexes with anti-Sp1 antiserum resulted in a supershifted band (upper arrow) in both uninduced (lane 9) and induced (lane 10) nuclear extracts. (C) EMSA supershift analysis was performed on the radiolabeled LF-C/EBP binding site (LF −74 bp to −51 bp) by using nuclear extracts from uninduced U937-C/EBPα cells and from U937-C/EBPα cells induced with zinc. C/EBPα bound to the LF-C/EBP site from both uninduced (lane 2) and induced (lane 5) U937-C/EBPα extracts (lower arrow). The protein-DNA complex was supershifted (upper arrow) by anti-C/EBPα antiserum (lanes 3 and 6) but not by anti-C/EBPβ antiserum (lanes 4 and 7).
Binding of Sp1 to the Sp1B probe (LF promoter −53 bp to −35 bp) was also determined by EMSA analysis. Sp1B probe was incubated with nuclear extracts prepared from both U937-C/EBPα cells (uninduced myeloid cells; Figure 2B, lane 2) and U937-C/EBPα cells induced with zinc for 3 days (induced myeloid cells; lane 6). The similar protein-DNA–complex pattern observed with the 2 cell types (Figure 2B, lanes 2 and 6) was eliminated competitively by the addition of a 100-fold molar excess of unlabeled Sp1B or a probe harboring a consensus Sp1 site (lanes 3, 4, 7, and 8) but not with a nonspecific probe (lanes 5 and 9). Preincubation of Sp1B-U937 protein-DNA complexes with an Sp1 antibody resulted in a supershifted band (Figure 2B, lanes 10 and 11, arrow). On the other hand, preincubation of the Sp1B-U937 protein-DNA complexes with preimmune serum did not result in a supershifted band (data not shown). These data confirm that Sp1 also recognizes and binds to the LF promoter between the −53-bp and −35-bp coordinates.
To confirm binding of C/EBPα to the C/EBP site in the LF promoter, an EMSA was performed using an LF89 fragment containing the C/EBP site (−74 bp to −51 bp of LF promoter) and nuclear extracts from uninduced U937-C/EBPα cells and from U937-C/EBPα cells induced with zinc for 3 days. Although C/EBPα bound to the LF-C/EBP probe in uninduced U937-C/EBPα extracts (because of either endogenous C/EBPα binding activity or the leakiness of the metallothionein promoter) (Figure 2C, lane 2), its binding was sharply increased in zinc-induced extracts (Figure 2C, lane 5). Supershift analysis confirmed that C/EBPα (Figure 2C, lanes 3 and 6, arrow) and not C/EBPβ (lanes 4 and 7) bound to the LF-C/EBP probe in this cell line.
Activation of the LF promoter by C/EBP in myeloid 32Dwt18 cells
Having established the ability of Sp1 and C/EBPα to bind to their respective sites in the LF89 fragment of the lactoferrin promoter, we next assessed the functional contribution of C/EBPα in LF gene expression in a myeloid cell line, 32Dwt18. This 32Dcl3 subline is dependent on IL-3 and constitutively expresses a stably transfected chimeric form of the granulocyte colony-stimulating factor (G-CSF) receptor that contains the extracellular domain of the erythropoietin receptor and the intracellular signaling domain of the G-CSF receptor. We previously showed that 32Dwt18 cells differentiate in response to erythropoietin induction in the absence of IL-3 and express the LF gene at levels comparable to those of the 32Dcl3–G-CSF system, without the 80% cell death associated with induction of 32Dcl3 with G-CSF.6
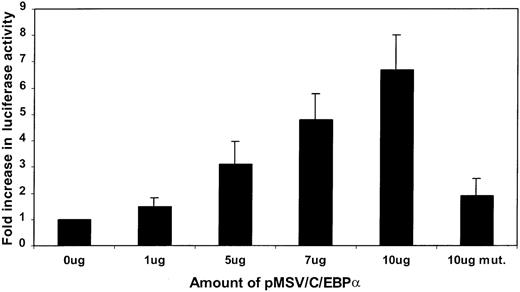
The 32Dwt18 cells were transiently cotransfected with LF89 or a C/EBP mutant of the LF89 plasmid and increasing concentrations (0, 1, 5, 7, and 10 μg) of an expression plasmid for C/EBPα (pMSVC/EBPα). Increasing concentrations of the C/EBPα expression plasmid correlated with an increase in luciferase activity of the LF89 (pGL3B-LF89) plasmid (Figure 3). However, 10 μg of C/EBPα expression vector was unable to transactivate the mutant LF89 plasmid to the same extent as wild-type LF89 (Figure 3). This experiment indicated that C/EBPα transactivates the LF promoter in 32Dwt18 cells and that its effect is mediated through the C/EBP binding site.
Cotransfection of LF89, mutant LF89, and an expression C/EBP plasmid (pMSVC/EBP) in 32Dwt18 cells.
32Dwt18 cells were transiently cotransfected with 10 μg of LF89 or a C/EBP mutant of the LF89 plasmid and increasing amounts (0-10 μg) of an expression plasmid for C/EBPα (pMSVC/EBPα). Two micrograms of pCMVβgal plasmid was included in each transfection. Cells were harvested 24 hours after transfection and reporter gene activity was measured. Normalized luciferase values are represented as a ratio of enzyme activity of LF89 plus pMSVC/EBPα to that of LF89 without pMSVC/EBPα. The mean ± SE value for 3 experiments performed in duplicate is represented.
Cotransfection of LF89, mutant LF89, and an expression C/EBP plasmid (pMSVC/EBP) in 32Dwt18 cells.
32Dwt18 cells were transiently cotransfected with 10 μg of LF89 or a C/EBP mutant of the LF89 plasmid and increasing amounts (0-10 μg) of an expression plasmid for C/EBPα (pMSVC/EBPα). Two micrograms of pCMVβgal plasmid was included in each transfection. Cells were harvested 24 hours after transfection and reporter gene activity was measured. Normalized luciferase values are represented as a ratio of enzyme activity of LF89 plus pMSVC/EBPα to that of LF89 without pMSVC/EBPα. The mean ± SE value for 3 experiments performed in duplicate is represented.
Role of Sp1 binding sites in the LF promoter in 32Dwt18 cells
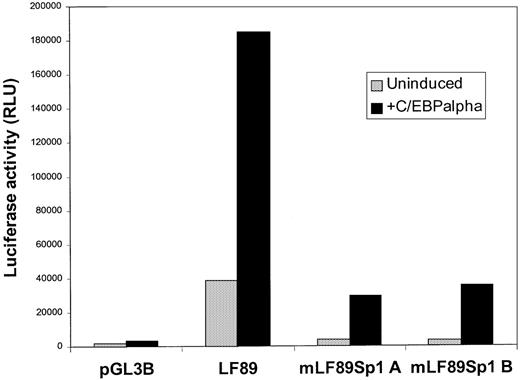
Because Sp1 is a ubiquitous transcription factor that is expressed abundantly in myeloid cells, a transient cotransfection experiment with increasing concentrations of an Sp1 expression vector and LF89 plasmid would not have been informative. Instead, 32Dwt18 cells were transiently transfected with LF89 or Sp1A and Sp1B mutant forms of the LF89 plasmid, with and without 10 μg of an expression plasmid for C/EBPα (pMSVC/EBPα). As shown in Figure4, wild-type LF89 plasmid was transactivated (4.8-fold) by C/EBPα as expected. However, both Sp1 mutants of the LF89 plasmid were unable to be transactivated by C/EBPα to the same extent as the wild-type LF89 plasmid, even though the C/EBP binding site in the Sp1 mutants A and B was intact. In fact, the level of wild-type LF89 transactivation was 5.7-fold greater than mutant Sp1 LF89 transactivation in the presence of exogenously expressed C/EBPα. This observation suggests that both Sp1 binding sites in the LF89 promoter fragment must be functional for efficient transactivation of the LF89 promoter fragment by C/EBPα.
Transient cotransfection of mutant LF Sp1 plasmids with a C/EBP expression plasmid in 32Dwt18 cells.
32Dwt18 cells were transiently cotransfected with 10 μg of LF89, SP1A, and SP1B mutant forms of the LF89 plasmid, with and without 10 μg of an expression plasmid for C/EBPα (pMSVC/EBPα); 2 μg of pCMVβgal plasmid was included in each transfection. Cells were harvested 24 hours after transfection and reporter gene activity was measured. Normalized luciferase values from 1 of 3 experiments performed in duplicate are represented.
Transient cotransfection of mutant LF Sp1 plasmids with a C/EBP expression plasmid in 32Dwt18 cells.
32Dwt18 cells were transiently cotransfected with 10 μg of LF89, SP1A, and SP1B mutant forms of the LF89 plasmid, with and without 10 μg of an expression plasmid for C/EBPα (pMSVC/EBPα); 2 μg of pCMVβgal plasmid was included in each transfection. Cells were harvested 24 hours after transfection and reporter gene activity was measured. Normalized luciferase values from 1 of 3 experiments performed in duplicate are represented.
Interestingly, a 5- to 12-fold increase in luciferase activity was observed in the presence of exogenous C/EBPα for LF89 and for the Sp1 mutant LF89 plasmids A and B; however, the overall reporter gene activity for the mutant Sp1 LF89 plasmids was clearly diminished compared with the results for wild-type LF89. Furthermore, the basal-level reporter gene activity of both Sp1 mutant plasmids was 11-fold lower than that of the wild-type LF89 plasmid. This may be because Sp1 may play a role in mediating basal-level transcription of the LF promoter in myeloid cells.
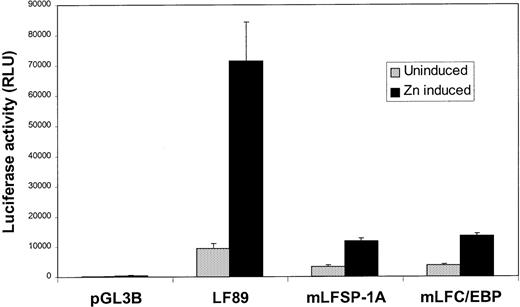
These observations were confirmed by using the U937-C/EBPα cell line, which can be made to undergo neutrophil maturation on induction with zinc ions.22 As shown in Figure5, induction of wild-type LF89 plasmid in the presence of zinc ions resulted in a 7.7-fold induction of reporter gene activity. A mutation in the C/EBP site in the LF89 promoter fragment resulted in a 5.3-fold reduction in luciferase activity compared with wild-type LF89 transactivation with C/EBPα. A mutation in the Sp1A site in the LF89 promoter fragment resulted in a reduction in reporter gene activity (6-fold) similar to that observed for the mutant C/EBP-LF89 plasmid. Mutant Sp1B showed a similar pattern of expression in this cell line (data not shown). Thus, it is evident that C/EBPα is incapable of transactivating LF89 promoter plasmids with mutations in either of the 2 Sp1 sites (A or B) or in the C/EBP site within the first 89 bps of the LF promoter to the same extent as the wild-type LF89 plasmid.
Transient transfection of wild-type and mutant LF89 plasmids in U937-C/EBP cells.
The C/EBPα gene under the control of a zinc-inducible metallothionein promoter was stably transfected into U937 cells. These cells can be made to undergo maturation along the granulocytic lineage after zinc induction, which results in high levels of C/EBPα expression. U937-C/EBPα cells were transiently transfected with LF89, a C/EBP mutant of LF89 (mLFC/EBP), or an SP1 (A) mutant of LF89 (mLFSP-1A) plasmids. One-half of the cells were induced with zinc for 48 hours after transfection (Zn induced), whereas the other one-half were incubated in medium without zinc (Uninduced); 2 μg of pCMVβgal plasmid was included in each transfection. Normalized luciferase values are represented as a ratio of luciferase activity with zinc to luciferase activity without zinc. The mean ± SE value for 3 experiments performed in duplicate are illustrated.
Transient transfection of wild-type and mutant LF89 plasmids in U937-C/EBP cells.
The C/EBPα gene under the control of a zinc-inducible metallothionein promoter was stably transfected into U937 cells. These cells can be made to undergo maturation along the granulocytic lineage after zinc induction, which results in high levels of C/EBPα expression. U937-C/EBPα cells were transiently transfected with LF89, a C/EBP mutant of LF89 (mLFC/EBP), or an SP1 (A) mutant of LF89 (mLFSP-1A) plasmids. One-half of the cells were induced with zinc for 48 hours after transfection (Zn induced), whereas the other one-half were incubated in medium without zinc (Uninduced); 2 μg of pCMVβgal plasmid was included in each transfection. Normalized luciferase values are represented as a ratio of luciferase activity with zinc to luciferase activity without zinc. The mean ± SE value for 3 experiments performed in duplicate are illustrated.
Activation of the LF promoter by Sp1 in Drosophila Schneider cells
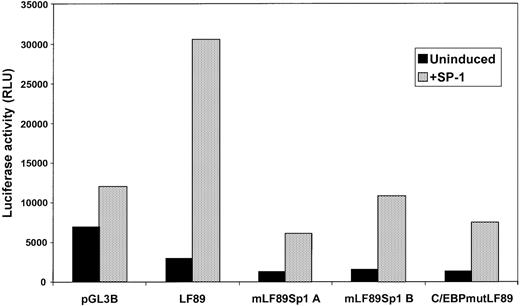
To further assess the role of Sp1 in activating the LF promoter, we transfected the LF89 promoter plasmids intoDrosophila Schneider cells, which lack endogenous Sp1. Wild-type, mutant Sp1A, mutant Sp1B, and mutant C/EBP-LF89 plasmids linked to the luciferase reporter gene were cotransfected intoDrosophila Schneider cells with an Sp1 expression plasmid (pPac-Sp1). The results shown in Figure 6indicate that cotransfection of wild-type LF89 (which contains the Sp1A site, the Sp1B site, and the C/EBP site) with pPac-Sp1 inDrosophila cells resulted in an increase in luciferase activity to 12-fold above that of LF89 alone. This observation demonstrates that Sp1 activates the LF promoter in cells that otherwise lack Sp1 activity. A decrease in reporter gene activity was observed when mutant Sp1A (one-fifth of wild-type LF89 levels) and mutant Sp1B LF89 (one-third of wild-type LF89 levels) plasmids were cotransfected with pPac-Sp1. Thus, disruption of either Sp1 site in the LF promoter significantly impaired the ability of pPac-Sp1 to activate the promoter. Of note, mutations in each of the Sp1 sites of the LF promoter (mutant Sp1A and mutant Sp1B LF89) did not completely abolish pPac-Sp1–mediated transactivation, presumably because Sp1 bound to the unmutated Sp1 site in each case. Interestingly, a mutated C/EBP site in the LF89 promoter fragment also interfered with the ability of pPac-Sp1 to activate the LF promoter in the Drosophila cell line. In this situation, reporter gene activity was decreased to one-fourth of wild-type LF89 levels. Thus we surmise that both Sp1 sites, as well as the C/EBP site, contribute to the activation of LF expression via Sp1.
Transient cotransfection analysis of LF promoter plasmids with an SP1 expression plasmid in Drosophila S2 cells.
Drosophila S2 cells (Sp1 negative) were transiently cotransfected with LF89 and Sp1A, Sp1B, and C/EBP mutant forms of the LF89 plasmid, with and without 5 μg of an expression plasmid for Sp1 (pPACSp1). Cells were harvested 48 hours after transfection and reporter gene activity was assessed. Luciferase values were normalized as per microgram of total protein. One representative experiment of 3 performed in duplicate is shown.
Transient cotransfection analysis of LF promoter plasmids with an SP1 expression plasmid in Drosophila S2 cells.
Drosophila S2 cells (Sp1 negative) were transiently cotransfected with LF89 and Sp1A, Sp1B, and C/EBP mutant forms of the LF89 plasmid, with and without 5 μg of an expression plasmid for Sp1 (pPACSp1). Cells were harvested 48 hours after transfection and reporter gene activity was assessed. Luciferase values were normalized as per microgram of total protein. One representative experiment of 3 performed in duplicate is shown.
Sp1 and C/EBP cooperate to activate the LF promoter
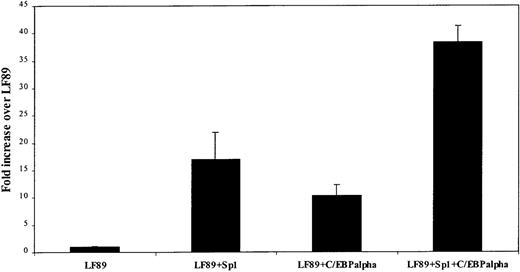
On the basis of the observations shown in Figure 5 and Figure 6, which suggested that both Sp1 and C/EBPα are necessary for LF expression, we hypothesized that Sp1 and C/EBPα must cooperate to activate the LF promoter at the transcriptional level. To test this hypothesis, we cotransfected the wild-type LF89 promoter plasmid with an Sp1 expression plasmid (pPac-Sp1), a C/EBPα expression plasmid (pPac-C/EBPα), or both expression plasmids simultaneously inDrosophila Schneider cells (Figure7). Under these conditions, Sp1 transactivated the LF89 plasmid to a level 17-fold above that observed with LF89 alone. C/EBPα also up-regulated LF89 luciferase activity but to a relatively lesser extent (10.3-fold above the level with LF89 alone). However, transfection with both Sp1 and C/EBPα activated the LF promoter 38-fold, an effect higher than the sum of the levels observed for the 2 individual transactivations. We therefore conclude that Sp1 and C/EBPα work cooperatively to activate the LF promoter.
Sp1 and C/EBP cotransfection with LF89 inDrosophila S2 cells.
Drosophila S2 cells were cotransfected with 10 μg of wild-type LF89 plasmid and 5 μg of an Sp1 expression plasmid (pPAC-Sp1), 5 μg of a C/EBPα expression plasmid (pPAC-C/EBPα), or 5 μg of each expression plasmid. Salmon-sperm DNA was used to normalize the total amount of DNA used in each transfection. Transfected cells were harvested 48 hours after transfection. Normalized luciferase values (per microgram of protein) are represented as a ratio of enzyme activity of LF89 plus expression plasmid to enzyme activity of LF89 without expression plasmid. The mean ± SE value for 3 experiments performed in duplicate is represented.
Sp1 and C/EBP cotransfection with LF89 inDrosophila S2 cells.
Drosophila S2 cells were cotransfected with 10 μg of wild-type LF89 plasmid and 5 μg of an Sp1 expression plasmid (pPAC-Sp1), 5 μg of a C/EBPα expression plasmid (pPAC-C/EBPα), or 5 μg of each expression plasmid. Salmon-sperm DNA was used to normalize the total amount of DNA used in each transfection. Transfected cells were harvested 48 hours after transfection. Normalized luciferase values (per microgram of protein) are represented as a ratio of enzyme activity of LF89 plus expression plasmid to enzyme activity of LF89 without expression plasmid. The mean ± SE value for 3 experiments performed in duplicate is represented.
Discussion
In this study, we demonstrated that a region spanning the first 89 bps of the LF promoter is sufficient to confer high-level reporter gene expression in myeloid cells (32Dwt18, U937, NB4 and EPRO cells) but not in nonmyeloid cells (MEL and Cos-7 cells). We also showed, by using transient transactivation analysis and site-directed mutagenesis, that a C/EBP site (located between −60 and −51 bp in the LF promoter) is essential for LF promoter activity. In addition, EMSA analysis demonstrated that C/EBPα specifically recognizes and binds to this region of the LF promoter. LF is thus among a growing number of myeloid-specific genes regulated by C/EBPα.
C/EBPα has been postulated to be a master regulator of the granulopoietic developmental program. It is expressed at high levels throughout myeloid differentiation and has been shown to bind to multiple myeloid-specific gene promoters thereby regulating gene expression at many different stages of myeloid maturation. C/EBPα−/−-mice die perinatally due to defects in gluconeogenesis that result in fatal hypoglycemia25; however, they also present a selective early block in the differentiation of granulocytes.26 Newborn C/EBPα knockout mice do not produce any mature neutrophils, although immature myeloid cells with an increased myeloid colony-forming unit potential are found in the peripheral blood.26 It should be noted, however, that since all members of the C/EBP family of transcription factors recognize the 5′ ATTGCACAAT 3′ consensus motif in the regulatory domains of target genes, other C/EBP family members may also contribute to myeloid-specific LF expression by means of the C/EBP site described. In this regard, we have demonstrated the ability of C/EBPε to transactivate the LF89 plasmid in 32Dwt18 cells (Khanna-Gupta A, Berliner N, unpublished data). The ability of C/EBPε to recognize and bind to the C/EBP site in the LF promoter in Cos cells overexpressing C/EBPε has been reported.27Additionally, since the C/EBP transcription factors are members of a bZip family of transcription factors capable of homodimerizing and heterodimerizing not only with other C/EBP family members but also with members of other bZip families (such as ATF/CREB and Fos/Jun), the possibility that such other bZip transcription factors contribute to LF expression by means of the C/EBP site cannot be ruled out.
Our data also demonstrate the importance of the 2 Sp1 binding sites, which flank the C/EBP site in the LF promoter, in mediating high-level LF expression. Mutations in either of the 2 Sp1 sites interfered with C/EBPα-mediated transactivation of the LF89 plasmid in myeloid cells. The converse was also true: a mutation in the C/EBP site in the LF promoter interfered with the Sp1-mediated up-regulation of the LF89 plasmid in Drosophila Schneider cells, which normally lack Sp1 activity. Taken together, these findings suggest that a functional interaction between C/EBPα and Sp1 is essential for maximal LF expression. In further analysis, a cooperative interaction between Sp1 and C/EBPα in mediating high-level LF expression was found. None of the sites appear to be absolutely essential for LF expression, since all the mutants (Sp1 and C/EBP) had reporter gene activity that exceeded basal luciferase activity. However, all of the sites contribute markedly to maximal LF expression.
The role of the ubiquitous transcription factor Sp1 in regulating myeloid lineage-specific gene expression is well documented for a host of myeloid-specific genes, including CD11b,28CD11c,20 CD14,29 CD18,19 neutrophil elastase,30 myeloperoxidase,31c-fes,32 myeloid cell nuclear differentiation antigen,33 and human hematopoietic cell kinase genes.34 Sp1 is a differentially phosphorylated and gylcosylated35,36 zinc finger–containing transcription factor that binds to GC or GT boxes.15 The mechanism by which Sp1 aids in effecting tissue- and stage-specific expression is still unknown. However, several explanations for the participation of this transcription factor in lineage-restricted gene expression have been proposed. Modulation of the levels of Sp1 is one such explanation. In this context, it is worth noting that Sp1 is abundant in hematopoietic cells.18 Differential glycosylation, phosphorylation, or both are modifications that may also contribute to the tissue-specific effects of Sp1. Additionally, protein-protein interactions between a ubiquitous transcription factor such as Sp1 and lineage-restricted factors such as C/EBPα may produce a tissue-specific effect. In this context, it is worth noting that Sp1 interacts both physically and functionally with several transcription factors. These include GATA-1,37,38 NFκB,39Erg-1,40 and C/EBPβ,41 among others.
Transcriptional synergy between Sp1 and C/EBP transcription factors was observed for the rat liver CYP2D5 P-450 gene. In that case, occupancy by Sp1 at its cognate site in the CYP2D5 promoter was a prerequisite to C/EBPβ binding.42 Although functional synergy between C/EBPα and Sp1 was demonstrated in the context of the CD11c integrin gene promoter, no such prerequisite for Sp1-site occupancy was observed for C/EBPα transactivation of the CD11c promoter in myeloid cells.20 In the case of the LF promoter, we showed that occupancy of both the 2 Sp1 sites seems to be necessary for C/EBPα-mediated transactivation. It is therefore likely that the mechanism underlying a functional interaction between Sp1 and members of the C/EBP family is unique to the gene promoter in question.
Although we demonstrated functional cooperation between Sp1 and C/EBPα in the context of the LF promoter in myeloid cells, we have not been able to show a direct physical interaction between the 2 transcription factors by using EMSA analysis (Khanna-Gupta A, Berliner N, unpublished data). The mechanism whereby C/EBPα and Sp1 cooperate to increase LF expression in myeloid cells remains undefined. However, it is tempting to speculate that the cooperative effect of the 2 transcription factors may be explained on the basis of the ability of Sp1 to directly contact components of the basal transcriptional machinery, including TBP43—an effect that may be facilitated in the presence of C/EBPα binding to the LF promoter at its cognate site. It is worth noting here that Sp1 is capable of forming homodimers when bound to distant sites in cis, thereby looping out the intervening DNA sequences and altering chromatin structure, which may in turn facilitate changes in DNA binding of other transcription factors.44 In this regard, it was found that Sp1 can be a target of histone deacetylase 1 (HDAC) 1–mediated transcriptional repression, and the HDAC inhibitor trichostatin A can activate the chromosomally integrated murine thymidine kinase promoter in an Sp1-dependent manner.45Alternatively, the binding of C/EBPα to the LF promoter may enhance the binding of Sp1 to the LF promoter, an effect that would lead to increased LF expression. Additionally, the binding of an unidentified accessory factor or factors to Sp1 and/or C/EBPα may contribute to their cooperative effect on LF expression. We believe that Sp1 may play a wider role than was previously thought in modulating the levels of C/EBP-regulated genes during myeloid differentiation.
Acknowledgments
We thank Dr Dan Tenen, Harvard Medical School, Boston, MA, for the U937-C/EBPα cells and helpful discussions, and Dr Robert Tjian, Berkeley, CA, for the pPAC and pPACSp1 plasmids.
Supported by National Institutes of Health grant R01DK53471 (N.B.) and a Swebilius Cancer Research Award (A.K.-G.).
Reprints:Nancy Berliner, Section of Hematology, WWW 428, Yale University School of Medicine, 333 Cedar Street, New Haven, CT 06510; e-mail: nancy.berliner@yale.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Fig. 1. Lactoferrin (LF) promoter transfection and nucleotide sequence. / (A) Transient transfection of LF promoter plasmids in 32Dwt18 (myeloid) and MEL (nonmyeloid) cells. LF promoter fragments (LF89, LF167, LF237, and LF 649) were cloned into the promoterless pGL3B vector and transiently transfected into 32Dwt18 and MEL cells, along with pCMVβgal plasmid. Cells were harvested 24 hours after transfection and luciferase reporter gene activity was measured. The figure represents the mean ± SE value from 3 independent experiments, each performed in duplicate. Normalized luciferase values are represented as a ratio of luciferase activity to pGL3B luciferase activity. (B) Nucleotide sequence of the human LF promoter and its 5′ flanking sequence. The position of the LF89 (−89 base pairs [bps]) is indicated. Putative binding sites for Sp1, Ets, C/EBP, and Myb transcription factors are indicated.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/12/10.1182_blood.v95.12.3734/8/m_bloo01227001x.jpeg?Expires=1769090849&Signature=wLa8-pTrhC6Bc2YobMJGMkgOxy8ni7VGIEkJLW9pGyEYAu1d~DsIiM84CQfyIZnpQGw6z-yVWIT6SSq4nk8azn3aeptS29P82ESZDlGiOtrGcAJa-3pTYQUdPVUOvQFWS-hBjqMJj4diABrOAeXFqkoHM4bDP0lBaEXY8GAjauwtQhsfEV0Er-3EeghUgYMXp8JzuMWsVFhj6StHZe1MFa~HdflWUt8xNYqbIEaDgzxPzqFSc0n8coeG27FKYPoXDuDvoLsjHWuYlgGA6pwwle3J9orWrGYPoAM1ruW2GrjlKGGT--Je~SdfQWpUVRckey1~txOqqvrtxycP5X3aHg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 2. Electrophoretic mobility shift assays (EMSAs) of fragments within the first 89 bps of the LF promoter. / (A) An EMSA was performed by using double-stranded phosphorus 32–labeled LF −89 bp to −66 bp (SP1A) as a probe with nuclear extracts from U937 cells. Binding of Sp1 (lower arrow) was subjected to competition with a 100-fold molar excess of self cold competitor (CC) (lane 3), Sp1 consensus probe (lane 4), and a CD18 Sp1 cold competitor (lane 5) but not a 100-fold molar excess of a nonspecific (n.s.) probe (lane 6). Preincubation of Sp1A probe and U937 nuclear extracts with antiserum to Sp1 resulted in a band shift (lane 7, upper arrow [supershift]). Purified Sp1 protein also bound to the Sp1A probe (lane 8). (B) Results of EMSA analysis using nuclear extracts from uninduced (U937/U) and zinc-induced (U937/I) U937-C/EBPα cells and LF −53 bp to −35 bp (SP1B) as a probe. In both uninduced and induced extracts, Sp1 was eliminated competitively by the addition of a 100-fold molar excess of Sp1B probe (self CC, lanes 3 and 7) and an Sp1 consensus probe (lanes 4 and 8) but not by a nonspecific probe (n.s., lanes 5 and 9, lower arrow). Preincubation of protein-DNA complexes with anti-Sp1 antiserum resulted in a supershifted band (upper arrow) in both uninduced (lane 9) and induced (lane 10) nuclear extracts. (C) EMSA supershift analysis was performed on the radiolabeled LF-C/EBP binding site (LF −74 bp to −51 bp) by using nuclear extracts from uninduced U937-C/EBPα cells and from U937-C/EBPα cells induced with zinc. C/EBPα bound to the LF-C/EBP site from both uninduced (lane 2) and induced (lane 5) U937-C/EBPα extracts (lower arrow). The protein-DNA complex was supershifted (upper arrow) by anti-C/EBPα antiserum (lanes 3 and 6) but not by anti-C/EBPβ antiserum (lanes 4 and 7).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/12/10.1182_blood.v95.12.3734/8/m_bloo01227002w.jpeg?Expires=1769090849&Signature=gtwenTGzl5CTpBaQZhYDvlFvD1Yo078NXa3OwqhGTaQehNO6aK9KP2b~mfB9mJJ1xpdmsgZPF7GsZ6VHXtz64GB2gZ0eOZiYPp-nE~ZKG-a0lVR~sbj6EISUPBttx3uTziinmFdU85RYK7f8P78GhenvEKmqN3OAThMFTiyzUxnzV~1ONlIX8juSYj7iDURuc0Xl6iygVM1isbHtKvWawaXWatkBnHtDoz4eRmCL9JPg6iXz4a6BUiFphPRSgNhmO~ny9NqxfJUVFdSRirEJoPWtpBLteuWGB8VeihjOhYIvy-DO1lXa3HIYLgXeruZhYNfjjSsCPANeJD2lPfQTTQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal