Abstract
Adenine deoxynucleosides induce apoptosis in quiescent lymphocytes and are thus useful drugs for the treatment of indolent lymphoproliferative diseases. To explain why deoxyadenosine and its analogs are toxic to a cell that is not undergoing replicative DNA synthesis, several mechanisms have been proposed, including the direct binding of dATP to the pro-apoptotic factor Apaf-1 and the activation of the caspase-9 and -3 pathways. In this study it is shown, by means of several assays on whole cells and isolated mitochondria, that 2-chloro-2′-deoxyadenosine (2CdA) and 2-choloro-2′-ara-fluorodeoxyadenosine (CaFdA) disrupt the integrity of mitochondria from primary chronic lymphocytic leukemia (B-CLL) cells. The nucleoside-induced damage leads to the release of the pro-apoptotic mitochondrial proteins cytochrome c and apoptosis-inducing factor. The other adenine deoxynucleosides tested displayed comparable DNA-damaging potency but did not affect mitochondrial function. Interference with mitochondrial integrity, thus, may be a factor in the potent cytotoxic effects of 2CdA and CaFdA toward nondividing lymphocytes.
Introduction
The deoxynucleoside analogs 2′-deoxyadenosine (dAdo), 2-chloro-2′-deoxyadenosine (2CdA, cladribine), 9-β-D-arabinofuranosyl-2-fluoroadenine (F-ara-A, fludarabine), and 2-chloro-2′fluorodeoxyadenosine (CaFdA) have achieved an important role in the treatment of indolent lymphoid malignancies, including hairy cell leukemia (HCL), chronic lymphocytic leukemia (CLL), and low-grade lymphoma, because of their ability to kill nondividing cells. The cytotoxicity of these drugs depends mainly on the accumulation of their triphosphate metabolites (dATP, 2CdATP, F-araATP, CaFdATP) in lymphocytes because of the high deoxycytidine kinase levels and the low 5′-deoxynucleotidase activities in these cells.1 Different concentrations of the 2 enzymes in malignant cells may explain resistance between patients.2 Once transformed into the triphosphate form, adenine deoxynucleosides can induce DNA damage2,3 and trigger the apoptotic cascade.4
Apoptosis is characterized by the activation of cysteine proteases called caspases, which cleave proteins essential for the survival of the cell. Furthermore, these caspases have been shown to activate the endonuclease responsible for the internucleosomal cleavage of genomic DNA.5
Mitochondria play a key role in the events leading to caspase activation in cells undergoing apoptosis. Alteration of their functions can induce the release of cytochrome c and apoptosis-inducing factor (AIF) from the intermembrane space into the cytosol, triggering activation of caspases and endonucleases.6-8 Bcl-2 family members, some of which are located in the mitochondria, control those events, by either inducing (eg, Bax, Bad, Bak, Bid, Bim) or inhibiting (eg, Bcl-2, Bcl-XL, Mcl-1) apoptosis.9 10
Cytochrome c released by the mitochondria and a cytosolic apoptosis protease activating factor (Apaf-1) oligomerize into a large multimeric complex, called the apoptosome.7 This complex itself is sufficient to recruit and activate procaspase-9. Once activated, caspase-9 cleaves the downstream caspases, such as caspase-3, and activates the programmed cell death cascade.11 In addition to cytochrome c, AIF is normally confined to mitochondria. The release of AIF to the cytosol is followed by its relocalization to the nucleus. Recombinant AIF causes chromatin condensation in isolated nuclei and large-scale fragmentation of DNA.12
Several anticancer drugs induce DNA damage, which can lead to the phosphorylation of p53 and the subsequent transcriptional activation of p53-dependent proteins such as p21, Bax, and Hdm-2, leading to growth arrest, repair, or apoptosis. One of the target genes involved in p53-dependent programmed cell death might be Bax, a pro-apoptotic Bcl-2 family member protein. Bax overexpression has been linked to cytochrome c release from the mitochondria13 and can induce apoptosis.14
The assembly of the apoptosome requires dATP, which binds to Apaf-1. In a previous study, we compared several clinically relevant adenine deoxynucleotide analogs for their capacity to bind APAF-1 and to activate caspase-9.15 In this study we analyzed and compared the effect of the corresponding nucleosides on both mitochondrial function and DNA integrity.
Materials and methods
Cell lines and drugs
Human fibroblasts HS-68 (ATCC CRL 1635) and human leukemia T lymphoblast CEM cell lines stably transfected with pZipneo vector (CEM/Neo) or with vector encoding BCL2 (CEM/Bcl-2)16 were cultured in complete medium (RPMI-1640 supplemented with 10% fetal bovine serum). The deoxynucleoside analogs dAdo and fludarabine were from Sigma (St Louis, MO); cladribine and CaFdA were synthesized by published procedure.17 F-AraATP and CaFdATP were kind gifts of Dr W. Plunkett (University of Texas M. D. Anderson Cancer Research Center, Houston, TX), and 2CdATP was synthesized in our laboratory by Dr Q. Chao.18
Cell isolation and analysis
Written informed consent was obtained to procure peripheral blood from all patients. Patients had to have B-CLL according to National Cancer Institute criteria of any Rai stage. Criteria for requiring therapy were as follows: disease-related symptoms; anemia, thrombocytopenia, or both; bulky lymphadenopathy, clinically relevant splenomegaly, or both. Heparinized peripheral blood samples from different patients with CLL and containing at least 90% malignant cells were fractionated by Ficoll-Hypaque sedimentation. Nonadherent mononuclear cells were resuspended in complete medium (RPMI-1640 supplemented with 10% fetal bovine serum) at a density of 1 to 2 × 106/mL. Cells were incubated at 37°C in an atmosphere of 5% CO2 with the indicated drugs. In some experiments, frozen cells were used. dAdo was used in cells that were pretreated for 1 hour with 50 μmol/L of the adenosine deaminase inhibitor deoxycoformycin.
Cellular assay for caspase activity
At the indicated time points, CLL cells were washed twice with phosphate-buffered saline (PBS), and the pellet was resuspended in caspase buffer (50 mmol/L HEPES, pH 7.4, 100 mmol/L NaCl, 1 mmol/L EDTA, 0.1% Chaps, and 5 mmol/L dithiothreitol [DTT]) for 10 minutes at 4°C. Lysates were then stored at −80°C. The caspase enzymatic assays were carried out in 96-well plates. Lysates (10-20 μg total protein) were mixed with 50 μL HEB buffer (PIPES 50 mmol/L, KCl 20 mmol/L, EGTA 5 mmol/L, MgCl2 2 mmol/L, and DTT 1 mmol/L, pH 7), and reactions were initiated by the addition of 100 μmol/L of the specific substrate. After a 1-hour incubation at 37°C, caspase-3-like protease activity was measured with the substrate Ac-DEVD-AMC, and caspase-9-like activity was measured using Ac-LEHD-AFC. Activity was measured by the release of 7-amino-4-trifluoromethyl-coumarin (AFC) or 7-amino-4-methyl-cumarin (AMC), monitoring fluorescence at excitation and emission wavelengths of 400 and 505 nm, and 380 and 460 nm respectively.
Cytofluorometric analysis of mitochondrial transmembrane potential (ΔΨm) by DiOC6 and cell membrane permeability by PI
CLL cells were treated with the indicated amount of the drugs either alone or with 50 μmol/L of the cell-permeable caspase inhibitor Z-VAD-fmk (Z-Val-Ala-Asp(Ome)-CH2F; Biomol, Plymouth Meeting, PA). Cells were then incubated for 10 minutes at 37°C in culture medium containing 40 nmol/L 3,3′ dihexyloxacarbocyanine iodide (DiOC6; Molecular Probes, Eugene, OR) and 5 μg/mL propidium iodide (PI; Molecular Probes), followed by analysis within 30 minutes of fluorochrome in a Becton Dickinson FACScalibur cytofluorometer. After suitable compensation, fluorescence was recorded at different wavelengths: DiOC6 at 525 nm (FL-1) and PI at 600 nm (FL-3).
Measurement of alkali-sensitive sites in DNA
DNA damage was assessed by alkaline unwinding and ethidium bromide fluorescence as previously described.19 Because ethidium bromide preferentially binds to double-strand DNA (ds-DNA) in alkali, the relative amounts of nonbroken ds-DNA and broken single-strand DNA can be assessed. Fluorescence was determined with a CytoFluor fluorescence plate reader (PerSeptive Biosystems, Framingham, MA) with excitation at 520 nm and emission at 585 nm. Results are expressed as [% ds-DNA = [(F − Fmin)/(Fmax − Fmin)] × 100], where F is the fluorescence of the experimental condition, Fmin is the background ethidium bromide fluorescence determined after conversion of all DNA into single-strand form by sonication, and Fmax is the fluorescence determined after the addition of the mercaptoethanol solution before the alkaline solution to maintain the pH at 11.0, which is well below that needed to augment unwinding of single-strand DNA. The amount of single-strand DNA in alkali after sonication is proportional to the number of DNA-strand breaks (either single or double), which varies directly as a function of the extent of DNA unwinding.
Neutral comet assay of double-strand DNA breaks
The single-cell comet electrophoresis assay was performed according to the Trevigen CometAssay (Trevigen, Gaithersburg, MD) reagent kit. CLL cells were suspended in complete medium (RPMI-1640 supplemented with 10% fetal bovine serum) at a density of 1 × 106/mL and treated with the indicated drugs for 6 hours at 37°C. Then, 0.5 mL aliquots of 0.5% low melting point agarose in PBS (Sigma, St Louis, MO) at 42°C were added to tubes containing 50 μL each of the cell suspensions. Fifty μL of the mixture was quickly pipetted onto a microscope slide, precovered with 0.5% agarose, and allowed to solidify at 4°C. Slides were immersed for 30 minutes in prechilled lysis buffer (2.5 mol/L sodium chloride, 100 mmol/L EDTA, pH 10, 10 mmol/L Tris base, 1% sodium lauryl sarcosinate, 0.01% Triton X-100) at 4°. After lysis, slides were incubated for 30 minutes in alkali buffer (0.3 mol/L NaOH, 1 mmol/L EDTA). TBE washed slides were placed in horizontal gel electrophoresis chambers containing 1 L 90 mmol/L Tris/90 mmol/L boric acid/2 mmol/L EDTA buffer (TBE) at 1 V/cm for 10 minutes. DNA was stained after SYBR green was added to the slides. A Zeiss Axioscope microscope (Carl Zeiss, Thornwood, NY) attached to a camera and image analysis system was used to quantify the different parameters of the comets. At least 100 cells were analyzed per slide. Results were expressed in terms of percentage of comet cells divided by total amount of cells.
Mitochondria isolation
The CLL cells were washed in ice-cold Dulbecco's phosphate-buffered saline for disruption by nitrogen cavitation.20 All subsequent steps were carried out on ice or at 4°C. The cells were washed with MA buffer (100 mmol/L sucrose, 1 mmol/L EGTA, 20 mmol/L MOPS pH 7.4, 1 g/L bovine serum albumin) then resuspended at 2 × 108 cells/mL in MB buffer (MA buffer plus 10 mmol/L triethanolamine, 5% Percoll, 1 mmol/L phenylmethylsulfonyl fluoride, and an antiproteinase mixture consisting of aprotinin, pepstatin A, and leupeptin, each at 10 μmol/L). The cells were then disrupted by cavitation, after which lysates were centrifuged twice at 2500g for 5 minutes to remove nuclei and unbroken cells then at 25 000g for 30 minutes to isolate mitochondria. The mitochondria were suspended in MC buffer (identical to MA buffer except that the sucrose concentration was 300 mmol/L and an antiprotease mixture was included that consisted of aprotinin, pepstatin A, and leupeptin, each at 10 μmol/L) and used for measuring membrane potential alterations with DiOC6.
Flow cytometry analysis of AIF release
Nondividing CLL cells were treated with the nucleosides as indicated. Cells were washed and permeabilized by 0.03% digitonin treatment for 15 minutes at room temperature. The concentration of digitonin used was obtained by titration and is enough to permeabilize the outer membrane of the cells without affecting the mitochondrial membrane. Cells were then fixed with 2% paraformaldehyde and incubated with the primary antibody to AIF (a kind gift from G. Kroemer, Institut Gustave Roussy, Villejuif, France), washed, and stained with the fluorescent-labeled secondary antibody (Alexa 488; Molecular Probes). An isotype-matched primary antibody was used as control. Cells were postfixed by resuspending the pellet in 2% paraformaldehyde and analyzed by flow cytometry.
Immunocytochemistry
One hundred microliters of the CLL cells at a concentration of 2 × 106/mL, treated for 24 hours with the indicated drugs, was stained for the detection of the mitochondrial membrane potential with 150 nmol/L Mitotracker Orange (Molecular Probes) for 30 minutes at 37°C. Then 2 × 105 cells were pelleted at 300g for 5 minutes on a slide. The cells were fixed for 10 minutes in 4% paraformaldehyde, washed in PBS, and incubated for 2 hours with the antibodies specific for AIF (supplied by G. Kroemer) and cytochrome c (clone 6H2.B4; Pharmingen, San Diego, CA), at a concentration of 2 to 10 μg/mL in I-Block blocking buffer (Tropix, Bedford, MA) supplemented with 0.05% Triton X-100. Alexa 488 (green) and Alexa 568 (red) served as species-specific secondary antibodies. The nuclear DNA was stained using DAPI. Coverslips were washed successively in PBS and deionized H2O for 5 minutes and mounted in Fluoromount (Fisher, CA). Images were obtained with a Zeiss Axioscope microscope.
Microinjection
HS-68 cells were grown on glass coverslips (Fisher, Pittsburgh, PA). The triphosphate forms of the nucleosides analogs were dissolved at a concentration of 5 mmol/L in microinjection buffer consisting of 5 mmol/L sodium phosphate and 100 mmol/L KCl (pH 7.4) containing a Dextran-FITC or Dextran-TRITC fluorescent dye (Sigma). Approximately 100 cells/analog were microinjected using glass capillary needles. The estimated final concentration in the cells was 500 μmol/L, based on an estimated injection volume of 50 fL. Cells were stained as described above.
Results
Cytotoxicity of the nucleoside analogs on CLL cells
For this study, we first screened 20 patients for their sensitivities to the nucleoside analogs 2CdA, F-Ara-A, CaFdA and dAdo (plus deoxycoformycin). We used equimolar concentrations of the nucleosides because the principal aim was to compare their effects on DNA and mitochondria integrity. CLL cells that were completely resistant to the nucleoside toxicity after 24 hours of incubation at 10 μmol/L (less than 20% specific killing) were excluded from analysis because these cells did not accumulate the active triphosphate analog intracellularly.21 Five of 20 patients with CLL were thus excluded. In the nucleoside-susceptible CLL populations, loss of viability measured by dye exclusion was already visible after a few hours (Figure 1A). Analysis of the apoptotic mechanisms triggered by the different nucleosides showed distinct peculiarities. Mitochondrial membrane potential measured by flow cytometry decreased in CLL cells treated with 2CdA and CaFdA already after 12 hours, but not after similar treatment with dAdo and F-Ara (Figure 1B). Similarly, the kinetics of enzymatic activation of caspase-9 and -3 (Figure 1 C,D) were different: 2CdA and CaFdA were able to activate the caspases already after 12 hours, whereas 24-hour incubation was required to see induction of the caspase activity by F-Ara-A. The intensity of caspase-3 (Figure 1C) and caspase-9 (Figure 1D) activation by F-AraA at 24 hours was also reduced by at least 2-fold when compared to 2CdA or CaFdA.
Nucleoside cytotoxicity on B-CLL cells.
(A) B-CLL cells were treated with 2CdA (▪), CaFdA (■), F-Ara-A (●), and dAdo (○) at 10 μmol/L for the indicated time points. Viability was assessed by counting the cells after staining with erythrosin B and is represented as percentage of the control. (B) The same cells were tested for mitochondria membrane potential by flow cytometry analysis of incorporation of DiOC6. (C) Cells were lysed in caspase buffer and incubated with 100 μmol/L of fluorometric substrate, and caspase-9 activity was measured by fluorometric analysis. (D) Caspase-3 enzymatic activity. These data (± SD) were obtained from a single patient studied in duplicate and are representative of 6 different patients.
Nucleoside cytotoxicity on B-CLL cells.
(A) B-CLL cells were treated with 2CdA (▪), CaFdA (■), F-Ara-A (●), and dAdo (○) at 10 μmol/L for the indicated time points. Viability was assessed by counting the cells after staining with erythrosin B and is represented as percentage of the control. (B) The same cells were tested for mitochondria membrane potential by flow cytometry analysis of incorporation of DiOC6. (C) Cells were lysed in caspase buffer and incubated with 100 μmol/L of fluorometric substrate, and caspase-9 activity was measured by fluorometric analysis. (D) Caspase-3 enzymatic activity. These data (± SD) were obtained from a single patient studied in duplicate and are representative of 6 different patients.
DNA damage analysis
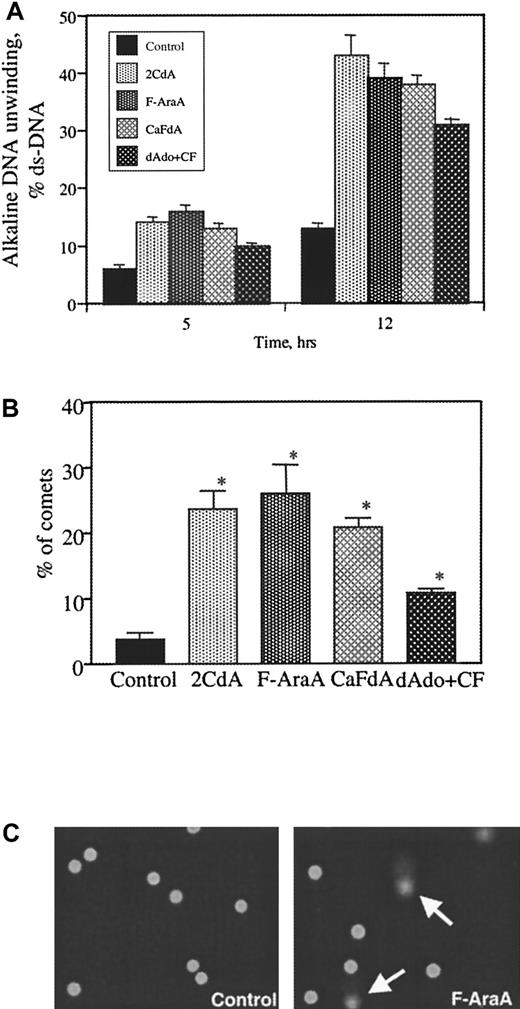
We used 2 different methods to study nucleoside effects on DNA integrity, alkaline unwinding and comet assay. As assessed by the unwinding assay, 2CdA, F-Ara-A and CaFdA at 10 μmol/L generate approximately equivalent DNA breaks after 12 hours (Figure2A). The broad-spectrum caspase inhibitor z-VAD-fmk did not protect the cells from early DNA damage (data not shown). dAdo was less potent. To confirm these results, single-cell analysis was performed. After 6 hours of continuous incubation with 10 μmol/L nucleoside, cells were placed on the slides in an agarose layer and lysed. Current was applied for 10 minutes, and DNA was stained with SYBR green. Damaged DNA migrated from the lysed cell membrane, forming a DNA tail in a comet shape (Figure 2C). Approximately 100 cells were counted, and the percentages of the cells displaying comets were enumerated (Figure 2B). More than 20% of the cells exposed to 2CdA, F-Ara-A, and CaFdA displayed a comet shape, whereas dAdo was able to affect only 12% of the cells.
Nucleoside-induced DNA damage of B-CLL cells.
(A) B-CLL cells from 4 different patients were treated at the indicated time points with 10 μmol/L of 2CdA, CaFdA, F-Ara-A, and dAdo. DNA damage by alkaline unwinding was assessed. Data are reported as percentage ds-DNA. (B) Single-cell DNA damage (comet assay) was performed after 6-hour treatment with 10-μmol/L concentrations of the drugs. Cells were plated on a slide in low-melting point agarose and lysed, and current was applied for 10 minutes at 1 V/cm. At least 100 cells were counted, and the percentages of the comets were determined. Columns represent the mean and error bars the standard deviation obtained from 4 patients. (*) Statistically increased from control untreated cells; P < .05 by the nonparametric Mann-Whitney test. (C) Example of comets after treatment with F-Ara-A compared to control cells. These data were obtained from a single patient and are representative for 4 patients.
Nucleoside-induced DNA damage of B-CLL cells.
(A) B-CLL cells from 4 different patients were treated at the indicated time points with 10 μmol/L of 2CdA, CaFdA, F-Ara-A, and dAdo. DNA damage by alkaline unwinding was assessed. Data are reported as percentage ds-DNA. (B) Single-cell DNA damage (comet assay) was performed after 6-hour treatment with 10-μmol/L concentrations of the drugs. Cells were plated on a slide in low-melting point agarose and lysed, and current was applied for 10 minutes at 1 V/cm. At least 100 cells were counted, and the percentages of the comets were determined. Columns represent the mean and error bars the standard deviation obtained from 4 patients. (*) Statistically increased from control untreated cells; P < .05 by the nonparametric Mann-Whitney test. (C) Example of comets after treatment with F-Ara-A compared to control cells. These data were obtained from a single patient and are representative for 4 patients.
Mitochondrial damage
Mitochondria are known to contain their own deoxynucleoside kinases.22 Mitochondria were isolated by cavitation from CLL cells and treated for 10 minutes with different concentrations of the nucleoside analogs. Both 2CdA and CaFdA were able to affect the membrane potential of the mitochondria by reducing the incorporation of the dye DiOC6 (Figure 3A). dAdo and F-Ara-A at the highest concentrations tested (1000 μmol/L) did not affect the membrane potential. We then analyzed the damage to mitochondria in whole CLL cells by measuring the cytoplasmic fluorescence of AIF, after treatment with 10 μmol/L of the analogs for 8 hours. Apparent relocalization of AIF was detected by flow cytometric analysis after permeabilization of the cells with 0.03% digitonin. This concentration of digitonin used had been shown in preliminary experiments to permeabilize the outer membrane of the cells without affecting the mitochondrial membrane. Only treatment with 2CdA, but not F-AraA, increased AIF fluorescence (Figure 3B). CaFdA behaved as 2CdA (data not shown). We believe that the increased AIF fluorescence is a result of its relocalization from mitochondria to the cytosol, as immunocytochemistry results also seem to indicate (data not shown). Pre-incubation of the CLL cells with 100 μmol/L Z-VAD-fmk did not alter the 2CdA-induced AIF relocalization.
Mitochondrial damage by nucleosides in B-CLL cells.
(A) Mitochondria from B-CLL cells were nitrogen isolated by cavitation, treated for 10 minutes with the indicated concentrations of 2CdATP (▪), CaFdATP (■), F-Ara-ATP (●), and dATP (○), stained with 40 nmol/L DiOC6 for 10 minutes at room temperature, and rapidly analyzed by flow cytometry. (B) B-CLL cells were treated with 10 μmol/L of the nucleosides for 8 hours, then permeabilized with 0.03% of digitonin and fixed with 4% paraformaldehyde. AIF release was detected by staining with specific antibody and with fluorescence-labeled secondary Alexa 488 and measured by flow cytometry. (C) Human leukemia T-lymphoblastoid CEM cell lines, transfected with bcl-2–expressing vector (CEM/bcl-2) (○) or control vector (CEM/NEO) (●), were treated with 1 μmol/L of the nucleosides. Membrane potential analyses and caspase analysis were performed at the indicated time points. Error bars represent the SD obtained from 3 independent experiments.
Mitochondrial damage by nucleosides in B-CLL cells.
(A) Mitochondria from B-CLL cells were nitrogen isolated by cavitation, treated for 10 minutes with the indicated concentrations of 2CdATP (▪), CaFdATP (■), F-Ara-ATP (●), and dATP (○), stained with 40 nmol/L DiOC6 for 10 minutes at room temperature, and rapidly analyzed by flow cytometry. (B) B-CLL cells were treated with 10 μmol/L of the nucleosides for 8 hours, then permeabilized with 0.03% of digitonin and fixed with 4% paraformaldehyde. AIF release was detected by staining with specific antibody and with fluorescence-labeled secondary Alexa 488 and measured by flow cytometry. (C) Human leukemia T-lymphoblastoid CEM cell lines, transfected with bcl-2–expressing vector (CEM/bcl-2) (○) or control vector (CEM/NEO) (●), were treated with 1 μmol/L of the nucleosides. Membrane potential analyses and caspase analysis were performed at the indicated time points. Error bars represent the SD obtained from 3 independent experiments.
Overexpression of the anti-apoptotic protein bcl-2 protects cells from direct mitochondrial damage.23 The CEM cell line overexpressing bcl-2 (CEM/Bcl-2) and the control mock-transfected cells (CEM/NES) were treated with 1 μmol/L 2CdA, CaFdA, and F-Ara-A. At different time points mitochondrial membrane potential and caspase activation were measured. There was no significant difference in the toxicity of F-Ara-A toward the 2 cell lines. In contrast, Bcl-2 overexpression significantly protected against early caspase activation and mitochondrial transmembrane alterations induced by 2CdA after 4 and 8 hours (Figure 3C). The protection was lost after 24 hours of incubation. CaFdA behaved like 2CdA (data not shown).
Immunocytochemistry
To corroborate the biochemical results, we performed immunocytochemical analysis of nucleoside-treated CLL cells. The effect of the nucleoside analogs on B-CLL mitochondria was analyzed after only 8 hours to exclude secondary effects related to caspase activation. Alteration of the mitochondria membrane potential was assessed by the incorporation of Mitotracker orange (MT orange) and the release of cytochrome c. In control cells, cytochrome c and MT orange colocalized in the mitochondria (Figure 4A). After treatment with 2CdA, the mitochondrial cytochrome c signal was strongly reduced, but the MT orange fluorescence remained constant, indicating that the mitochondria may have released cytochrome c but maintained the transmembrane potential (Figure 4B). F-Ara-A and dAdo did not visibly affect the mitochondria (Figure 4C; dAdo data not shown). Finally, experiments performed using the pan-specific caspase inhibitor Z-VAD-fmk indicated that the nucleoside-induced release of cytochrome c was not caspase dependent (data not shown).
Immunocytochemistry of mitochondrial damage.
B-CLL cells were treated with the indicated drugs for 8 hours. Cells were stained for mitochondria membrane potential with MT orange (red, A) for 30 minutes, pelleted on a slide at 300g, fixed in 4% paraformaldehyde, and stained for cytochrome c (green, B) with monoclonal antibody and secondary antibody Alexa 488. DNA was visualized with DAPI (blue, C). (D) Composite image. Arrows show cells affected by the drug, where cytochrome c release is noticeable. Images were obtained with a Zeiss Axioscope microscope, with CCD camera.
Immunocytochemistry of mitochondrial damage.
B-CLL cells were treated with the indicated drugs for 8 hours. Cells were stained for mitochondria membrane potential with MT orange (red, A) for 30 minutes, pelleted on a slide at 300g, fixed in 4% paraformaldehyde, and stained for cytochrome c (green, B) with monoclonal antibody and secondary antibody Alexa 488. DNA was visualized with DAPI (blue, C). (D) Composite image. Arrows show cells affected by the drug, where cytochrome c release is noticeable. Images were obtained with a Zeiss Axioscope microscope, with CCD camera.
Microinjection
To confirm, in a separate system, the direct role of nucleotides in mitochondrial damage, we microinjected triphosphates into human fibroblasts. Because of their size, HS-68 cells were relatively easy to microinject. The results show that the triphosphates of 2CdA (and CaFdA; data not shown) induced a change in the subcellular distribution of mitochondria (Figure 5). To detect the mitochondria in the cells, both MT orange staining and labeling using cytochrome c antibodies were used, but only results using MT orange are shown. The microinjected cells were identified by the co-injection of florigenically labeled dextran (in green). The mitochondria were arranged along microtubules, forming a fibrillar array radiating toward the cell periphery. After microinjection with 2CdA (Figure 5C,D), but not with F-AraA (Figure 5E,F), the mitochondria retracted from the cell periphery and clumped around the nucleus. Interestingly, this mitochondrial redistribution was not accompanied by a dramatic change in cell shape or by cytoplasmic cytochrome c release.
Nucleotide microinjection experiments.
Human HS-68 fibroblasts were grown on coverslips. Nucleotide triphosphates were microinjected at an estimated final concentration of 500 μmo/L. To identify the microinjected cells, fluorescein isothiocyanate-labeled dextran was co-injected with the nucleotides; therefore, the microinjected cells appear in green (C, E). Cells were incubated with 150 nmol/L MT orange (red stain) for 30 minutes to stain for mitochondria membranes, and fixed in 4% paraformaldehyde, and DNA was dyed with DAPI stain (blue, A, C, E). Left panels represent composite pictures that include DNA (blue), mitochondria (red), and dextran (green for microinjected cells). Right panels (B, D, F) represent the stained mitochondria alone. Cells treated with 2CdATP show a rearrangement of the mitochondria. Cells treated with the same concentration of F-AraATP do not display the same morphologic changes. Images were obtained with a Zeiss Axioscope microscope, equipped with CCD camera, using a 40 × 1.3-na oil objective.
Nucleotide microinjection experiments.
Human HS-68 fibroblasts were grown on coverslips. Nucleotide triphosphates were microinjected at an estimated final concentration of 500 μmo/L. To identify the microinjected cells, fluorescein isothiocyanate-labeled dextran was co-injected with the nucleotides; therefore, the microinjected cells appear in green (C, E). Cells were incubated with 150 nmol/L MT orange (red stain) for 30 minutes to stain for mitochondria membranes, and fixed in 4% paraformaldehyde, and DNA was dyed with DAPI stain (blue, A, C, E). Left panels represent composite pictures that include DNA (blue), mitochondria (red), and dextran (green for microinjected cells). Right panels (B, D, F) represent the stained mitochondria alone. Cells treated with 2CdATP show a rearrangement of the mitochondria. Cells treated with the same concentration of F-AraATP do not display the same morphologic changes. Images were obtained with a Zeiss Axioscope microscope, equipped with CCD camera, using a 40 × 1.3-na oil objective.
Discussion
Adenine deoxynucleoside analogs, such as F-Ara-A and 2CdA, are used for the treatment of B-CLL. However, the mechanisms of action of these drugs are not understood precisely. In this study, we confirm with 2 different assays that all the cytotoxic adenine deoxynucleosides induce DNA damage in CLL, as previously reported.24,25 At equimolar concentrations, the DNA strand break formation was comparable, and was at the early time points, when the damage was a result of the direct effect of the drugs on DNA. After 24 hours, a big difference between 2CdA/CaFdA and F-Ara-A was observed. This was probably a consequence of the activation of the caspases, leading to endonuclease-mediated cleavage of the DNA,26 because caspase activity at 24 hours was higher in 2CdA- and CaFdA-treated cells.
The data indicate that 2CdA/CaFdA and F-Ara-A have distinct effects on mitochondria. Time-course analyses of mitochondrial membrane potential in whole cells, release of AIF by flow cytometry, cytochrome c release by immunofluorescence, and studies of purified mitochondria demonstrated that 2CdA/CaFdA, but not F-Ara-A, disrupted the mitochondrial integrity. These conclusions were supported by analyses of fibroblasts microinjected with the respective triphosphates.
Only 2CdA and CaFdA reduced the incorporation of DiOC6 in mitochondria at 8 to 12 hours, induced AIF release (probably as a consequence of the opening of the PT pore12), stimulated the release of cytochrome c, and disrupted the structural filamentous arrangement of mitochondria, leaving the organelles dispersed in the cytoplasm. The immunofluorescence images suggest that both cytochrome c and AIF release preceded the loss of mitochondrial transmembrane potential. Caspase activation also anticipated the loss of the mitochondrial membrane potential, suggesting that the 2 events may be independent, as already demonstrated in staurosporine-treated human tumor cells.27
Several explanations are possible for the severe and apparently direct mitochondrial damage observed with 2CdA- and CaFdA-treated CLL cells. First, the corresponding nucleotides could directly bind to proteins that are known to regulate mitochondrial functions, such as the F1-F0 ATPase (mitochondrial adenosine triphosphate [ATP] synthase or complex V) or the adenine nucleotide translocator (ANT). Both proteins are situated on the mitochondrial inner membrane and are actively involved in the process of oxidative phosphorylation that produces most of the cellular energy. The F1-F0 ATPase contains ATP and adenosine diphosphate binding sites, and it has been shown to bind dATP with an affinity similar to that of ATP.28 We recently demonstrated that the nucleotide analogs can substitute for dATP in Apaf-1–mediated caspase-9 activation,29 and it is thus conceivable that these analogs could also bind the F1-F0 ATPase. The ANT, with the voltage-dependent anion channel (VDAC, or porin), Bax, and cyclophilin D, are thought to come together at the mitochondrial inner and outer membrane contact points to create the mitochondrial permeability transition pore (mtPTP).30,31 Recently, the mtPTP and VDAC alone have been associated with the release of cytochrome c and AIF that precedes caspase activation.32 It is conceivable that the adenine nucleotide analogs, by binding to ANT, could affect the opening of the mtPTP, allowing the release of AIF and cytochrome c. Results with purified mitochondria indicate that the mitochondrial transmembrane potential is rapidly reduced. Unfortunately, our analysis of AIF and cytochrome c release from isolated mitochondria produced inconsistent results (data not shown), but dATP-induced cytochrome c release from isolated mitochondria has been demonstrated.33 The hypothesis of nucleotide-induced mtPTP opening is also consistent with the protection observed in Bcl-2–overexpressing cells, early after incubation with the 2CdA and CaFdA.
A second explanation for the observed mitochondrial damage may be the intramitochondrial accumulation of the triphosphate analogs, leading to mitochondrial DNA damage followed by a breakdown of metabolic functions. 2CdATP and CaFdATP may accumulate more rapidly than F-AraATP in the mitochondria because they are converted to their triphosphate forms with a 10-fold higher efficiency by the mitochondrial deoxyguanosine kinase.34 Mitochondrial deoxyguanosine kinase-overexpressing cells showed increased sensitivity to 2CdA compared to untransfected cells.35 It is also conceivable that the 2-chloro derivatives are slightly more hydrophobic than the corresponding 2-fluoro derivatives and therefore may be able to insert more easily into the mitochondrial membrane, affecting their integrity.
F-Ara-A also affected mitochondria, but only as a late event, suggesting an indirect mechanism. DNA damage is known to activate the p53 pathway by inducing phosphorylation of p53 and transcriptional activation of pro-apoptotic genes. This leads to the release of cytochrome c and the activation of the intrinsic apoptotic machinery, resulting in caspase activation.13 36
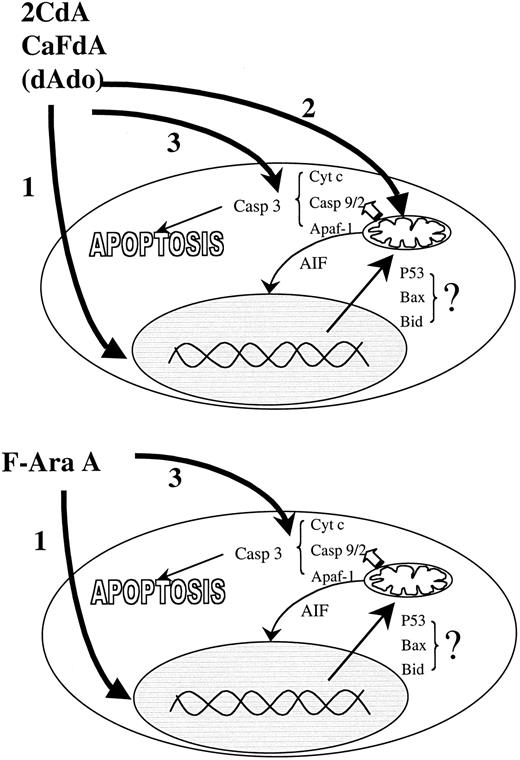
In a previous study,29 we demonstrated that F-Ara-ATP is the most potent nucleotide activator of Apaf-1, as assessed by procaspase-9 and -3 cleavage. The current study indicates that APAF-1 binding is not the only determinant in the adenosine deoxynucleoside cytotoxicity pathway in B-CLL cells. 2CdA and CaFdA are much more toxic to CLL cells than F-Ara-A, and only the chlorinated analogs cause direct mitochondrial damage. Thus, the mitochondrial effects of 2CdA and CaFdA may explain why these drugs are toxic to CLL cells at concentrations 5- to 10-fold lower than F-Ara-A. The different mechanisms for the toxicity of the nucleoside analogs are summarized in Figure 6.
Mechanisms of action.
There are 3 modes of action of the adenine deoxynucleoside analogs in nonproliferating CLL cells. In the cells the drugs are transformed into their triphosphate form, which induce DNA damage, mitochondrial dysfunction, and binding to APAF-1 and activation of the caspase pathway. F-ara-A kills cells mainly by DNA damage and Apaf-1 activation. 2CdA and CaFdA also interfere with mitochondrial function. The different effects of the adenine deoxynucleosides in various lymphoid malignancies may reflect the relative importance of the 3 mechanisms in apoptosis regulation.
Mechanisms of action.
There are 3 modes of action of the adenine deoxynucleoside analogs in nonproliferating CLL cells. In the cells the drugs are transformed into their triphosphate form, which induce DNA damage, mitochondrial dysfunction, and binding to APAF-1 and activation of the caspase pathway. F-ara-A kills cells mainly by DNA damage and Apaf-1 activation. 2CdA and CaFdA also interfere with mitochondrial function. The different effects of the adenine deoxynucleosides in various lymphoid malignancies may reflect the relative importance of the 3 mechanisms in apoptosis regulation.
Supported in part by National Institutes of Health grants GM23200 and CA81534 and by a grant from the DOD Breast Cancer Research Program (L.M.L.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Lorenzo M. Leoni, Dept of Medicine, University of California San Diego, 9500 Gilman Dr, La Jolla, CA 92093-0663; e-mail:lleoni@ucsd.edu.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal