Abstract
The t(8;21) translocation is one of the most frequent chromosomal abnormalities associated with acute myeloid leukemia (AML). In this translocation, the AML1 (CBFA2/PEBP2aB) gene is disrupted and fused to the MTG8 (ETO) gene. The ectopic expression of the resulting AML1-MTG8 fusion gene product in L-G and 32Dcl3 murine myeloid precursor cells stimulates cell proliferation without inducing morphologic terminal differentiation into mature granulocytes in response to granulocyte-colony stimulating factor (G-CSF). This study found that the ectopic expression of AML1-MTG8 elevates the expression of the G-CSF receptor (G-CSFR). Analysis of the promoter region of the G-CSFR gene revealed that up-regulation of G-CSFR expression by AML1-MTG8 does not depend on the AML1-binding sequence, but on the C/EBP (CCAAT/enhancer binding protein) binding site. The results suggest that the overproduction of G-CSFR is at least partly mediated by C/EBPɛ, whose expression is activated by AML1-MTG8. The ectopic expression of G-CSFR in L-G cells induced cell proliferation in response to G-CSF, but did not inhibit cell differentiation into mature neutrophils. Overexpression of C/EBPɛ in L-G cells also stimulated G-CSF–dependent cell proliferation. High expression levels of G-CSFR were also found in the leukemic cells of AML patients with t(8;21). Therefore, G-CSF–dependent cell proliferation of myeloid precursor cells may be implicated in leukemogenesis.
The molecular analyses of the recurrent chromosomal translocations associated with leukemia or lymphoma have provided insights into the mechanism of malignant transformation and have led to the identification of several transcription factors that participate in the regulation of normal hematopoiesis.1,2 We previously showed that the AML1 gene3 is located at the translocation breakpoint of chromosome 21 in the t(8;21) translocation, which is frequently found in a subtype of acute myeloid leukemia (AML) (M2 according to the French-American-British classification)4 but rarely in myelodysplasia.5Biallelic and heterozygous point mutations in the Runt domain of theAML1 gene are suggested to be responsible for AML.6AML1 directly binds the enhancer core DNA (cDNA) sequence TGT/cGGT, which is present in several viral and cellular promoters and enhancers, and AML1 DNA binding activity is stabilized by heterodimerization through the Runt domain with CBFβ/PEBP2β.7 Studies on AML1-deficient mice have suggested that AML1-regulated genes are essential for the definitive hematopoiesis of all lineages,8,9 and that AML1 functions as a transcriptional regulator at a high level of hierarchy in hematopoietic lineages.10 It is also believed that AML1 requires cooperative transcription factors to regulate expression of its target genes. AML1 interacts with many transcription factors or modulates the activities of those known to be important for hematopoiesis, including ETS1, c-MYB, ALY, PU.1, TLE, YAP, and C/EBP.7 We also found that AML1 specifically interacts and functionally cooperates with p300, a transcriptional coactivator possessing histone acetyltransferase activity.11 AML1 also interacts with mSin3A and represses expression of p21/WAF1/CIP1.12
The AML1/CBFβ transcription factor complex is most frequently targeted in leukemia-associated chromosomal aberrations such as t(8;21), t(12;21), t(3;21), t(16;21), and inv(16).7 These translocations result in the expression of chimeric transcription factors such as AML1-MTG8/ETO, TEL-AML1, AML1-EVI1, AML1-MTG16, and CBFβ-SMH11. AML1-MTG8 can function as a complex with other MTG8 family members, such as MTGR113 or MTG16 (I. Kitabayashi, unpublished data). The recent finding that the MTG8 portion of AML1-MTG8 interacts with the histone deacetylase complex involved in transcriptional repression of target genes is consistent with the idea that AML1-MTG8 interferes with AML1-dependent transactivation.7 However, the mechanism of gene regulation by AML1-MTG8 varies depending on the target gene. AML1-MTG8 was also shown to induce expression of the BCL2 gene.14Several experiments of ectopic expression of AML1-MTG8 were done to demonstrate the transforming activity of the AML1-MTG8 chimeric protein. AML1-MTG8 induces granulocyte colony-stimulating factor (G-CSF)-dependent cell proliferation of murine hematopoietic precursor L-G or 32D cells and inhibits their terminal differentiation into segmented neutrophils.13,15 16 These results provide a sound basis for further studies on the function of AML1-MTG8. However, in these experiments, the mechanism of G-CSF–dependent proliferation was not revealed. At present, the target genes of AML1-MTG8 responsible for leukemogenic transformation remain to be identified.
G-CSF stimulates both the proliferation and differentiation of neutrophilic precursor cells. It also stimulates a variety of responses in mature neutrophils, including prolonged survival, phagocytosis, and superoxide production.17-19 G-CSF binds to its receptor (G-CSFR) on the cell surface to form a homodimeric receptor complex.20 The membrane-proximal region of G-CSFR has been shown to be essential for the transmission of growth signals21,22 via JAK2 activation, STAT3 and STAT5 phosphorylation, and MAP kinase phosphorylation.23,24 Because point mutations in theG-CSFR gene were identified in some patients with AML25,26 and in severe congenital neutropenia,27 the signal transduction pathway via G-CSFR is likely to be important for normal hematopoiesis. In addition, because of the huge production (120 billion/d) and rapid turnover (48 hours) of neutrophils, tight regulation of this signaling pathway is probably critical.21 Thus, the accumulated evidence indicates that G-CSF and its receptor are essential for hematopoietic development and that G-CSF may be implicated in malignant disease. However, the precise role played by these proteins in the early commitment to a particular cell lineage, whether it be to direct multipotent cells down a particular cellular pathway or to merely support the survival of cells that have intrinsically selected a specific hematopoietic lineage, remains controversial.28 29
We show here that AML1-MTG8 induces the expression of G-CSFR that accompanies G-CSF–dependent cell proliferation in murine myeloid precursor cell lines by activating the promoter of the G-CSFRgene. We also show that this activation is indirectly mediated by AML1-MTG8 up-regulation of C/EBPε.
Materials and methods
Cells, viruses, and expression plasmids
The L-G30 and 32Dcl331 cells were cultured in RPMI1640 medium supplemented with 10% fetal calf serum (FCS) and recombinant mouse interleukin-3 (IL-3; 0.25 ng/mL; a generous gift from Kirin Brewery) with or without 50 mmol/L β-mercaptoethanol, respectively. BOSC2332 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FCS. Leukemic cell fractions of bone marrow or of peripheral blood from AML-M2 patients were prepared by centrifugation with Ficol Paque. Granulocytic fractions from healthy individuals were prepared with Mono-poly Resolving Medium. LNSX and LXSH retrovirus vectors33 were generously provided by Dr D. Miller. AML1b cDNA,34AML1-MTG8 cDNA35 and mouse G-CSFR cDNA36 (a generous gift from Dr S. Nagata) were inserted between the StuI site and Cla I site of the LNSX vector as described previously.13 C/EBPε cDNA with the HA-tag sequence was generated by the polymerase chain reaction (PCR) with specific primers; 5′-TGAATTCACCATGGCATACCCATACGACGTGCC- TGACTACGCCTCCTCCCCAGGGGACCTA-3′ and 5′-TTCGGCCGTCAGCTGCAACCCCCCAC-3′ to generate cDNA encoding 32-kd protein37 and cloned into the LNSX vector. All of the viruses were transiently produced in BOSC23 cells after transfection of each vector by the calcium phosphate precipitation method. After L-G cells were infected with LNSX viruses for 2 days, the cells were transferred to G418 (1 mg/mL) containing medium and used for analyses. For LXSH virus infection, cells were selected with hygromycin B (2.5 mg/mL). The 32Dcl3 cells were transfected with the expression plasmid pRc/CMV (Invitrogen) carrying murine G-CSFR by electroporation, and after 2 days, the cells were transferred to medium containing 400 μg/mL G418. For time-course experiments, L-G infectants maintained in the presence of IL-3 were washed once with IL-3–free medium and incubated in medium containing human recombinant G-CSF (10 ng/mL) (a generous gift from Chugai Pharmaceutical Company). Viable cells were counted at the indicated times by the trypan blue exclusion method or with a Coulter counter. For cell morphology, cells were stained with May Gruenwald-Giemsa solution.
Northern blotting
Procedures for Northern blotting were the same as those described previously.38 Poly (A) + RNA (1 μg) or total RNA (5 μg) was denatured and fractionated on a 1.0% agarose gel containing formaldehyde. The probe used was mouse or human G-CSFR cDNA (generous gifts from Dr S. Nagata), mouse or human myeloperoxidase (MPO) cDNA (generous gifts from Dr K. Morishita), or human glyceraldehyde-3-phosphate-dehydrogenase (G3PDH) cDNA. The probe for C/EBPα cDNA was a generous gift from Dr A. D. Friedman. The probe for mouse C/EBPε was generated by PCR. Primers used for C/EBPε were F5′-ACATGTGTGAGCATGAGGCC-3′) and R5′-TGTGCCACTTGGTACTGCAG-3′).37
Western blotting
Cell lysates were fractionated on sodium dodecyl sulfate-10% polyacrylamide gels (SDS-PAGE) and transferred to a membrane filter by electroblotting. Immunodetection using an anti-HA antiserum was performed as described elsewhere13 except that all of the washing and hybridization procedures were performed in tris-buffered saline (TBS) containing 0.2% Tween-20. The immune complexes were visualized by an alkaline phosphatase detection system (Bio-Rad).
Ligand-binding assay for G-CSFR
Specific G-CSFR activity was determined as the difference between total and nonspecific binding.20 The cells were incubated, with rotation, for 4 hours at 4°C with [125I]G-CSF (Amersham) in the presence (nonspecific binding) or absence (total binding) of 50 nM unlabeled G-CSF. One arbitrary unit is defined as 1 active site bound by 1 G-CSF molecule per cell in the presence of 1.5 nmol/L radiolabeled G-CSF.
Isolation of the human G-CSFR promoter and C/EBPɛ promoter
On the basis of the published sequence,39 the sequences of the 5′ promoter region of the human G-CSFR gene were amplified and cloned from human genomic DNA by the PCR with primers F 5′-AACGCGTCCAAAGGGCTTTGACTTTG-3′) and R 5′ GAAGCTTACTCACGTTGGCACCTCT-3′). This PCR fragment was sequenced and confirmed to be identical to the published sequence. The synthesized fragment represents bp −1360 to +10, with the major transcription start site designated +1. Deletion mutants were constructed by PCR to generate fragments containing nucleotides +10 to −60, +10 to −188, +10 to −243. The forward primers were 5′-AGCTTTGAGCTCAGGAAATC-3′ and R, 5′-TAAGACCCCCAAGGCAGGAA- 3′ and R, and 5′-CGGAAGGTGTTGCAATCC-3 and R, respectively. The mutant carrying 2 base substitutions in the consensus site for AML1 binding at −390 to −385 was also constructed by PCR. The PCR products were sequenced and cloned into the luciferase reporter plasmid pGL3-Basic (Promega).
For isolation of the promoter region of C/EBPε gene, human PAC library was screened. C/EBPε genomic primers,37 F: 5′-CTACAATCCCCTGCAGTAC-3′ and R: 5′-CATTTCACAGAGCGACACCA-3′ were used for screening by PCR. An EcoRI fragment of approximately 12 kb containing the C/EBPε gene was subcloned into a plasmid vector and the C/EBPε gene promoter region was sequenced. A region of 1280 bp from the promoter region was cloned into pGL3-Basic.
Transcriptional analysis
For the analysis of G-CSFR promoter activity, L-G cells infected with AML1b or AML1-MTG8 were transiently transfected by diethylaminoethanol (DEAE)-dextran treatment (300 μg/mL, 30 minutes). Cells were cultured in the presence of IL-3 for 66 to 72 hours. Cells were harvested by centrifugation. Activity in cell lysates (10 μL) was detected with the Dual Luciferase Assay Kit (Promega) and luciferase activity was measured by Lumat LB9507 (Berthold). In all assays of luciferase activity, pRL-TK (Promega) (0.2 μg) was cotransfected as an internal control.
Electrophoretic mobility shift assays
The L-G cells were infected either with LNSX, LNSA-AML1b, or LNSX-AML1-MTG8. Whole cell lysates were prepared as described elsewhere.40 The cells were washed in cold phosphate-buffered saline (PBS) and suspended in cold buffer (10 mmol/L HEPES-KOH, pH7.9, 1.5 mmol/L MgCl2, 10 mmol/L KCl, 0.5 mmol/L DTT, 0.2 mm phenylmethylsulfonyl fluoride [PMSF]). The cells were allowed to swell on ice for 10 minutes and vortexed for 10 seconds. After centrifugation at 12 000 rpm for 10 seconds, the pellet was resuspended in a buffer containing 20 mmol/L HEPES-KOH, pH7.9, 25% glycerol, 420 mmol/L KCl, 1.5 mmol/L MgCl2, 0.2 mmol/L EDTA, 0.5 mmol/L DTT, and 0.2 mmol/L PMSF. The suspension was incubated on ice for 20 minutes and centrifuged at 12 000 rpm for 20 minutes. The supernatant was used for the assays. For synthesis of C/EBPε protein, the cDNA was cloned into the pSP64poly(A) vector. In vitro transcription/translation reactions were carried out with the TnT SP6 quick coupled transcription/translation system (Promega).
Electrophoretic mobility shift assays (EMSA) were performed in 20 mL of binding buffer (20 mmol/L HEPES, pH 7.9, 0.1 M KCl, 5 mmol/L MgCl2, 2 mmol/L EDTA, 1 mmol/L DTT, 20% glycerol) containing 1 μg of poly(dI-dC), 32P-labeled oligonucleotide (5 × 103 cpm) and the cell extract on ice for 30 minutes. DNA-protein complexes were resolved on 5% polyacrylamide gels.
Results
AML1-MTG8 induces and AML1b represses the expression of G-CSFR
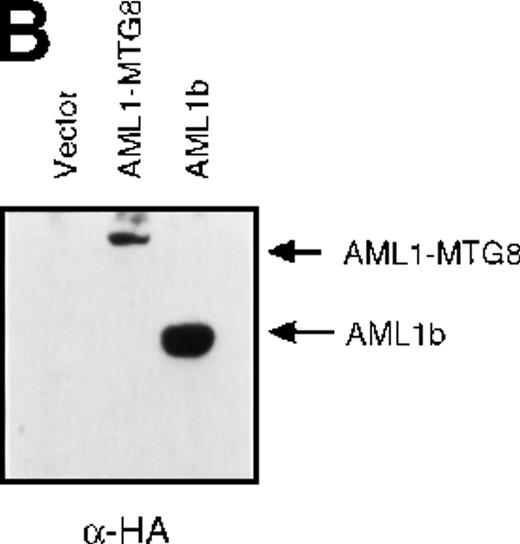
The murine IL-3–dependent hematopoietic precursor cell lines L-G and 32Dcl3 differentiate into mature neutrophils in response to G-CSF.30,31 We previously reported ectopic expression of AML1b, a wild-type full-length protein of AML1 and AML1-MTG8, in the L-G cell line using a retrovirus vector. In these experiments, cells were not cloned after selection with drugs to avoid clonal diversity of the cells. Like the parental L-G cells, the cells infected with LNSX-AML1b or the control LNSX-vector did not proliferate in the presence of G-CSF during the 7-day culture period and underwent terminal differentiation into mature neutrophils. In contrast, cells that expressed AML1-MTG8 proliferated exponentially in response to G-CSF without differentiating into mature granulocytes, as judged from the changes in cell morphology.13
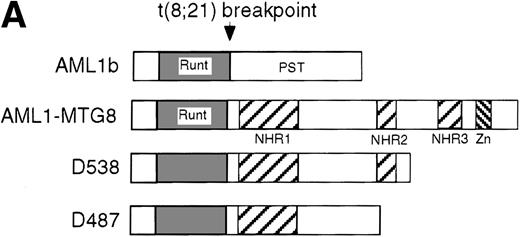
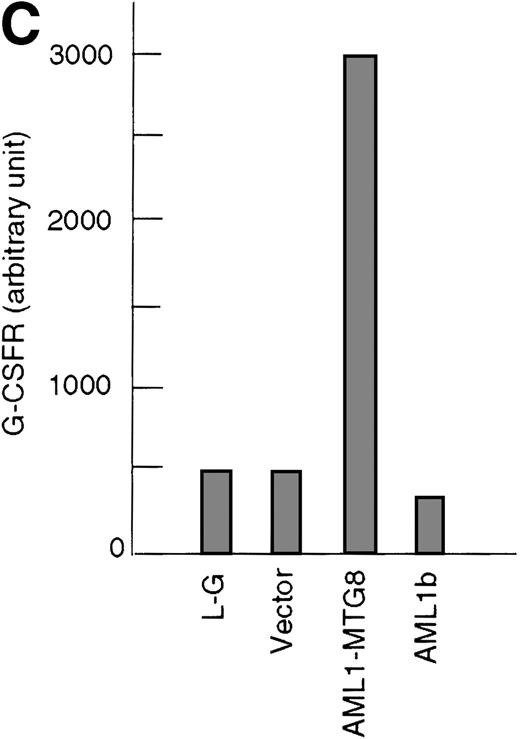
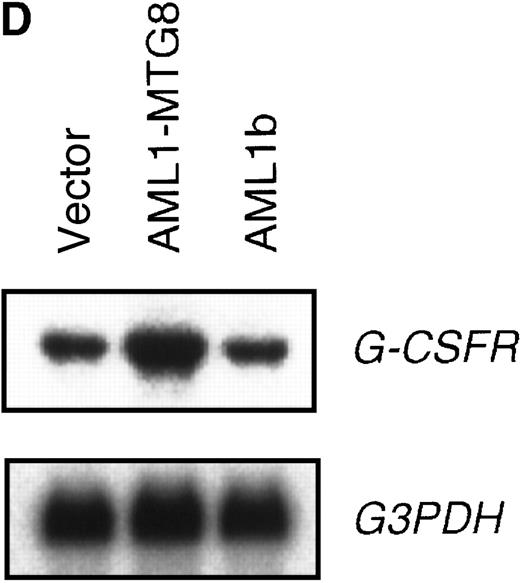
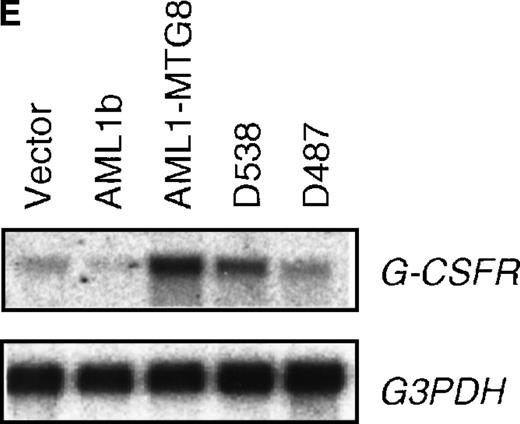
These results suggest that AML1-MTG8 might modulate cell responses to G-CSF. To identify the molecule responsible for this modulation, we initially measured the expression of G-CSFR in L-G infectants expressing either AML1b or AML1-MTG8. A ligand-binding assay using radiolabeled recombinant G-CSF showed elevated levels of G-CSFR activity (about 6-fold) after infection of the cells with a retrovirus containing AML1-MTG8 (Figure 1C). On the other hand, the number of functional G-CSFRs slightly decreased in infectants expressing AML1b (Figure 1C). Northern blot analyses showed consistently that the G-CSFR transcript level was also increased in cells expressing AML1-MTG8 (about 3-fold), whereas slight decreases were observed in cells expressing AML1b (Figure 1D). We previously found that the NHR2 region of AML1-MTG8, which is required to form complexes with MTG8 family members, is essential for the induction of G-CSF–dependent cell proliferation.13 To clarify the relationship between G-CSF–dependent proliferation and the G-CSFR transcript level, Northern blot analysis was carried out using cells infected with a series of C-terminal deletion mutants of AML1-MTG8 (Figure 1E). The C-terminal deletion mutant D538 containing NHR2 (Figure 1A) as well as wild-type AML1-MTG8 stimulated the synthesis of G-CSFR transcripts. However, further deletion to residue 487 eliminated this activity. These results indicated that the activation of G-CSFR expression coincided well with the proliferation response of L-G cells to G-CSF.
Up-regulation of G-CSFR by AML1-MTG8 and down-regulation by AML1b in L-G infectants.
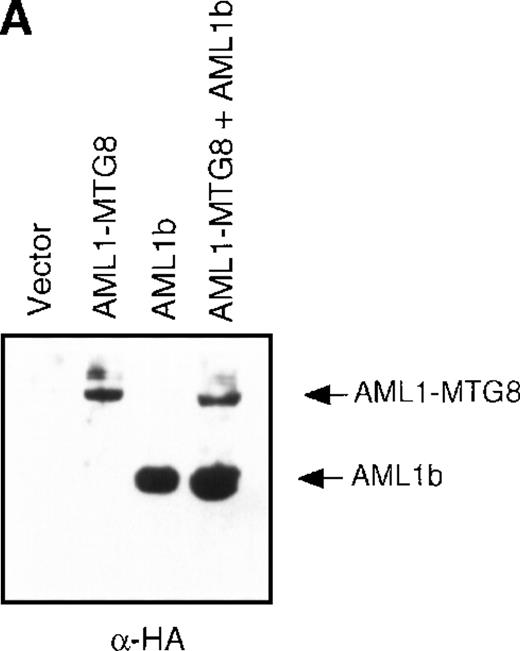
(A) Schematic representation of AML1b protein, AML1-MTG8 protein, and deletion mutants of AML1-MTG8 proteins, D538 and D487. Runt indicates the runt homology region; PST, proline, serine, threonine-rich region; NHR, nervy homology region; Zn, zinc finger motifs. Note that AML1-MTG8 retains the runt homology region and most of the MTG8 protein. (B) Expression of AML1b and AML1-MTG8 protein in L-G cells detected by Western blot analyses. Proteins from the L-G infectants with viruses encoding AML1b and AML1 MTG8 or with a control virus were analyzed by immunodetection using anti-HA antibody. The positions of AML1 and AML1-MTG8 are indicated by arrows. (C) Ligand binding assay for G-CSFR activity of L-G infectants. The activity was measured as described in “Materials and methods.” (D) Northern blot analyses showing the expression of G-CSFR mRNA. Total RNAs (5 μg of RNA) were prepared from L-G infectants cultured with IL-3, blotted, and hybridized with32P-labeled mouse G-CSFR cDNA and human G3PDH cDNA. (E) Northern blot analyses showing the expression of G-CSFR mRNA in deletion mutants of AML1-MTG8. Total RNAs were prepared from L-G infectants cultured with IL-3, blotted, and hybridized with32P-labeled mouse G-CSFR cDNA and human G3PDH.
Up-regulation of G-CSFR by AML1-MTG8 and down-regulation by AML1b in L-G infectants.
(A) Schematic representation of AML1b protein, AML1-MTG8 protein, and deletion mutants of AML1-MTG8 proteins, D538 and D487. Runt indicates the runt homology region; PST, proline, serine, threonine-rich region; NHR, nervy homology region; Zn, zinc finger motifs. Note that AML1-MTG8 retains the runt homology region and most of the MTG8 protein. (B) Expression of AML1b and AML1-MTG8 protein in L-G cells detected by Western blot analyses. Proteins from the L-G infectants with viruses encoding AML1b and AML1 MTG8 or with a control virus were analyzed by immunodetection using anti-HA antibody. The positions of AML1 and AML1-MTG8 are indicated by arrows. (C) Ligand binding assay for G-CSFR activity of L-G infectants. The activity was measured as described in “Materials and methods.” (D) Northern blot analyses showing the expression of G-CSFR mRNA. Total RNAs (5 μg of RNA) were prepared from L-G infectants cultured with IL-3, blotted, and hybridized with32P-labeled mouse G-CSFR cDNA and human G3PDH cDNA. (E) Northern blot analyses showing the expression of G-CSFR mRNA in deletion mutants of AML1-MTG8. Total RNAs were prepared from L-G infectants cultured with IL-3, blotted, and hybridized with32P-labeled mouse G-CSFR cDNA and human G3PDH.
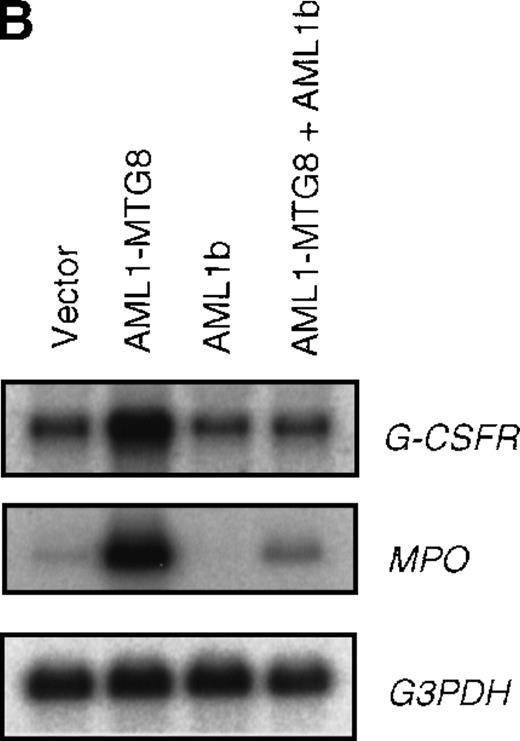
The effect of AML1-MTG8 on cell proliferation and the differentiation block in response to G-CSF can be completely rescued by overexpressing AML1b.11 This suggests that the induction of G-CSFR expression by AML1-MTG8 might be due to AML1-MTG8 alleviation of the repressive activity of AML1b on G-CSFR expression. The AML1b-dependent decrease in the level of G-CSFR was apparent when the cells expressing AML1-MTG8 were further infected with a virus expressing AML1b. Up-regulated expression of G-CSFR by AML1-MTG8 was almost completely inhibited by overexpressing AML1b (Figure 2).
Inhibition of AML1-MTG8–induced up-regulation of G-CSFR by AML1b.
(A) Expression of AML1b and AML1-MTG8 protein detected by Western blot analyses. L-G cells infected with LXSH or LXSH-AML1-MTG8 were further infected with LNSX-AML1b viruses and the protein extracts from the infectants were analyzed by immunodetection using anti-HA antibody. The positions of AML1 and AML1-MTG8 are indicated by arrows. (B) Northern blot analyses showing the expression of G-CSFR mRNA. Total RNAs (5 μg of RNA) were prepared from L-G infectants cultured with IL-3, blotted, and hybridized with 32P-labeled mouse G-CSFR cDNA, mouse MPO cDNA, and human G3PDH cDNA.
Inhibition of AML1-MTG8–induced up-regulation of G-CSFR by AML1b.
(A) Expression of AML1b and AML1-MTG8 protein detected by Western blot analyses. L-G cells infected with LXSH or LXSH-AML1-MTG8 were further infected with LNSX-AML1b viruses and the protein extracts from the infectants were analyzed by immunodetection using anti-HA antibody. The positions of AML1 and AML1-MTG8 are indicated by arrows. (B) Northern blot analyses showing the expression of G-CSFR mRNA. Total RNAs (5 μg of RNA) were prepared from L-G infectants cultured with IL-3, blotted, and hybridized with 32P-labeled mouse G-CSFR cDNA, mouse MPO cDNA, and human G3PDH cDNA.
AML1-MTG8 activates and AML1b represses the promoter of G-CSFR
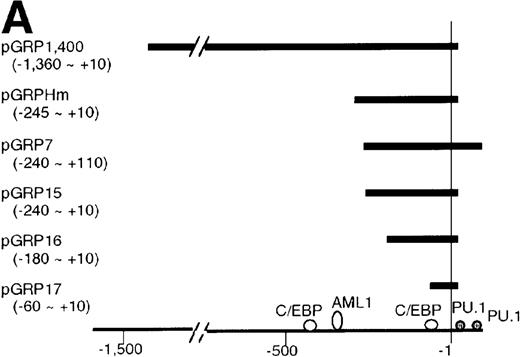
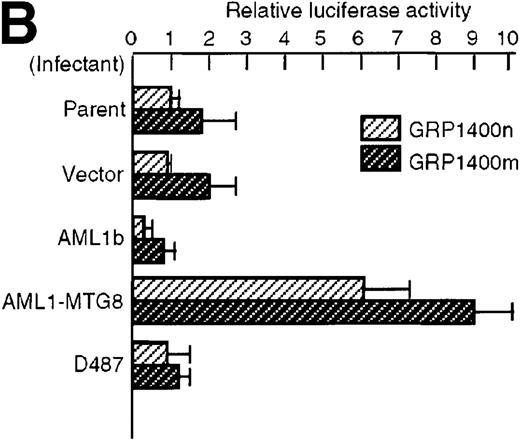
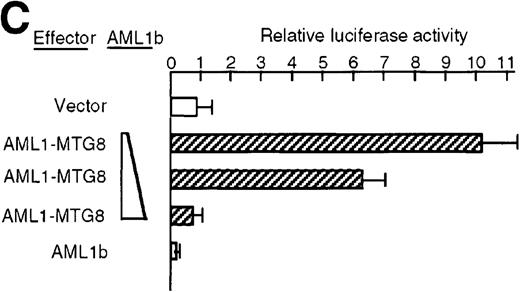
To understand the molecular mechanism of transcriptional regulation of the G-CSFR gene by AML1-MTG8, fusion reporter constructs comprising about 1.4-kb of DNA from the 5′ upstream region of the human G-CSFR promoter and the luciferase gene (pGRP1400n) were transfected into L-G cells ectopically expressing AML1b, AML1-MTG8, or a deletion mutant of AML1-MTG8, D478. TheG-CSFR promoter was significantly activated in cells expressing AML1-MTG8 (about 4-fold) (Figure 3B). On the other hand, the activity in cells expressing AML1b was suppressed (about 1/2-fold) compared to cells infected with the vector. In addition, the activation was not induced by a deletion mutant of AML1-MTG8, D478, which did not induce expression of G-CSFR. Because a consensus sequence for AML1-binding (−391 to −385) is present in the upstream region of the G-CSFR gene, we initially constructed a mutant reporter with 2 base substitutions in this sequence (pGRP1400m). However, these mutations in the AML1 site did not affect the transactivation of the promoter by AML1-MTG8 or AML1b. This suggests that up- or down-regulated expression is not mediated by direct binding of AML1-MTG8 or AML1b to the AML1 site (Figure 3B). Compared to the native promoter, the mutant promoter pGRP1400m was slightly activated in all infectants, suggesting that the AML1 site might function as a negative regulatory element. Cotransfection of theG-CSFR promoter with effector plasmids expressing AML1b or AML1-MTG8 or both also consistently showed that AML1-MTG8 activates theG-CSFR promoter and that AML1 inhibits the AML1-MTG-8 activation effect in a dose-dependent manner (Figure 3C).
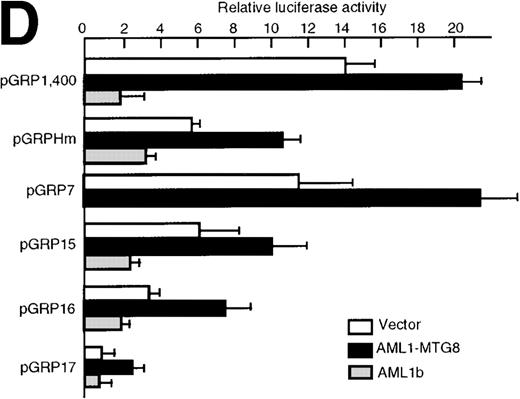
Activation of the G-CSFR promoter by AML1-MTG8.
(A) Schematic representation of the promoter region of G-CSFRand its deletion mutants. (B) Activation of the G-CSFR promoter by AML1-MTG8. LG cells expressing either vector, AML1b, AML1-MTG8 or a deletion mutant of AML1-MTG8, D487, were transfected with G-CSFR–luciferase (pGL3) using DEAE-dextran. Transfection efficiency was normalized by cotransfection with pRL-TK as an internal control. pGRP1400n; normal 1370 bp promoter, pGRP1400m; a mutant (2 base substitution) of the AML1 consensus site of pGRP1400n. (C) AML1b interferes with the enhancing activity of AML1-MTG8 on theG-CSFR promoter. Cells were cotransfected with pGRP1400n and either LNSX, LNSX-AML1b, or LNSX-AML1-MTG8. LNSX-AML1b was added in addition to the effector plasmid AML1-MTG8 at 0.3 μg, 1 μg, and 3 μg in lanes 2, 3, and 4, respectively. The total amount of effector DNA was 6 μg each. LNSX was used to adjust the total amount of DNA. (D) Activity of 5′ deletion mutants of the G-CSFRpromoter in L-G cells. Cells were cotransfected with each mutant and either 3 μg of LNSX, LNSX-AML1b, or LNSX-AML1-MTG8. The activity of the control vacant reporter with LNSX was designated as being equal to 1.
Activation of the G-CSFR promoter by AML1-MTG8.
(A) Schematic representation of the promoter region of G-CSFRand its deletion mutants. (B) Activation of the G-CSFR promoter by AML1-MTG8. LG cells expressing either vector, AML1b, AML1-MTG8 or a deletion mutant of AML1-MTG8, D487, were transfected with G-CSFR–luciferase (pGL3) using DEAE-dextran. Transfection efficiency was normalized by cotransfection with pRL-TK as an internal control. pGRP1400n; normal 1370 bp promoter, pGRP1400m; a mutant (2 base substitution) of the AML1 consensus site of pGRP1400n. (C) AML1b interferes with the enhancing activity of AML1-MTG8 on theG-CSFR promoter. Cells were cotransfected with pGRP1400n and either LNSX, LNSX-AML1b, or LNSX-AML1-MTG8. LNSX-AML1b was added in addition to the effector plasmid AML1-MTG8 at 0.3 μg, 1 μg, and 3 μg in lanes 2, 3, and 4, respectively. The total amount of effector DNA was 6 μg each. LNSX was used to adjust the total amount of DNA. (D) Activity of 5′ deletion mutants of the G-CSFRpromoter in L-G cells. Cells were cotransfected with each mutant and either 3 μg of LNSX, LNSX-AML1b, or LNSX-AML1-MTG8. The activity of the control vacant reporter with LNSX was designated as being equal to 1.
The C/EBP binding site is required for activation of the G-CSFR promoter by AML1-MTG8
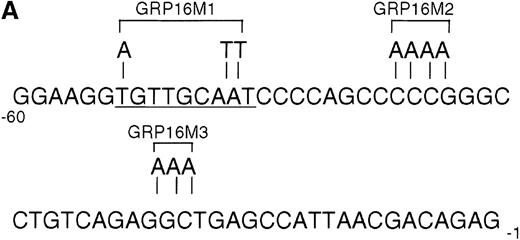
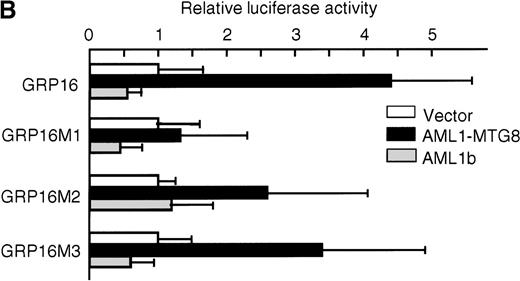
To determine the functionally critical regions of the G-CSFRpromoter for up-regulation by AML1-MTG8, we created a series of 5′ truncation mutants by PCR. As shown in Figure 3D, AML1-MTG8 significantly activated the constructs pGRP1400n, pGRP7 (−243 to +100), pGRP15 (−243 to +10), pGRP16 (−188 to +10) and pGRP17 (−60 to +10). The shortest construct pGRP17 contains no other transcriptional factor binding site besides the potential sites for C/EBPs and Sp1. The C/EBP family, especially C/EBPα and C/EBPε, are important for myeloid cell proliferation and differentiation and for the regulation of myeloid-specific proteins.25,41-43C/EBPα activates promoters for the cytokine receptors of granulocyte/macrophage colony-stimulating factor (GM-CSF), macrophage colony-stimulating factor (M-CSF) and G-CSF.44-46 C/EBPε activates G-CSFR37 and myeloperoxidase.47 We created mutants of the C/EBP binding site and Sp1 binding site in pGRP16 (Figure 4A). AML1-MTG8 stimulation of G-CSFR promoter activity was significantly reduced by mutations of the C/EBP site (pGRP16M1) but not by mutations of the Sp1 site (pGRP16M2/M3) (Figure 4B).
Requirement for the C/EBP binding site for activation of the G-CSFR promoter by AML1-MTG8.
(A) Schematic representation of the base substitution of the mutants around the C/EBP site in the G-CSFR promoter. GRP16M1 is a mutant of the C/EBP consensus site. GRP16M2 is a mutant of the putative Sp1 site. GRP16M3 is a mutant of the Sp1 consensus site. (B) Effects of AML1-MTG8 on the activity of GRP16 mutants in L-G cells. Cells were cotransfected with each mutant reporter and either LNSX, LNSX-AML1b, or LNSX-AML1-MTG8.
Requirement for the C/EBP binding site for activation of the G-CSFR promoter by AML1-MTG8.
(A) Schematic representation of the base substitution of the mutants around the C/EBP site in the G-CSFR promoter. GRP16M1 is a mutant of the C/EBP consensus site. GRP16M2 is a mutant of the putative Sp1 site. GRP16M3 is a mutant of the Sp1 consensus site. (B) Effects of AML1-MTG8 on the activity of GRP16 mutants in L-G cells. Cells were cotransfected with each mutant reporter and either LNSX, LNSX-AML1b, or LNSX-AML1-MTG8.
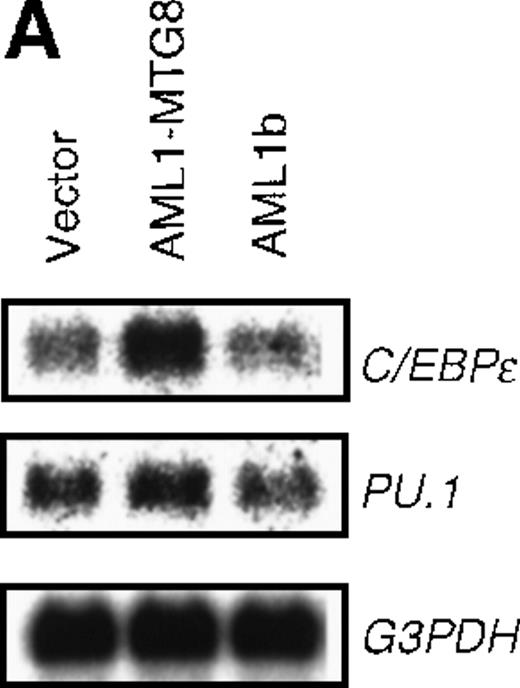
AML1-MTG8 induces expression of C/EBPɛ
Analysis of the G-CSFR promoter using mutated sequences indicated the possible involvement of C/EBPs in the regulation ofG-CSFR expression. Among C/EBP family members, C/EBPα, C/EBPβ, and C/EBPε were shown to be important for hematopoiesis. We therefore examined their expression levels to determine which one was responsible for the stimulation of the G-CSFR promoter by AML1-MTG8. Northern analysis indicated that the expression of C/EBPε was significantly up-regulated in the cells ectopically expressing AML1-MTG8 (Figure 5A), whereas the expression of C/EBPα and C/EBPβ were neither detected in L-G cells expressing AML1-MTG8 nor in the parental L-G cells (data not shown). No significant change was observed in the expression level of PU.1 (Figure 5A), a transcription factor known to be important for the activation of G-CSFR.46
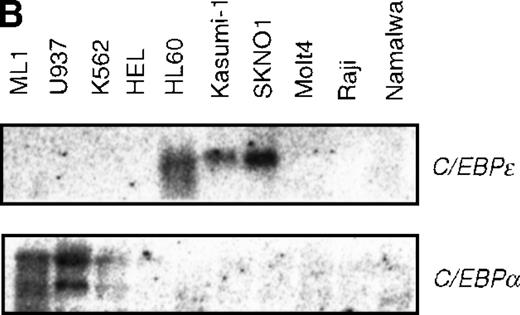
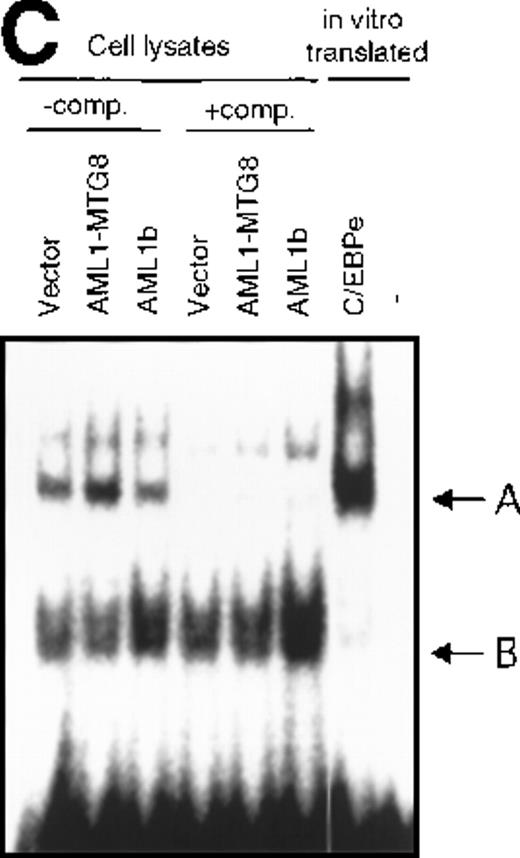
Induced expression of C/EBPε by AML1-MTG8.
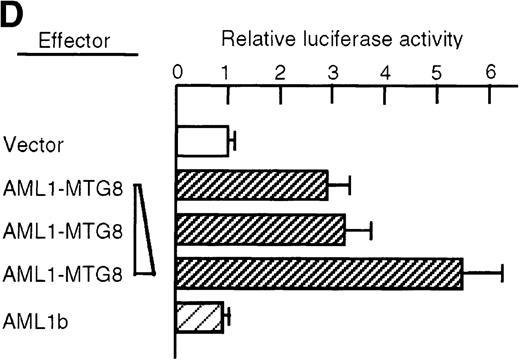
(A) Northern blot analyses showing the expression of C-EBPε mRNA. Total RNAs (5 μg of RNA) were prepared from L-G infectants cultured with IL-3, blotted, and hybridized with 32P- labeled human C/EBPε cDNA, mouse PU.1 cDNA, and human G3PDH cDNA. (B) Northern blot analyses of poly(A)+RNAs from hematopoietic cell lines. Northern blots were hybridized with human C/EBPα cDNA and human C/EBPε cDNA. (C) Gel mobility shift assay showing the binding activities of cell extracts from L-G infectants and in vitro synthesized C/EBPε protein. (D) AML1-MTG8 activates the promoter of the C/EBPε gene. Cells were cotransfected with pGL3-C/EBPε (1280 bp) and either 0.2 μg of LNSX, 0.2 μg of LNSX-AML1b, or various concentrations of LNSX-AML1-MTG8. LNSX-AML1-MTG8 was added at 0.05 μg, 0.1 μg, and 0.2 μg in lanes 2, 3, and 4, respectively.
Induced expression of C/EBPε by AML1-MTG8.
(A) Northern blot analyses showing the expression of C-EBPε mRNA. Total RNAs (5 μg of RNA) were prepared from L-G infectants cultured with IL-3, blotted, and hybridized with 32P- labeled human C/EBPε cDNA, mouse PU.1 cDNA, and human G3PDH cDNA. (B) Northern blot analyses of poly(A)+RNAs from hematopoietic cell lines. Northern blots were hybridized with human C/EBPα cDNA and human C/EBPε cDNA. (C) Gel mobility shift assay showing the binding activities of cell extracts from L-G infectants and in vitro synthesized C/EBPε protein. (D) AML1-MTG8 activates the promoter of the C/EBPε gene. Cells were cotransfected with pGL3-C/EBPε (1280 bp) and either 0.2 μg of LNSX, 0.2 μg of LNSX-AML1b, or various concentrations of LNSX-AML1-MTG8. LNSX-AML1-MTG8 was added at 0.05 μg, 0.1 μg, and 0.2 μg in lanes 2, 3, and 4, respectively.
Expression of C/EBPε was observed in leukemia cell lines carrying t(8;21), SKNO1 and Kasumi-1, and in HL60 cells established from the M2 AML patients (Figure 5B). No detectable transcripts were seen in the other hematopoietic cell lines tested. These observations corroborate a report that showed the expression of C/EBPε is restricted to the granulocytic lineage and the T-cell lineage and that C/EBPε is induced along with granulocytic differentiation.37 48C/EBPα is expressed in ML1 and U937, but weakly in K562, and not at all in cells expressing C/EBPε (Figure 5B).
To confirm the involvement of C/EBPε in the regulation of the G-CSFR expression, EMSA was carried out using a 23-mer oligonucleotides probe N1 that contains the C/EBP binding sequence (−62 to −39) of the G-CSFR promoter and lysates from L-G cells overexpressing AML1b or AML1-MTG8. Complex A (Figure 5C) was found to be more abundant in the lysate from AML1-MTG8 expressing cells. Because complex A migrated at the same position as the complex formed with in vitro translated C/EBPε, complex A is likely to contain C/EBPε. The differences in the amount of complex A between the lysates were compatible with the variations in the messenger RNA (mRNA) levels of C/EBPε in the infectants used to make the lysates. These results suggested that C/EBPε was involved in the regulation of G-CSFR transcription by AML1-MTG8. Band B was considered to be a nonspecific complex, because its intensity was not diminished by adding excess competitor (Figure 5C). These results indicated that C/EBPε mediates the transcriptional regulation of G-CSFR by AML1-MTG8.
To determine the possible mechanism of enhanced expression of C/EBPε by AML1-MTG8, a PAC clone containing genomic DNA coding for the human C/EBPε gene was isolated and sequenced. The promoter region of 1280 bp, which contains the 130 bp reported sequence37 of C/EBPε was fused to the luciferasegene and the reporter was cotransfected into L-G cells with either AML1b or AML1-MTG8. As shown in Figure 5D, AML1-MTG8 significantly activated the promoter of the C/EBPε gene in a dose-dependent manner. In contrast, AML1b did not affect the promoter. These results suggest that AML1-MTG8 stimulates transcription of the C/EBPεgene. However, no consensus binding motif for AML1 is present in the 1280 bp promoter region, suggesting that additional steps might be involved in transcriptional activation of G-CSFR by AML1-MTG8.
Overexpression of C/EBPɛ induces expression of G-CSFR and G-CSF–dependent cell proliferation
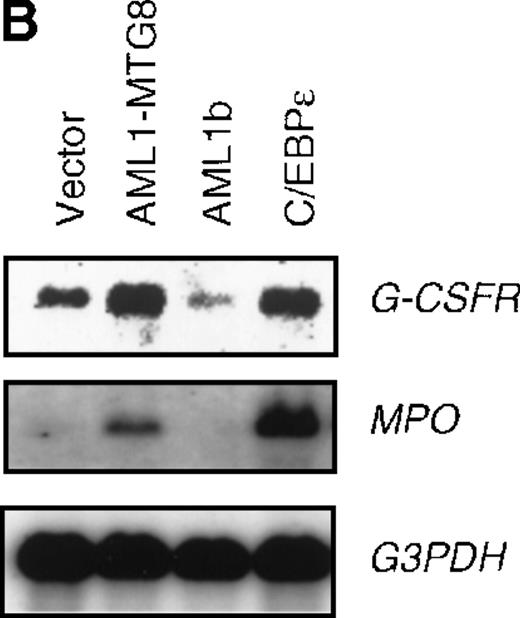
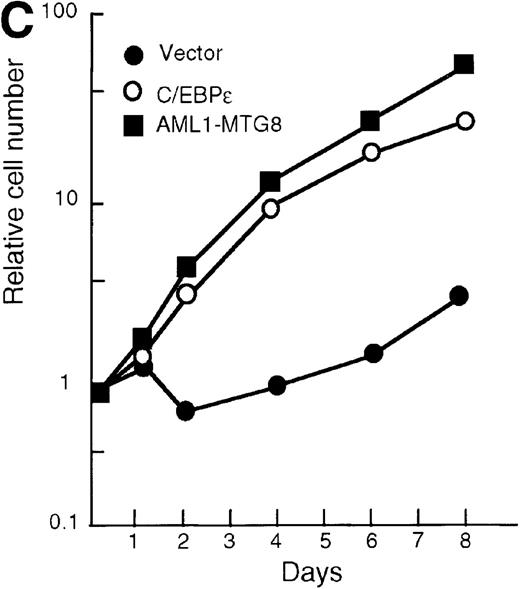
To confirm the finding that AML1-MTG8 enhanced expression of C/EBPε, L-G cells were infected with a C/EBPε-carrying retrovirus. Northern blot analysis showed that the expression of G-CSFR was induced by C/EBPε as in the case of AML1-MTG8 (Figure6B). The introduction of C/EBPε also induced MPO expression like that observed after the introduction of AML1-MTG8, although induction by C/EBPε was stronger than that by AML1-MTG8. The similar effects of AML1-MTG8 and C/EBPε on MPO expression suggest that the induction of MPO expression by AML1-MTG8 is also mediated by C/EBPε. The cells expressing C/EBPε proliferated in response to G-CSF for at least 8 days after exposure to G-CSF (Figure 6C). However, after 7 days of culture in the presence of G-CSF, a significant percentage of the cells (about 80%) showed morphologic differentiation into mature neutrophils with segmented nuclei (data not shown).
Induced expression of G-CSFR and growth stimulation by C/EBPε.
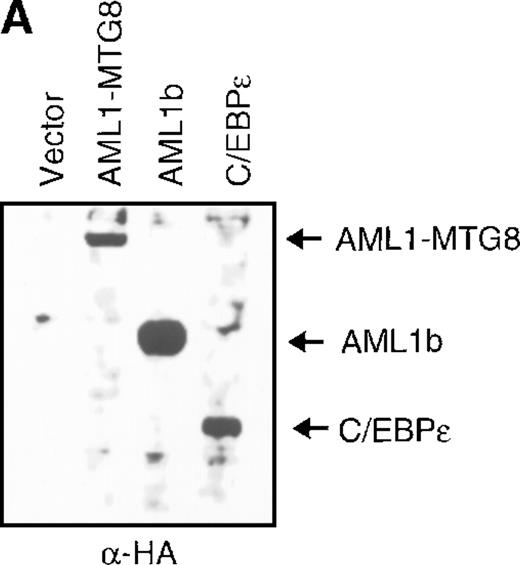
(A) Expression of C/EBPε protein in L-G cells detected by Western blot analyses. Proteins from the L-G infectants with viruses encoding AML1b, AML1-MTG8, or C/EBPε were analyzed for immunodetection using anti-HA antibody. The positions of AML1b, AML1-MTG8, and C/EBPε are indicated by arrows. (B) Total RNAs (5 μg of RNA) were prepared from L-G infectants cultured with IL-3, blotted, and hybridized with32P-labeled mouse G-CSFR cDNA, mouse MPO cDNA, and human G3PDH cDNA. (C) Growth curves of L-G cells infected with LNSX retroviruses carrying human C/EBPε cDNA or AML1-MTG8 cDNA in the presence of G-CSF.
Induced expression of G-CSFR and growth stimulation by C/EBPε.
(A) Expression of C/EBPε protein in L-G cells detected by Western blot analyses. Proteins from the L-G infectants with viruses encoding AML1b, AML1-MTG8, or C/EBPε were analyzed for immunodetection using anti-HA antibody. The positions of AML1b, AML1-MTG8, and C/EBPε are indicated by arrows. (B) Total RNAs (5 μg of RNA) were prepared from L-G infectants cultured with IL-3, blotted, and hybridized with32P-labeled mouse G-CSFR cDNA, mouse MPO cDNA, and human G3PDH cDNA. (C) Growth curves of L-G cells infected with LNSX retroviruses carrying human C/EBPε cDNA or AML1-MTG8 cDNA in the presence of G-CSF.
The effect of overexpression of G-CSFR on G-CSF–induced cell proliferation and differentiation
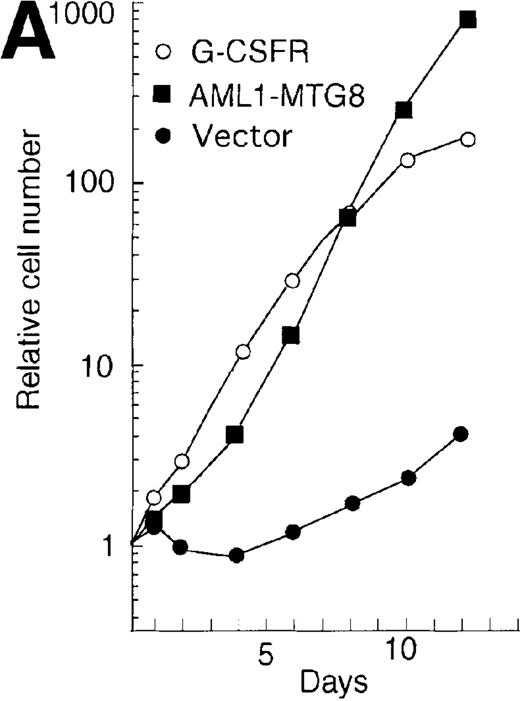
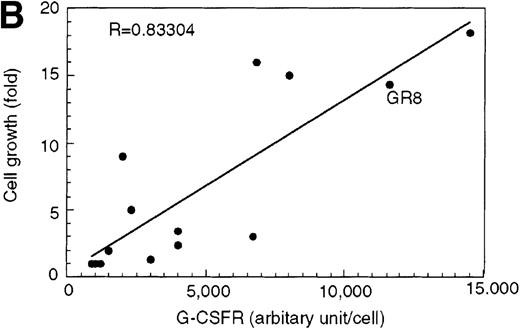
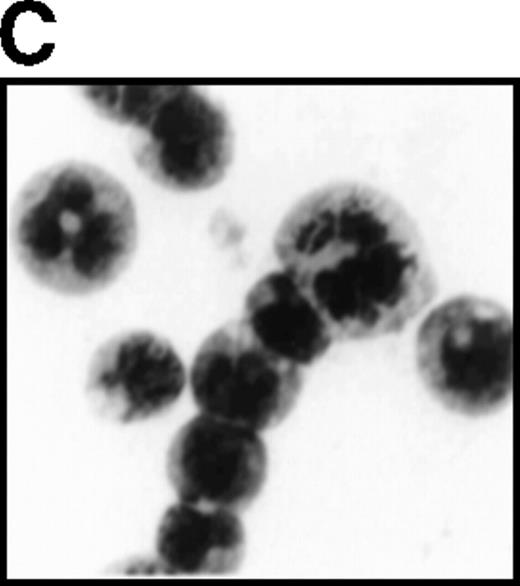
The high levels of G-CSFR expression and the high proliferative responses to G-CSF in the cells expressing AML1-MTG8 suggest that the proliferative responses of these cells may be associated with the up-regulation of G-CSFR. To evaluate this notion, the murine G-CSFR gene was introduced into L-G cells and 32Dcl3 cells by virus infection and transfection, respectively. L-G infectants, which overexpressed G-CSFR (about 20 000 sites/cell) proliferated in response to G-CSF for at least 10 days after exposure to G-CSF (Figure7A). To test the relationship between G-CSFR levels and G-CSF–dependent cell proliferation, 13 clones, which stably expressed various levels of G-CSFR (900∼15 000 sites/cell), were isolated after transfection of 32Dcl3 cell. The transfectants also showed growth response to G-CSF, and their growth rate over a period of 5 days in the presence of G-CSF was well correlated with the level of G-CSFR expression (Figure 7B). These results indicate that the elevated expression of G-CSFR stimulates cell proliferation in response to G-CSF. However, the growth rate of L-G and 32Dcl3 transformants overexpressing G-CSFR gradually decreased with time. After 9 to 12 days of culture in the presence of G-CSF, a significant percentage of the cells (20-80%) showed morphologic differentiation into mature neutrophils with segmented nuclei. However, other cells were still present that continued to grow without terminal differentiation (Figure7C). In contrast, the cells expressing AML1-MTG8 proliferated continuously in response to G-CSF without terminal differentiation.13 This difference in response to G-CSF between AML1-MTG8–expressing cells and G-CSFR–expressing cells suggests that additional factor(s) may be required for the continuous proliferation of cells and for the inhibition of morphologic terminal differentiation.
Overexpression of G-CSFR stimulates cell proliferation in response to G-CSF.
(A) Growth curves of L-G cells infected with LNSX retroviruses carrying mouse G-CSFR cDNA or AML1-MTG8 cDNA in the presence of G-CSF. (B) Correlation between G-CSFR activity and the rate of proliferation in the presence of G-CSF. The 32Dcl3 transfected cells were inoculated into medium supplemented with 10 ng/ml of G-CSF at a cell density of 2 × 104 cells/mL, and cell numbers were counted with a Coulter counter after 5 days of incubation. (C) Cell morphology of G-CSFR transformed GR8 cells when cultured with G-CSF for 10 days. GR8 is a representative clone that expresses G-CSFR at a high level. Cells were stained with May-Gruenwald-Giemsa solution.
Overexpression of G-CSFR stimulates cell proliferation in response to G-CSF.
(A) Growth curves of L-G cells infected with LNSX retroviruses carrying mouse G-CSFR cDNA or AML1-MTG8 cDNA in the presence of G-CSF. (B) Correlation between G-CSFR activity and the rate of proliferation in the presence of G-CSF. The 32Dcl3 transfected cells were inoculated into medium supplemented with 10 ng/ml of G-CSF at a cell density of 2 × 104 cells/mL, and cell numbers were counted with a Coulter counter after 5 days of incubation. (C) Cell morphology of G-CSFR transformed GR8 cells when cultured with G-CSF for 10 days. GR8 is a representative clone that expresses G-CSFR at a high level. Cells were stained with May-Gruenwald-Giemsa solution.
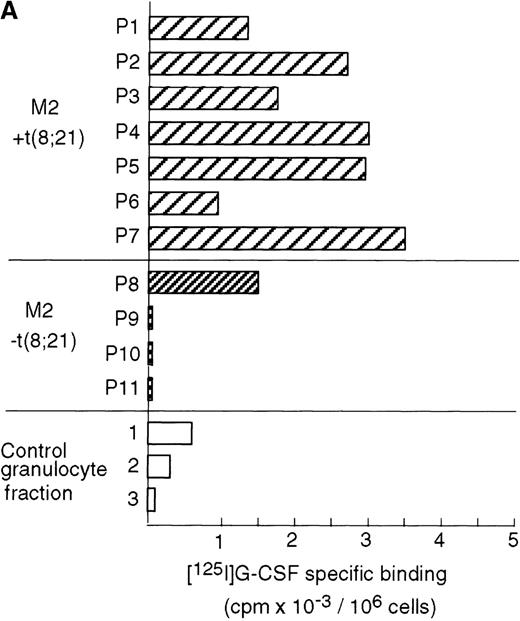
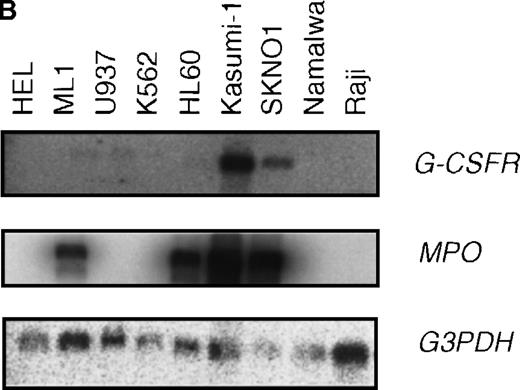
t(8;21) leukemic cells express G-CSFR
Case reports have shown that high expression levels of G-CSFR can be found in leukemic cells from M2 AML patients.49,50 To determine the relationship between AML with a t(8;21) translocation and G-CSFR expression, we measured G-CSFR activity in leukemic cells from M2 AML patients who were positive or negative for t(8;21). A ligand-binding assay showed that elevated levels of G-CSFR (1000∼3500 sites/cell) were present in leukemic cells from all of the AML patients positive for t(8;21), but not in the cells of AML patients negative for t(8;21), except for 1 case (about 1500 sites/cell) (Figure8A). G-CSFR is mainly expressed during the differentiation of myeloblasts into mature neutrophils, and its expression level increases during granulocytic differentiation.22 The increased activity of G-CSF binding seen in t(8;21) cells was not due to contamination by mature granulocytes because large numbers of G-CSF receptors were not detected in fractions enriched with granulocytes from healthy volunteers.
High expression levels of G-CSFR in leukemic cells from patients with t(8;21) and hematopoietic cell lines with t(8;21).
(A) Ligand binding assays for G-CSFR activities in leukemic cells with or without t(8;21) in M2, and in normal granulocytes. (B) Northern blot analyses of poly(A)+RNAs from hematopoietic cell lines. Northern blots were hybridized with human G-CSFR, MPO, and G3PDH cDNA.
High expression levels of G-CSFR in leukemic cells from patients with t(8;21) and hematopoietic cell lines with t(8;21).
(A) Ligand binding assays for G-CSFR activities in leukemic cells with or without t(8;21) in M2, and in normal granulocytes. (B) Northern blot analyses of poly(A)+RNAs from hematopoietic cell lines. Northern blots were hybridized with human G-CSFR, MPO, and G3PDH cDNA.
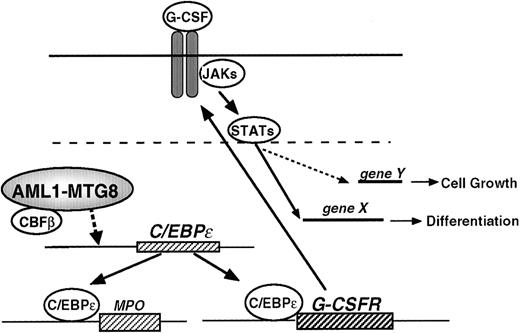
Model for the regulation of G-CSFR expression, cell proliferation, and cell differentiation by AML1-MTG8.
Model for the regulation of G-CSFR expression, cell proliferation, and cell differentiation by AML1-MTG8.
High expression levels of the G-CSFR transcript were also observed in leukemia cell lines carrying t(8;21), SKNO1 and Kasumi-1 cell lines (Figure 8B). None of the other hematopoietic cell lines tested, which did not carry t(8;21), expressed such a high level of G-CSFR. These results indicate that AML with t(8;21) is specifically accompanied by high levels of G-CSFR expression, and the results are consistent with those obtained using the L-G cell system.
Discussion
Transcription factor genes are frequently involved in the chromosomal translocations associated with human leukemia. The resulting deregulation of downstream target genes probably contributes to differentiation arrest and the aberrant proliferation of hematopoietic progenitor cells. Accordingly, studies to identify the downstream gene products that confer the neoplastic nature are a prerequisite for a full understanding of the mechanism of leukemogenesis. However, reports identifying the target genes of chimeric transcription factors are rare, and so far the genes directly responsible for the leukemogenetic process have not been identified for AML1-MTG8 or for other chimeric transcription factors resulting from various chromosome translocations. In the present study, we identified the G-CSFR gene as a candidate downstream target gene involved in the leukemogenic process induced by the chimeric transcription factor AML1-MTG8.
AML1-MTG8 induces expression of G-CSFR through a transcription cascade
To understand the tumorigenic potential of these chimeric proteins, the most commonly accepted explanation is that AML1 chimeric proteins retain the ability to interact with the enhancer core DNA sequence, thereby interfering with AML1-dependent transactivation in a dominant negative manner.7 Here we demonstrated that deregulation, caused by AML1-MTG8, is indirect and mediated by C/EBPε synthesis (Figure 9). It has been reported that PU.1 and C/EBPα are involved in the positive regulation of G-CSFR expression.46 However, our results indicate that the expression of PU.1 and C/EBPα are unlikely to be involved in this regulation, because no changes in the PU.1 transcript or C/EBPα transcript level were observed in L-G cells. The C/EBP family is assumed to be important for hematopoiesis. C/EBPε is almost exclusively expressed in the granulocytic lineage and preferentially up-regulated during granulocytic differentiation.37,48 C/EBPε −/− mice fail to generate functional neutrophils and eosinophils.42 C/EBPε can activate the MIM-1, neutrophil elastase, and G-CSFR promoters.47 C/EBPε and C/EBPα have similar affinity for the consensus binding sites within G-CSFR promoter. C/EBPε is able to repress the transactivation of proteinase 3 by C/EBPα and c-MYB in a dose-dependent manner.51 These findings show that C/EBPε can act either as a transcriptional activator or a repressor depending on the target promoter.
AML1 also functions as both an activator and repressor depending on the presence of cooperative factors. Analysis of the truncated promoter suggested that AML1 might be involved in the regulation of G-CSFR in 2 different ways. First, it directly inactivates the G-CSFRpromoter by binding to an AML1 sequence locating at −391 to −385. Second, it inactivates the promoter by repressing the expression of C/EBPε that is essential for the activation ofG-CSFR. Although the molecular basis of the regulation of the C/EBPε expression has not been well elucidated, the present data show that AML1-MTG8 activated the promoter of the C/EBPε gene. This activation again seems to be indirect, because the AML1 binding motif is not present in the promoter region analyzed.
Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by AML1-MTG8 is suggested to be responsible for leukemogenesis. However, the mechanism through which the histone deacetylase complex represses the expression of the target genes, which seems to be important for myeloid cell growth and differentiation, is still unknown. The preliminary gene expression profiles obtained by mRNA differential display analyses showed that AML1-MTG8 rather than AML1 up-regulates many genes relating to myeloid cell differentiation in L-G myeloid precursor cells.57 If AML1-MTG8 always acts as a transcriptional repressor, it might repress the expression of possible repressors that might be under the positive control of AML1. Alternatively, up-regulation by AML1-MTG8 could depend on changes in AML1 or AML1-MTG8 complexes that occur during certain stages of differentiation. The changes in AML1 or AML1-MTG8 complexes may specifically regulate the target genes.
Up-regulation of G-CSFR induces cell proliferation of myeloid precursor cells
We found the overexpression of G-CSFR can mimic certain leukemogenic activities of AML1-MTG8. However, the overexpression of G-CSFR does not completely explain all the effects of AML1-MTG8. Unlike AML1-MTG8–expressing cells, G-CSFR–expressing cells could differentiate into mature neutrophils. It is possible that additional factors, whose expression would be deregulated by AML1-MTG8, are required to fully express the transforming activity of AML1-MTG8.
We also showed that the number of functional G-CSFRs is high in leukemic specimens from AML patients with t(8;21). There are several possible explanations of how elevated G-CSFR levels might contribute to leukemogenesis. First, high levels of G-CSFR expression might provide leukemic cells with a growth advantage. Our results show that growth stimulation is proportional to the level of G-CSFR expression. Supporting evidence shows that overexpressed G-CSFR may activate STAT3 and induce self-renewal of hematopoietic stem cells. Self-renewal of pluripotent embryonic stem cells is mediated via ectopic expression of STAT3.52 The stimulation of cell proliferation induced by the overexpression of G-CSFR may provide a clue about malignant transformation. Second, a high level of G-CSFR may suppress apoptosis, which may contribute to leukemogenesis. In fact, ectopic expression of G-CSFR inhibits apoptosis of L-G cells in the presence of G-CSF. When parental L-G cells are cultured in the presence of G-CSF, a significant proportion of the cell population dies before and after maturation. However, the L-G cells that ectopically expressed high levels of G-CSFR did not die before maturation (data not shown). Third, the high level of G-CSFR might specify the phenotype of leukemic cells with t(8;21). AML with t(8;21) represents a unique phenotype with features of granulocytic differentiation.53 Elevated expression of G-CSFR may induce commitment of leukemic cells to the granulocytic lineage and may be associated with this unique differentiation phenotype. However, in contradiction to this notion, G-CSF stimulates the development of primitive multipotential progenitors both in vitro and in vivo in transgenic mice ubiquitously expressing G-CSFR, but does not induce exclusive commitment to the myeloid lineage.54Other reports indicate that ectopic expression of G-CSFR in hematopoietic progenitor cells permits multilineage differentiation.29 Thus, overexpression of G-CSFR is not sufficient for commitment to the myeloid lineage. The present results suggest that other factors such as C/EBPε may mediate induction of the differentiated phenotype of t(8;21).
AML1-MTG8 might induce functional differentiation through C/EBPɛ
We showed that cells expressing AML1-MTG8 induce the expression of MPO when cultured in the presence of IL-3 and that this induction is likely to be due to up-regulation of C/EBPε. Morphologic analysis showed that many granules were present in cells expressing AML1-MTG8 (data not shown). The expression of G-CSFR, C/EBPε, and MPO are up-regulated as the cell differentiates into the granulocytic lineage. Thus, AML1-MTG8 may induce some of the phenotypes of granulocytic differentiation. These considerations seem to run contrary to the results obtained with L-G cells, where AML1-MTG8 blocked terminal differentiation into segmented neutrophils in response to G-CSF. However, the induced expression of C/EBPε might enhance the functional differentiation of t(8;21)-positive precursor cells. In addition, because C/EBPε is critical for eosinopoeisis and granulopoiesis, the increased number of eosinophils in t(8;21) leukemia could be explained by enhanced synthesis of C/EBPε. It is probable that the nature of the fusion gene in this unique type of leukemia specifies the tumor phenotype. Because t(8;21) is exclusively associated with M2 AML, AML1-MTG8 might not block the differentiation of pluripotent stem cells, but rather specify them for the myeloid lineage. In the case of chimeric mice harboring knocked-in embryonic stem cells expressing MLL-AF9, the resulting acute myeloid leukemias were preceded by effects on differentiation, which resulted in myeloproliferation and the accumulation of Mac-1/Gr-1 double-positive mature myeloid cells.55 56 AML1-MTG8 might have a potential analogous to that of MLL-AF9. At present, there is no evidence to show that hematopoietic malignancy is present in AML1-MTG8 knock-in chimeric mice.
Acknowledgments
We thank Mrs M. Mori and Mrs C. Hatanaka for technical assistance.
Supported in part by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science and Culture, Japan; by a grant from the Special Coordination Funds for the Promotion of Science and Technology from the Science and Technology Agency, Japan; by a Grant-in-Aid for the 2nd Term Comprehensive 10-year Strategy for Cancer Control and the Grant for Research on Aging and Health from the Ministry of Health and Welfare, Japan; and by the program for Promotion of Fundamental Studies in Health Sciences of the Organization for Drug ADR Relief, R&D Promotion and Product Review of Japan.
Reprints:Kimiko Shimizu, Radiobiology Division, National Cancer Center Research Institute, Tsukiji 5-1-1, Chuo-ku, Tokyo 104-0045, Japan; e-mail: kkikuchi@ncc.go.jp.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal