Abstract
The ability of lentiviral vectors to transfer genes into human hematopoietic stem cells was studied, using a human immunodeficiency virus 1 (HIV-1)–derived vector expressing the green fluorescence protein (GFP) downstream of the phosphoglycerate kinase (PGK) promoter and pseudotyped with the G protein of vesicular stomatitis virus (VSV). High-efficiency transduction of human cord blood CD34+cells was achieved after overnight incubation with vector particles. Sixteen to 28 percent of individual colony-forming units granulocyte-macrophage (CFU-GM) colonies derived from cord blood CD34+ cells were positive by polymerase chain reaction (PCR) for the GFP gene. The transduction efficiency of SCID-repopulating cells (SRC) within the cord blood CD34+population was assessed by serial transplantation into nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice. When 400 000 cord blood CD34+ cells were transplanted into primary recipients, all primary and secondary recipients contained and expressed the transgene. Over 50% of CFU-GM colonies derived from the bone marrow of these primary and secondary recipients contained the vector on average as determined by PCR. Transplantation of transduced cells in limiting dilution generated GFP+ lymphoid and myeloid progeny cells that may have arisen from a single SRC. Inverse PCR analysis was used to amplify vector-chromosomal junctional fragments in colonies derived from SRC and confirmed that the vector was integrated. These results show that lentiviral vectors can efficiently transduce very primitive human hematopoietic progenitor and stem cells.
Introduction
The development of gene therapy strategies to correct hematopoietic and genetic disorders has been hampered by the low level of gene transfer into human hematopoietic stem cells (HSC) using vectors derived from oncoretroviruses such as the Moloney murine leukemia virus.1-5 Oncoretroviruses indeed require cell division for integration and because repopulating HSC are largely quiescent; oncoretroviral vectors are largely inefficient in these targets.6,7 Thus, much interest has recently been focused on vectors derived from lentiviruses such as human immunodeficiency virus 1 (HIV-1), which have been shown to transduce a variety of nondividing cells, including hematopoietic cells.8-12Furthermore, lentiviral vector particles pseudotyped with vesicular stomatitis virus glycoprotein G (VSV-G) can enter a large variety of targets due to the ubiquity of the VSV-G phospholipidic receptor and can be easily concentrated by ultracentrifugation.13 14
Several recent studies have shown that HIV-1 vectors pseudotyped with VSV-G can infect hematopoietic cells.10,15-20 Using a 2-plasmid vector system where the vector plasmid doubles as a packaging construct encoding most of the viral genes, Akkina and coworkers transferred the luciferase gene into human CD34+ cells and demonstrated efficient gene transfer into colony-forming units granulocyte-macrophage (CFU-GM) colonies.10 Similarly, Reiser and colleagues transferred the HSA gene into quiescent CD34+ cells from peripheral blood and demonstrated expression of the transgene 3 days after transduction, but the functional potential of the cells was not assessed.15Highly purified quiescent human hematopoietic progenitors can also be transduced with lentiviral vectors.17,19 Using a 3-plasmid system with a second-generation packaging construct that lacks the accessory genes of HIV, quiescent CD34+, CD38−, Thy1+ human hematopoietic progenitors could be transduced. Stable expression of the transgene was seen in progeny cells from these purified progenitors, sorted initially as single cells, after growth on stromal cells for 5 to 6 weeks.17 In similar studies, Case and coworkers demonstrated stable transduction for 15 weeks in progeny cells from nondividing CD34+, CD38− cells. The gene transfer efficiency into extended long-term culture-initiating cells was approximately 10% to 30%.19 One recent study has reported efficient gene transfer of severe combined immunodeficiency (SCID) repopulating cells (SRC).18 The CD34+ cells were transduced with a green fluorescent protein (GFP) gene containing HIV vector and transplanted into nonobese diabetic (NOD)/SCID mice. Transgene expression was seen in peripheral blood of the mice up to 22 weeks after transplantation and 30% to 40% of the human hematopoietic colonies from the bone marrow cells contained the transgene. Despite these encouraging results, a recent study demonstrated that even though CD34+cells in G0 can be transduced with HIV vectors, CD34+ cells in G1 or S/G2/M are much more effectively transduced than when they are in G0.16
We wanted to explore further the use of HIV-based lentiviral vectors to transduce human hematopoietic cells by asking the following questions: (1) Does lentiviral transduction of HSC lead to sustained multilineage expression of the transgene, not only in primary, but also in secondary NOD/SCID recipients after serial transplantation? (2) Is the transduction efficiency and expression profile in different lineages similar in primary and secondary NOD/SCID recipients? (3) Does lentiviral transduction lead to integration of the transgene in the genome of primitive hematopoietic progenitor and candidate stem cells? To answer these questions we used a replication-defective, second-generation HIV-1–based vector with GFP as reporter gene and pseudotyped with VSV-G to assess the lentiviral transduction efficiency into cells that can sustain long-term lymphomyelopoiesis by serial transplantation into NOD/SCID mice.21-23 Transplantation of human hematopoietic cells into sublethally irradiated NOD/SCID mice provides the best assay system for candidate human HSC currently available. Indications suggest that the engrafting cells, defined as SRC, are biologically distinct from and more primitive than the hematopoietic cells that can be assayed in vitro.24-26 Our studies suggest efficient transduction of primary and secondary SRC. Correspondingly, the use of an “inverted” polymerase chain reaction (PCR) technique confirmed the integration of the vector provirus in the chromosomal DNA of hematopoietic progenitor-derived colonies, including those derived from primary and secondary transplanted NOD/SCID mice.
Materials and methods
Lentiviral vectors
The 3-plasmid expression system used to generate lentiviral vectors by transient transfection was used as previously described.8 The 3 plasmids were the packaging plasmid pCMVΔR8.91 designed to provide the Gag, Pol, Tat, and Rev proteins to produce the virus particle, the envelope-coding plasmid pMD.G. for pseudotyping the virion with VSV-G, and the transfer vector DNA.12 The transfer vector plasmid, pHR′ PGK GFP, contains the enhanced GFP marker gene driven by the phosphoglycerate kinase promoter (PGK) and has been described before.8 12
Preparation of high-titer vector stocks
Stocks of the lentiviral vector were generated by transient transfection of the 3-plasmid system into 293T cells as previously described8 and stored at −80°C. Where large amounts of virus were required (as in the NOD/SCID transplantation experiments), the transfection protocol was scaled upward to make use of 15-cm dishes or the 6000-cm2 Nunc cell factory (Nalge Nunc, Rochester, NY). Vector supernatants consisting of virion particles and Dulbecco modified Eagle medium (DMEM) supplemented with 100 IU/mL penicillin, 100 μg/mL streptomycin (Gibco BRL, Cleveland, OH), and 10% fetal bovine serum (FBS) (Gibco BRL) were harvested as previously described.27 Titration of concentrated supernatants was performed by 5 serial dilutions 5 × 1:2 of 1 μL of concentrated virus on 100 000 HeLa cells incubated 96 hours followed by fluorescence-activated cytometric analysis for GFP using the FL1 channel on the fluorescence-activated cell sorting (FACS) Calibur instrument (Becton Dickinson Immunocytometry Systems [BDIS], San Jose, CA). Titers achieved often exceeded 108 TU/mL. The concentration was approximately 200-fold with 70% recovery of transducing units. As a control for retroviral transduction into primitive hematopoietic cells, an MGIN retrovirus pseudotyped with the VSV-G protein, VSV-G/MGIN, was used. To generate this vector, the MGIN vector plasmid28 (obtained from Robert Hawley, Toronto Hospital, Toronto, ON, Canada) was transfected into the GFP+ env AM12 packaging cell line. Supernatants from this cell line were used to transduce 293GPG cells with tetracycline-inducible VSV-G expression29 (obtained from R. Mulligan, Children's Hospital, Boston, MA) to obtain VSV-G pseudotyped MGIN vector (concentrated as described above).
Preparation of CD34+ cells
Umbilical cord blood samples were collected at the Department of Obstetrics and Gynecology, Malmö General Hospital, Malmö, Sweden, from consensual mothers having normal births. Samples were stored for less than 24 hours at 4°C in flasks containing DMEM (Gibco BRL), 0.1% detoxified bovine serum albumin (BSA) (Stem Cell Technologies, Vancouver, BC, Canada), the anticoagulant heparin at 150 IU/mL (Pharmacia AB, Stockholm, Sweden), 100 IU/mL penicillin, and 100 μg/mL streptomycin (Gibco BRL) before mononuclear cell isolation using Lymphoprep density gradient (Nycomed, Oslo, Norway) according to the manufacturer's instructions. CD34+ cell enrichment was performed using Midi MACS LS+ separation columns and cell isolation kit (Miltenyi Biotec, Auburn, CA) according to the manufacturer's instructions. CD34+ cells for transduction experiments were washed and resuspended in X-Vivo 15 medium (Bio Whittaker) supplemented with 1% BSA (Stem Cell Technologies), 2 mmol/L l-glutamine (Gibco BRL), 10−3 mmol/L 2-mercaptoethanol (Sigma Chemical AB, Stockholm, Sweden), 100 IU/mL penicillin, and 100 μg/mL streptomycin (Gibco BRL). From the initial collection of hematopoietic cells to CD34+ cell isolation, serum-free conditions were maintained.
Transduction of CD34+ cells
Lentiviral transduction of CD34+ cells was performed overnight under serum-free conditions in X-Vivo 15 medium (Bio Whittaker) supplemented with the cytokine megakaryocyte growth and development factor (MGDF) at 50 ng/mL (a gift from Amgen, Thousand Oaks, CA). X-Vivo 15 medium here and hereafter mentioned is supplemented with 1% BSA (Stem Cell Technologies), 2 mmol/Ll-glutamine (Gibco BRL), 10−3 mmol/L 2-mercaptoethanol (Sigma Chemical AB), 100 IU/mL penicillin, and 100 μg/mL streptomycin (Gibco BRL). Transduction of up to 400 000 CD34+ cells per well (1.0 mL) was performed on a RetroNectin CH296 (10 μg/cm2) (Takara Shuzo, Otsu, Japan) coated 24-well plate with a multiplicity of infection (MOI = 20-100).30 For FACS analysis, cells were incubated for a further 96 hours in X-Vivo 15 medium supplemented with cytokines MGDF at 50 ng/mL and stem cell factor (SCF) at 100 ng/mL (both cytokines were a gift from Amgen, Inc) before flow cytometric analysis for GFP expression. Colony assays were performed in methylcellulose medium H4230 (Stem Cell Technologies) supplemented with the cytokines SCF, granulocyte colony-stimulating factor (G-CSF; a gift from Amgen), granulocyte-macrophage colony-stimulating factor (GM-CSF; a gift from Novartis, East Hanover, NJ), and interleukin (IL)-3 (also a gift from Novartis) with final concentrations of 10 ng/mL each. Colonies were picked into 25 μL of lysis buffer (105 mmol/L KCl, 14 mmol/L Tris HCl2, 2.5 mmol/L MgCl2 [aq], 0.3 mg/mL gelatin, 0.45% NP 40 (IGEPAL), 0.45% Tween 20, 60 μg/mL proteinase K), vortexing and incubating for 1 hour at 56°C followed by 96°C for 15 minutes. PCR was performed on the lysed cells to detect the viral genome using a GFP/lentiviral long terminal repeat (LTR) primer pair (GFP 56F: 5′-GAG CTG GAC GGC GAC GTA AAC G and LTR 86R: 5′-ATC CCT GGC CCT GGT GTG TAG TTC), with a primer annealing temperature of 60°C for 1 minute, an Mg++concentration of 1.5 mmol/L (Gibco BRL) and 30 cycles run (Peltier Thermal Cycler 200, MJ Research Inc, Watertown, MA). Control transduction experiments using the VSV-G/MGIN oncoretroviral vector on CD34+ cells were performed using our aforementioned lentiviral transduction protocol. A positive control transduction with the VSV-G/MGIN was done by stimulating the CD34+ cells for 48 to 96 hours in 50 ng/mL MGDF, 100 ng/mL SCF, and 50 ng/mL flt3-ligand (FL) (a gift from Immunex, Seattle, WA) prior to transduction to test the transduction competency of the purified VSV-G/MGIN vector. The determination of the MGIN transduction efficiency into hematopoietic cells was performed as described above using the GFP primers, GFP56F: 5′-GAG CTG GAC GGC GAC GTA AAC G, and GFP629R: 5′-CGC TTC TCG TTG GGG TCT TTG CT and 32 cycles.
Cell cycle analysis
To determine cell cycle status of CD34+ cells before and after the transduction period a combination of staining for DNA with 7-aminoactinomycin D (7-AAD) and nuclear antigen Ki67 was used as previously described.31 Actively proliferating HeLa cells were used as positive control.
NOD/SCID mice
The immunocompromised NOD/SCID mice were handled under sterile conditions and maintained in microisolators. Irradiated mice were treated with the antibiotic ciprofloxacin at 100 μg/mL in drinking water to prevent infectious death. Transplant recipients (aged 6-8 weeks) were treated with an irradiation dose of 350 cGy administered from a 137Cs source. Transplantation of 5000 to 400 000 transduced and mock-transduced human CD34+ cells followed within 24 hours in 0.5 mL volume of phosphate-buffered saline (PBS) supplemented with 1% BSA by tail vein injections. Mice were killed 5 to 6 weeks after transplantation and bone marrow from the femurs, tibiae, and fibulae was harvested. Each secondary transplant recipient received 12 to 50 million bone marrow mononuclear cells from each primary transplant recipient mouse. Secondary transplant recipients were killed and analyzed 5 to 6 weeks after transplantation as described above.
Flow cytometric analysis
Flow cytometric analysis was used for determination of human cell engraftment in transplanted NOD/SCID mice. Aliquots of bone marrow cells were stained with antihuman CD45 antibody (anti-Hle-1) conjugated to PerCP. Mouse IgG1 conjugated to PerCP-stained bone marrow was used as a control. Bone marrow cells were incubated with ChromPure mouse IgG whole molecule (Jackson ImmunoResearch Laboratories, Westgrove, PA) in PBS (Gibco BRL) supplemented with 1% FBS (Gibco BRL) prior to CD45 staining to prevent unspecific binding. Erythrocytes were lysed with BDIS lysis buffer (BDIS) or ammonium chloride (Stem Cell Technologies) prior to FACS analysis. Lineage analysis was performed by double staining using antihuman CD45 PerCP antibody with each of the following antibodies conjugated to allophycocyanine (APC); CD34 (anti-HPCA-2), CD38, CD33, CD19 (SJ25C1), CD14, (BDIS).
Southern blot analysis
The PCR-amplified DNA from hematopoietic colonies was run on a 1.5% agarose gel and transferred to Hybond N+ membrane (Amersham Pharmacia, Buckinghamshire, England) and hybridized with an oligonucleotide probe specific for the lentiviral LTR sequence (LTR 71R: 5′-GTA GCC TTG TGT GTG GTA GAT CCA), elongated with terminal deoxynucleotidyl transferase (Promega, Madison, WI), and labeled using the enhanced chemiluminescence (ECL) kit (Amersham Pharmacia Biotech) according to the manufacturer's instructions.
Inverse polymerase chain reaction
Single hematopoietic colonies were lysed in Eppendorf tubes in 200 μL of lysis buffer (100 mmol/L Tris-HCl pH 8.5, 0.5 mmol/L EDTA, 200 mmol/L NaCl, 0.2% sodium dodecyl sulfate [SDS], and 100 μg of proteinase K/mL) for 16 hours at 37°C. One volume of isopropanol was added to the cell lysate and incubated at −20°C for at least 2 hours. After precipitation and centrifugation of genomic DNA, the pellet was resuspended in 10 mmol/L Tris-Cl 1 mmol/L EDTA pH = 8 (TE) buffer. Digestion ensued with BamHI or a combination of BamHI and BglII and DNA was purified by Wizard DNA Clean-Up System according to the manufacturer's instructions (Promega). DNA was then self-ligated using T4 DNA ligase (New England Biolabs, Beverly, MA) at 16°C for 16 hours. The first round of amplification of 5 μL of the circularized DNA used the primers GFP245R 5′-TTG AAG AAG TCG TGC TGC TTC A) and LTR417F 5′-AGA TCC TGC ATA TAAG CAG CTG C) in eLONGase (Gibco BRL) reaction mixture with a final Mg++ concentration of 1.25 mmol/L. After preheating at 94°C for 30 seconds, the first cycle was 94°C denaturation for 30 seconds, 55°C annealing for 30 seconds, and 68°C extension for 1 minute. The subsequent 32 cycles were identical. Nested PCR was then performed on 2 μL of the amplified product with primers GFP195R: 5′-CTG AAG CAC TGC ACG CCG TAG G and LTR469F: 5′-GAC CAG ATC TGA GCC TGG GAG CTC by using the same reaction conditions and cycles. The resulting PCR products were electrophoresed on 1.5% agarose gel, transferred to Hybond N+ membrane (Amersham Pharmacia), and hybridized with an oligonucleotide probe (5′-TGG CTA ACT AGG GAA CCC ACT G) specific for LTR sequences, labeled with32P by 5′end labeling kit.
Results
Transduction of CD34+ cells by lentiviral vectors
The transduction conditions were designed to maintain the in vivo repopulating ability and viability of the HSC while minimally stimulating the cells into the S/G2/M phase of the cell cycle. Freshly isolated CD34+ bone marrow or cord blood cells were transduced overnight in serum-free conditions in the presence of MGDF. To determine cell cycle status before and after the transduction period, a combination of staining for DNA with 7-AAD and nuclear protein antigen Ki67 was used.31 In both freshly isolated cord blood CD34+ cells and cord blood CD34+ cells incubated 18 hours in the presence of MGDF, less than 2% of the cells reside in the S/G2/M phase (7-AAD high and Ki67 high) of the cell cycle (n = 2). Thus, there appears to be little or no transition of cells into the S phase following overnight incubation in MGDF. However, after treatment with MGDF, a lower fraction of cells is present in G0 (7-AAD low and Ki67 low) (21% versus 36%) and a higher fraction is in G1 (7-AAD low and Ki67 high) (62% versus 77%) (P = .0044, n = 2). Therefore, the transduction conditions used appear to promote some activation of the cells into G1, but they do not enter the S/G2/M phase of the cell cycle during transduction for 18 hours with MGDF.
The transduction efficiency with the lentiviral vector as scored by FACS was compared with the transduction efficiency of colony-forming cells (CFC). CD34+ cells were transduced, an aliquot grown for 4 days and subjected to FACS analysis, and the remainder of the cells plated in methylcellulose immediately following the overnight transduction. After 14 days, GFP+ colonies were scored by fluorescence microscopy and the presence of the GFP gene in individual randomly picked CFU-GM colonies was analyzed by PCR. When transduction was performed with an MOI of 50, 11% to 17% of the CFU-GM colonies were GFP+ by fluorescence microscopy. PCR analysis showed the presence of the GFP gene in 16% to 28% of randomly picked colonies (Table 1). Table 1, columns 5 and 6, show transduction efficiency as determined by FACS analysis and PCR on colonies before transplantation into NOD/SCID mice for experimental groups III and IV. In group III, FACS showed 20% of the cells GFP+ and 16% of the colonies were vector positive by PCR (Table 1). Similarly, 34% of the cells were positive by FACS in experimental group IV and 28% of the colonies were positive by PCR. There were no detectable GFP+ colonies when transduction was performed with the MGIN vector nor were any of these colonies PCR positive.
Transduction of SRC with lentiviral and oncoretroviral vectors
| Group . | Mouse . | Cell dose . | MOI . | TE FACS (%) . | 1° CFU-GM PCR (%) . | Primary recipients . | Secondary recipients . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CD45+ FACS (%) . | GFP+ FACS (%) . | 2° CFU-GM PCR (%) . | CD45+ FACS (%) . | GFP+ FACS (%) . | 3° CFU-GM PCR (%) . | ||||||
| Lentiviral vector | |||||||||||
| I | 1.1 | 100 000 | 100 | 37 | ND | 5 | 14 | ND | |||
| 1.2 | 8 | 9 | ND | ||||||||
| II | 2.1 | 100 000 | 70 | 33 | ND | 13 | 11 | ND | |||
| 2.2 | 3 | 13 | ND | ||||||||
| 2.3 | 6 | 7 | ND | ||||||||
| III | 3.1 | 400 000 | 50 | 20 | 16 | 35 | 6 | 62 (20/32) | 3 | 18 | 74 (23/31) |
| 3.2 | 37 | 10 | 80 (24/30) | 2 | 33 | 44 (7/16) | |||||
| 3.3 | 62 | 9 | 59 (19/32) | 6 | 21 | 59 (19/32) | |||||
| IV | 4.1 | 400 000 | 50 | 34 | 28 | 31 | 9 | 38 (10/26) | 4 | 27 | 71 (22/31) |
| 4.2 | 25 | 12 | 33 (7/21) | 4 | 42 | 78 (25/32) | |||||
| 4.3 | 30 | 13 | ND | Dead | |||||||
| 4.4 | 2 | 15 | 16 (4/25) | < 1 | NA | 56 (9/16) | |||||
| MGIN vector | |||||||||||
| I | 1.3 | 100 000 | 60 | < 1 | ND | 5 | < 1 | ND | |||
| 1.4 | 70 | < 1 | ND | ||||||||
| IV | 4.5 | 100 000 | 20 | < 1 | 0 | 16 | < 1 | 0 | |||
| 4.6 | 15 | < 1 | 0 | ||||||||
| 4.7 | 11 | < 1 | 0 | ||||||||
| 4.8 | 29 | < 1 | 0 | ||||||||
| Group . | Mouse . | Cell dose . | MOI . | TE FACS (%) . | 1° CFU-GM PCR (%) . | Primary recipients . | Secondary recipients . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CD45+ FACS (%) . | GFP+ FACS (%) . | 2° CFU-GM PCR (%) . | CD45+ FACS (%) . | GFP+ FACS (%) . | 3° CFU-GM PCR (%) . | ||||||
| Lentiviral vector | |||||||||||
| I | 1.1 | 100 000 | 100 | 37 | ND | 5 | 14 | ND | |||
| 1.2 | 8 | 9 | ND | ||||||||
| II | 2.1 | 100 000 | 70 | 33 | ND | 13 | 11 | ND | |||
| 2.2 | 3 | 13 | ND | ||||||||
| 2.3 | 6 | 7 | ND | ||||||||
| III | 3.1 | 400 000 | 50 | 20 | 16 | 35 | 6 | 62 (20/32) | 3 | 18 | 74 (23/31) |
| 3.2 | 37 | 10 | 80 (24/30) | 2 | 33 | 44 (7/16) | |||||
| 3.3 | 62 | 9 | 59 (19/32) | 6 | 21 | 59 (19/32) | |||||
| IV | 4.1 | 400 000 | 50 | 34 | 28 | 31 | 9 | 38 (10/26) | 4 | 27 | 71 (22/31) |
| 4.2 | 25 | 12 | 33 (7/21) | 4 | 42 | 78 (25/32) | |||||
| 4.3 | 30 | 13 | ND | Dead | |||||||
| 4.4 | 2 | 15 | 16 (4/25) | < 1 | NA | 56 (9/16) | |||||
| MGIN vector | |||||||||||
| I | 1.3 | 100 000 | 60 | < 1 | ND | 5 | < 1 | ND | |||
| 1.4 | 70 | < 1 | ND | ||||||||
| IV | 4.5 | 100 000 | 20 | < 1 | 0 | 16 | < 1 | 0 | |||
| 4.6 | 15 | < 1 | 0 | ||||||||
| 4.7 | 11 | < 1 | 0 | ||||||||
| 4.8 | 29 | < 1 | 0 | ||||||||
Lentiviral but not MGIN-based retroviral vectors can transduce human CD34+ cells that can repopulate primary and secondary NOD/SCID transplant recipients. The results from 4 different experiments are shown. TE indicates transduction efficiency assessed by FACS 4 days after transduction. 1° CFU-GM are colonies derived from CD34+ cells harvested directly following overnight transduction. 2° CFU-GM are colonies derived from the BM of primary recipients and 3° CFU-GM are colonies derived from the bone marrow of secondary recipients. The presence of the GFP gene was confirmed by PCR analysis of 32 individual CFU-GM colonies from 1° CFU-GM and 32 individual colonies from each recipient mouse. The large discrepancy between the percentages of vector-positive CFU-GM and GFP expressing CD45+ cells as determined by FACS may be attributed in part to the use of Becton Dickinson lysis buffer, which results in loss of GFP expression during the lysis procedure. This can be avoided by the use of ammonium chloride to lyse the red cells.
ND indicates not determined; NA, not available.
Engraftment of transduced CD34+ cells in primary NOD/SCID recipients
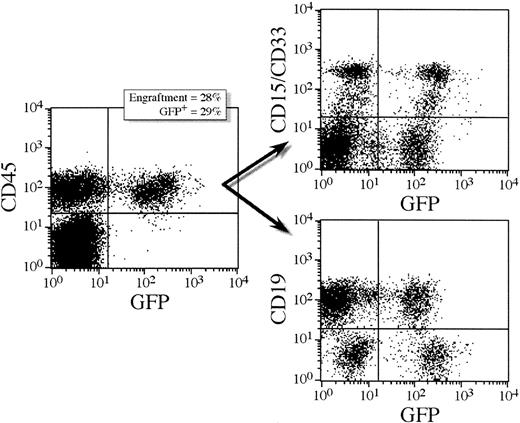
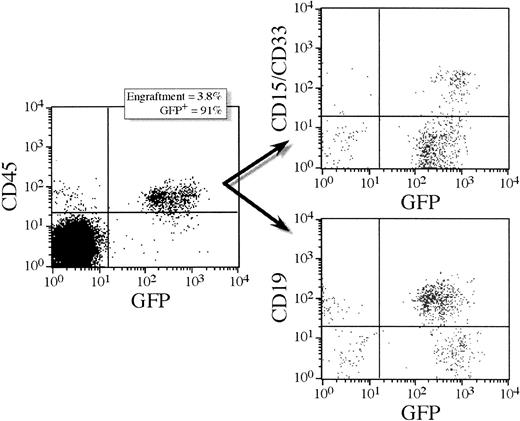
To assess the transduction efficiency of candidate human stem cells in the CD34+ population, CD34+ cells transduced overnight in the presence of MGDF were transplanted into sublethally irradiated NOD/SCID mice. After 5 to 6 weeks, the mice were killed and the bone marrow analyzed by FACS for engraftment of human cells by staining with a monoclonal antibody directed against human CD45.25,32 Figure 1shows a representative FACS staining profile for a mouse that received a transplantation dose of 400 000 cells (mouse 5.3). Here 28% of the cells found in the bone marrow were of human origin of which 29% expressed GFP. Further staining with CD19, CD33, and CD15 confirms the presence of both lymphoid and myeloid cells. Table 1 shows engraftment levels and GFP expression in 18 mice receiving transplants with either 100 000 or 400 000 lentivirally transduced CD34+ cells. The level of engraftment reflected the number of cells transplanted. In mice receiving a transplantation dose of 100 000 cells, the number of CD45+ cells ranged from 3% to 13%. At the higher cell dose, the number of CD45+ cells ranged from 25% to 62% except in one animal, which engrafted at only 2%. GFP expression was assessed by FACS analysis of the CD45+ cell population and 6% to 15% of the CD45+ cells were found to express GFP (Table 1). Multilineage hematopoiesis was obtained in all the animals. GFP+ cells were found in all lineages tested and at comparable frequencies (Table 2). GFP+ CD34+ cells were detected in all animals suggesting that transduced progenitor cells were maintained in the bone marrow of NOD/SCID mice. The results from 4 separate NOD/SCID experiments using the lentiviral vectors are summarized in Table 1. The percentages of GFP+ cells in clonogenic myeloid progenitors was assessed in methylcellulose cultures that only support outgrowth of human progenitors.26 When scoring by fluorescence microscopy, 23% to 48% of the colonies were GFP+. By PCR analysis of randomly picked individual colonies the GFP gene was detected in 46% ± 23% (range, 16%-80%) of the CFU-GM colonies (Table 1). Thus, the percentage of GFP+ colonies, as analyzed by PCR, increased compared with the primary CFU-GM colonies, that is, colonies obtained from CD34+ cells cultured directly after the initial transduction period (Table 1). In the mice transplanted with CD34+ cells that were transduced overnight with VSV-G/MGIN, engraftment ranged from 5% to 70% but there were no detectable GFP+ cells by FACS or PCR of colonies (Table 1).
Multilineage reconstitution and GFP expression in human CD45+ cells from a representative NOD/SCID transplant recipient.
This mouse (5.3) received a cell dose of 400 000 lentivirally transduced CD34+ cells. Transduction was performed overnight in serum-free medium supplemented with MGDF, MOI = 100. Lymphoid and myeloid differentiation of human SRC in NOD/SCID mice is shown by CD19 and CD33/15 antibody staining, respectively (71% of human cells were CD19+ and 28% were CD15/33+). Both lymphoid and myeloid populations expressed GFP; 23% of the CD19+ cells expressed GFP and 41% of the CD15/33+ cells expressed GFP.
Multilineage reconstitution and GFP expression in human CD45+ cells from a representative NOD/SCID transplant recipient.
This mouse (5.3) received a cell dose of 400 000 lentivirally transduced CD34+ cells. Transduction was performed overnight in serum-free medium supplemented with MGDF, MOI = 100. Lymphoid and myeloid differentiation of human SRC in NOD/SCID mice is shown by CD19 and CD33/15 antibody staining, respectively (71% of human cells were CD19+ and 28% were CD15/33+). Both lymphoid and myeloid populations expressed GFP; 23% of the CD19+ cells expressed GFP and 41% of the CD15/33+ cells expressed GFP.
Lineage analysis of and GFP expression in bone marrow cells from primary and secondary recipient NOD/SCID mice
| . | Engraftment in recipients (%) . | GFP expression in recipients (%) . | ||
|---|---|---|---|---|
| Primary . | Secondary . | Primary . | Secondary . | |
| CD34 | 26 ± 2 | 10 ± 5 | 9 ± 2 | 27 ± 9 |
| CD38 | 81 ± 17 | 67 ± 10 | 8 ± 2 | 22 ± 8 |
| CD33 | 4 ± 2 | 14 ± 4 | 14 ± 3 | 39 ± 5 |
| CD19 | 32 ± 6 | 40 ± 8 | 9 ± 1 | 19 ± 3 |
| CD14 | 3 ± 1 | 15 ± 2 | 14 ± 6 | 23 ± 5 |
| . | Engraftment in recipients (%) . | GFP expression in recipients (%) . | ||
|---|---|---|---|---|
| Primary . | Secondary . | Primary . | Secondary . | |
| CD34 | 26 ± 2 | 10 ± 5 | 9 ± 2 | 27 ± 9 |
| CD38 | 81 ± 17 | 67 ± 10 | 8 ± 2 | 22 ± 8 |
| CD33 | 4 ± 2 | 14 ± 4 | 14 ± 3 | 39 ± 5 |
| CD19 | 32 ± 6 | 40 ± 8 | 9 ± 1 | 19 ± 3 |
| CD14 | 3 ± 1 | 15 ± 2 | 14 ± 6 | 23 ± 5 |
Multilineage engraftment of human cells and expression of GFP in the bone marrow of primary and secondary NOD/SCID recipient mice transplanted with lentivirally transduced cord blood CD34+cells. Bone marrow from 6 mice from groups III and IV were stained with the following human specific monoclonal antibodies: CD34, CD38, CD33, CD19, and CD14 and analyzed by flow cytometry. For all antibodies the negative control used was IgG1. Double staining with CD45 was done with each lineage marker and the fraction of GFP-expressing cells in the varying lineages was assessed. The results are presented as mean ± SD.
Engraftment of secondary NOD/SCID mice with cells from primary recipients
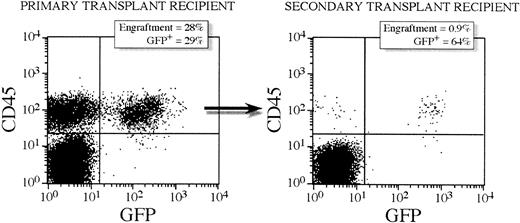
Serial transplantation was performed to assess the transduction efficiency into cells that can sustain long-term lymphomyelopoiesis. Bone marrow mononuclear cells from 7 primary recipients were transplanted into 7 secondary sublethally irradiated animals. No growth factors were administered to any of the primary or secondary recipients. The 6 mice (1 mouse died prematurely) were killed at 6 weeks following transplantation and the level of engraftment determined by FACS analysis. As shown in Table 1, all 6 secondary transplant recipient mice engrafted with human cells in the range of 1% to 6%. FACS analysis of GFP expression in the CD45+ population showed that 26% ± 10% of the human cells in the secondary recipients were GFP+ whereas 10% ± 3% of CD45+ cells in the primary recipients expressed GFP. Multilineage engraftment was obtained in all secondary transplant recipients and the transgene expressed in all the following tested lineages CD19, CD33, CD14, CD38, and CD34 (Table 2). Figure2 shows FACS profiles for a representative secondary transplant recipient and primary bone marrow donor with 28% engraftment and 29% expressing GFP in primary donor mouse 5.3 and 0.9% engraftment and 64% expressing GFP in the corresponding secondary recipient 5.3.3. Clonogenic progenitor cells from the secondary transplant recipients were assessed in methylcellulose cultures and scored for presence of the GFPgene by PCR. A very high number of colonies was positive for the vector (64% ± 13%, range 56%-78%, n = 6 [Table 1]). Figure3 shows a representative Southern blot analysis for the PCR-amplified GFP gene from CFU-GM prior to transplantation, from a primary transplant recipient and from a secondary transplant recipient. Statistical comparisons of transduction efficiencies between CFU-GM prior to transplantation and from primary and secondary transplant recipients show that there is a statistical difference between vector-positive CFU-GM prior to transplantation and from primary transplant recipients (P = .0396). That is, the percent of vector-positive CFU-GM before transplantation was lower than that of CFU-GM after transplantation. There was, however, no significant difference between the percent vector-positive CFU-GM from primary and secondary transplant recipients (P = .27).
GFP gene expression in human CD45+ cells from a representative primary and corresponding secondary transplant recipient mouse.
The primary recipient (mouse 5.3) received a transplantation dose of 400 000 CD34+ lentivirally transduced cells. Six weeks after transplantation, bone marrow was harvested and 12 million mononuclear cells were transplanted into a secondary transplant recipient (mouse 5.3.3). A decrease in engraftment from primary to secondary recipients is seen but a high proportion of GFP+ cells are detected in this secondary recipient by FACS.
GFP gene expression in human CD45+ cells from a representative primary and corresponding secondary transplant recipient mouse.
The primary recipient (mouse 5.3) received a transplantation dose of 400 000 CD34+ lentivirally transduced cells. Six weeks after transplantation, bone marrow was harvested and 12 million mononuclear cells were transplanted into a secondary transplant recipient (mouse 5.3.3). A decrease in engraftment from primary to secondary recipients is seen but a high proportion of GFP+ cells are detected in this secondary recipient by FACS.
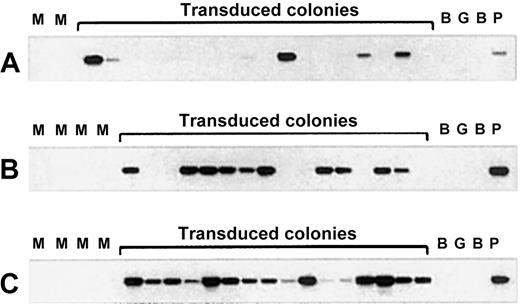
Transduction efficiency of CFU-GM colonies as detected by PCR analysis of individual colonies derived from cord blood CD34+ cells transduced with the PGK-GFP vector.
(A) PCR analysis of randomly picked individual CFU-GM colonies derived from CD34+ cells directly after overnight transduction. (B) PCR analysis of CFU-GM colonies from the bone marrow of primary NOD/SCID mice engrafted with PGK-GFP transduced CD34+cells. (C) PCR analysis of CFU-GM colonies derived from secondary transplant NOD/SCID mice transduced with the same vector. M indicates mock transduced; B, blank; G, human genomic DNA; P, positive control.
Transduction efficiency of CFU-GM colonies as detected by PCR analysis of individual colonies derived from cord blood CD34+ cells transduced with the PGK-GFP vector.
(A) PCR analysis of randomly picked individual CFU-GM colonies derived from CD34+ cells directly after overnight transduction. (B) PCR analysis of CFU-GM colonies from the bone marrow of primary NOD/SCID mice engrafted with PGK-GFP transduced CD34+cells. (C) PCR analysis of CFU-GM colonies derived from secondary transplant NOD/SCID mice transduced with the same vector. M indicates mock transduced; B, blank; G, human genomic DNA; P, positive control.
Cell-dose–limiting dilution assay in primary and secondary NOD/SCID recipients
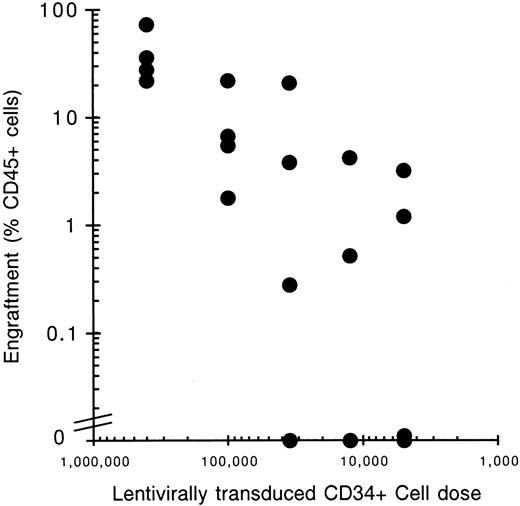
To address the issue of clonality in the reconstitution of transplanted NOD/SCID mice a cell-dose–limiting dilution experiment was performed. Mice were transplanted with reducing numbers (400 000-5000) of lentivirally transduced CD34+ cells (Table 3). Only the groups with 100 000 and 400 000 cells transplanted had all 4 mice positive for engraftment, defined as the presence of 5 lymphoid (CD19) and 5 myeloid (CD33/15) cells present per 20 000 mouse mononuclear cells and the generation of CFU-GM colonies in methylcellulose. Groups with lower cells doses had reduced numbers of positive engrafting mice with the group with only 5000 cells transplanted having no positive engrafting mice (Figure 4, Table 3). A linear relationship exists between the cell transplantation dose and engraftment level obtained by percent CD45+ cells. Poisson statistics suggest that positive-engrafting mice receiving low cell doses, where more than 63% of the mice are in fact negative for engraftment, are positive due to expansion of one cell clone. Groups of mice given a cell dose of 12 500 or 35 000 CD34+ cells had 50% (2 of 4) and 75% (3 of 4) of the mice within each group positive for engraftment (Table 3). This suggests that either one or a low number of SRC reconstituted the positive-engrafting mice in these groups. FACS analysis for GFP expression of these low cell-dose recipients reveals that in all mice the lentiviral vector is present (58.4% ± 28.3%, range 16%-91%). Interestingly, one mouse (7.23) had 91% GFP-expressing cells and all bone marrow-derived CFU-GM colonies visually green. Lineage analysis of this unique mouse showed lymphoid and myeloid cells both expressing GFP at 91% and 93%, respectively (Figure 5). This mouse may have been repopulated by a single transduced SRC whose myeloid and lymphoid progeny cells are expressing the transgene. CFU-GM colony plating of bone marrow from positive-engrafting mice and further analysis by fluorescence microscopy revealed a high degree of correlation between the percent of GFP expressing CD45+cells and the percent GFP+ CFU-GM as determined by microscopy (Table 3) (P = .893). To analyze clonal reconstitution in secondary mice, we performed cell–dose-limiting dilution transplantations of bone marrow from primary transplant recipients receiving 400 000 and 100 000 transduced CD34+cells into secondary transplant recipients. From one primary mouse receiving 400 000 cells, 3 secondary transplant recipients were obtained, and one primary mouse receiving 100 000 cells was transplanted into one secondary transplant recipient. In total 16 mice were transplanted of which only 2 were positive for engraftment (mouse 5.3.2 and mouse 5.3.3, Table 3); 4 mice died prematurely. These data indicate that one or a very few SRC are engrafting these secondary transplant recipients. FACS analysis of the 2 positive-engrafting mice showed high gene transfer efficiency in mouse 5.3.3 with 64% of the human cells expressing GFP, and low gene transfer efficiency in mouse 5.3.2 with 5% of human cells expressing the transgene.
Polyclonal and mono/oligoclonal reconstitution of NOD/SCID mice by lentivirally transduced SRC
| Limiting dilution . | Primary recipients . | Secondary recipients . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Group . | 1° mouse . | Cell dose . | CD45+FACS (%) . | GFP+ FACS (%) . | Multilineage engraftment . | 2° CFU-GM GFP (%) . | 2° mouse . | CD45+FACS (%) . | GFP+ FACS (%) . | Multilineage engraftment . | 3° CFU-GM GFP (%) . |
| V | 5.1 | 400 000 | 73 | 14 | yes | 10 | 5.1.1 | 0.05 | 0.5 | no | NA |
| 5.2 | 22 | 11 | yes | ND | 5.1.2 | 0.04 | 0 | no | NA | ||
| 5.3 | 28 | 29 | yes | 30 | 5.1.3 | dead | NA | ||||
| 5.4 | 36 | 14 | yes | ND | |||||||
| 5.2.1 | 0.05 | 10 | no | NA | |||||||
| VI | 6.5 | 100 000 | 1.8 | 1.6 | yes | ND | 5.2.2 | 0.02 | 0 | no | NA |
| 6.6 | 6.7 | 21 | yes | 0 | 5.2.3 | dead | NA | ||||
| 6.7 | 5.5 | 22 | yes | ND | |||||||
| 6.8 | 22 | 9.3 | yes | ND | 5.3.1 | 0.05 | 0 | no | NA | ||
| 5.3.2 | 0.5 | 5 | yes | 0 | |||||||
| VII | 7.21 | 35 000 | 0.28 | 52 | yes | 100 | 5.3.3 | 0.1 | 64 | yes | 74 |
| 7.22 | 21 | 76 | yes | 67 | |||||||
| 7.23 | 3.8 | 91 | yes | 100 | 5.4.1 | 0.04 | 0 | no | NA | ||
| 7.24 | 0.01 | 0 | no | NA | 5.4.2 | 0.03 | 0 | no | NA | ||
| 5.4.3 | dead | NA | |||||||||
| VIII | 8.25 | 12 500 | 4.2 | 16 | yes | 13 | |||||
| 8.26 | 0.52 | 57 | yes | 0 | 6.5.1 | 0.03 | 0 | no | NA | ||
| 8.27 | 0.01 | 0 | no | NA | 6.6.1 | 0.2 | 22 | no | NA | ||
| 8.28 | 0 | 0 | no | NA | 6.7.1 | 0.05 | 0 | no | NA | ||
| IX | 9.29 | 5000 | 1.2 | 0 | no | NA | 6.8.1 | 0.2 | 0 | no | NA |
| 9.30 | 3.2 | 0 | no | NA | |||||||
| 9.31 | 0.11 | 0 | no | NA | |||||||
| 9.32 | 0.01 | 0 | no | NA | |||||||
| Limiting dilution . | Primary recipients . | Secondary recipients . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Group . | 1° mouse . | Cell dose . | CD45+FACS (%) . | GFP+ FACS (%) . | Multilineage engraftment . | 2° CFU-GM GFP (%) . | 2° mouse . | CD45+FACS (%) . | GFP+ FACS (%) . | Multilineage engraftment . | 3° CFU-GM GFP (%) . |
| V | 5.1 | 400 000 | 73 | 14 | yes | 10 | 5.1.1 | 0.05 | 0.5 | no | NA |
| 5.2 | 22 | 11 | yes | ND | 5.1.2 | 0.04 | 0 | no | NA | ||
| 5.3 | 28 | 29 | yes | 30 | 5.1.3 | dead | NA | ||||
| 5.4 | 36 | 14 | yes | ND | |||||||
| 5.2.1 | 0.05 | 10 | no | NA | |||||||
| VI | 6.5 | 100 000 | 1.8 | 1.6 | yes | ND | 5.2.2 | 0.02 | 0 | no | NA |
| 6.6 | 6.7 | 21 | yes | 0 | 5.2.3 | dead | NA | ||||
| 6.7 | 5.5 | 22 | yes | ND | |||||||
| 6.8 | 22 | 9.3 | yes | ND | 5.3.1 | 0.05 | 0 | no | NA | ||
| 5.3.2 | 0.5 | 5 | yes | 0 | |||||||
| VII | 7.21 | 35 000 | 0.28 | 52 | yes | 100 | 5.3.3 | 0.1 | 64 | yes | 74 |
| 7.22 | 21 | 76 | yes | 67 | |||||||
| 7.23 | 3.8 | 91 | yes | 100 | 5.4.1 | 0.04 | 0 | no | NA | ||
| 7.24 | 0.01 | 0 | no | NA | 5.4.2 | 0.03 | 0 | no | NA | ||
| 5.4.3 | dead | NA | |||||||||
| VIII | 8.25 | 12 500 | 4.2 | 16 | yes | 13 | |||||
| 8.26 | 0.52 | 57 | yes | 0 | 6.5.1 | 0.03 | 0 | no | NA | ||
| 8.27 | 0.01 | 0 | no | NA | 6.6.1 | 0.2 | 22 | no | NA | ||
| 8.28 | 0 | 0 | no | NA | 6.7.1 | 0.05 | 0 | no | NA | ||
| IX | 9.29 | 5000 | 1.2 | 0 | no | NA | 6.8.1 | 0.2 | 0 | no | NA |
| 9.30 | 3.2 | 0 | no | NA | |||||||
| 9.31 | 0.11 | 0 | no | NA | |||||||
| 9.32 | 0.01 | 0 | no | NA | |||||||
NA indicates not applicable due to absence of human CFU; ND, not done.
A cell-dose–limiting dilution of transduced CD34+ cells (MOI = 200) transplanted into NOD/SCID recipients shows successful lentiviral vector gene transfer into both primary and secondary SRC. Bone marrow from high-cell-dose primary transplant recipients (groups V and VI) were transplanted into secondary recipients with one donor for one recipient mouse for group VI donors and one donor for three recipient mice for group V donors. FACS analysis for engraftment (CD45) and GFP expression was performed along with colony analysis for GFP expression by microscopy. Multilineage engraftment was seen as defined by the presence of CFU-GM and a minimum of 5 human lymphoid cells (CD19) and 5 human myeloid cells (CD15 or CD33) per 20 000 mouse bone marrow mononuclear cells.
Relationship between cell dose and engraftment.
The degree of cell engraftment in NOD/SCID mice is proportional to the number of transplanted human CD34+ cells. A cell–dose-limiting dilution experiment into NOD/SCID mice shows a linear relationship between cell dose and engraftment levels. Lentivirally transduced CD34+ cells were transplanted into NOD/SCID recipients and the bone marrow mononuclear cells analyzed 6 weeks later by FACS using antibodies directed against human CD45 to detect human cells. Four mice underwent transplantation with the following cell doses and the percentage of CD45+ cell plotted against cell dose: 400 000, 100 000, 35 000, 12 500, and 5000.
Relationship between cell dose and engraftment.
The degree of cell engraftment in NOD/SCID mice is proportional to the number of transplanted human CD34+ cells. A cell–dose-limiting dilution experiment into NOD/SCID mice shows a linear relationship between cell dose and engraftment levels. Lentivirally transduced CD34+ cells were transplanted into NOD/SCID recipients and the bone marrow mononuclear cells analyzed 6 weeks later by FACS using antibodies directed against human CD45 to detect human cells. Four mice underwent transplantation with the following cell doses and the percentage of CD45+ cell plotted against cell dose: 400 000, 100 000, 35 000, 12 500, and 5000.
Lineage analysis of a transplant recipient with practically all human cells transduced.
A unique mouse receiving a low cell transplantation dose showing high-level transduction efficiency as determined by FACS for GFP. This mouse (7.23) received 35 000 lentivirally transduced CD34+cells. Six weeks after transplantation, bone marrow was harvested and analyzed for percent GFP+ human CD45+ cells. High-level transduction efficiency had been achieved with 91% of human cells expressing GFP. Lineage analysis for lymphoid (CD19) and myeloid (CD15/33) cells showed similar high-level transduction efficiencies in both lineages (ie, 91% expressing GFP in the lymphoid lineage and 93% in the myeloid lineage). All the human CFU-GM colonies derived from this mouse expressed the transgene.
Lineage analysis of a transplant recipient with practically all human cells transduced.
A unique mouse receiving a low cell transplantation dose showing high-level transduction efficiency as determined by FACS for GFP. This mouse (7.23) received 35 000 lentivirally transduced CD34+cells. Six weeks after transplantation, bone marrow was harvested and analyzed for percent GFP+ human CD45+ cells. High-level transduction efficiency had been achieved with 91% of human cells expressing GFP. Lineage analysis for lymphoid (CD19) and myeloid (CD15/33) cells showed similar high-level transduction efficiencies in both lineages (ie, 91% expressing GFP in the lymphoid lineage and 93% in the myeloid lineage). All the human CFU-GM colonies derived from this mouse expressed the transgene.
Lentiviral transgene integration in colony-forming unit granulocyte-macrophage colonies
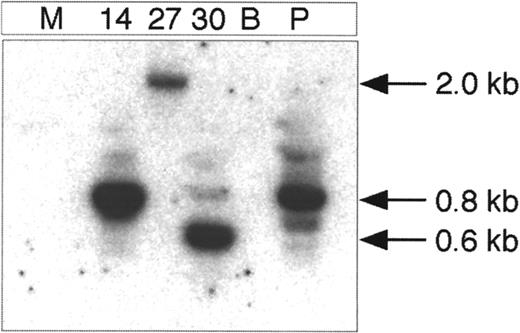
Because the hematopoietic colonies derived from lentivirally transduced cells contain too few cells to generate adequate amounts of DNA for Southern blot analysis to show proviral integration, we developed an “inverted” PCR assay to detect integrated vector copies in individual colonies. Similar assays have been used to detect integrated retroviral vector copies in hematopoietic colonies.33 The restriction enzymes BamHI or a combination of BamHI and BglII were used to cut the DNA, which is subsequently self-ligated to amplify the fragment from the circular ligated DNA with primers that go in opposite directions in the linear DNA. The resulting junctional vector-genomic fragment is unique for each integration site. Figure6 demonstrates vector integration into visually green CFU-GM colonies derived from the 3 primary recipient NOD/SCID mice in experiment III. Vector genomic junctional fragments from a few colonies have been sequenced and demonstrate the sequence of the HIV-1 LTR in tandem with sequences from human genomic DNA.
Integration analysis of the provirus in lentivirally transduced bone marrow cells of primary NOD/SCID recipient mice.
DNA from lentivirally transduced individual visually green CFU-GM colonies from 3 different mice was digested with BamHI, self-ligated at a low DNA concentration, and used as template in the PCR reaction. GFP and LTR primer sets (GFP 195R and LTR 469) were amplified. The colonies are numbered 14, 27, and 30. The figure shows the membrane of the inverse PCR reaction hybridized with the LTR specific oligo probe. The minimal size of the inverse PCR product is approximately 450 bp. M indicates mock-transduced control colony; B, blank; and P, positive control. The P colony is derived from lentivirally transduced ES cells that are known to contain integrated PGK-GFP vector as determined by Southern blot analysis.33
Integration analysis of the provirus in lentivirally transduced bone marrow cells of primary NOD/SCID recipient mice.
DNA from lentivirally transduced individual visually green CFU-GM colonies from 3 different mice was digested with BamHI, self-ligated at a low DNA concentration, and used as template in the PCR reaction. GFP and LTR primer sets (GFP 195R and LTR 469) were amplified. The colonies are numbered 14, 27, and 30. The figure shows the membrane of the inverse PCR reaction hybridized with the LTR specific oligo probe. The minimal size of the inverse PCR product is approximately 450 bp. M indicates mock-transduced control colony; B, blank; and P, positive control. The P colony is derived from lentivirally transduced ES cells that are known to contain integrated PGK-GFP vector as determined by Southern blot analysis.33
Discussion
In this study we have transduced very primitive human hematopoietic progenitors or candidate stem cells that are capable of repopulating NOD/SCID mice after serial transplantation (secondary transplant SRC). Interestingly, it looks like the gene transfer efficiency into these SCID repopulating cells is higher than the transfer efficiency into clonogenic progenitors. In 2 experiments (experiments III and IV), CFU-GM progenitors were transduced with an efficiency of 22% ± 8%, whereas the efficiency into CFU-GM colonies derived from primary NOD/SCID mice was 48% ± 23% and 64% ± 13% into clonogenic progenitors derived from secondary transplant mice. There was no significant difference between the gene transfer efficiency into CFU-GM colonies from primary and secondary mice and the results from these were therefore pooled (56% ± 19%) and defined as the average gene transfer efficiency into SRC. The difference between gene transfer efficiency into CFU-GM colonies (22% ± 8%) and SRC was found to be statistically significant (P < .02, Student t test). These data agree in principle with the report by Miyoshi and coworkers18 who found gene transfer into clonogenic progenitors to be 12% to 17%, whereas the progenitor colonies from transplanted NOD/SCID mice (primary transplants) were 28% to 38% vector positive. A recent report has presented data indicating that dividing hematopoietic cells are more easily transduced than quiescent ones.16 Because clonogenic progenitors tend to have a higher proportion of cells in active cell cycle than more primitive HSC, our data and the results of Miyoshi and colleagues18 may seem surprising. However, the transduction conditions used in both studies were short (5-18 hours) and the cells were not stimulated to divide with a cocktail of growth factors. Therefore, most of the clonogenic progenitors would be expected to be quiescent under these conditions. It is also interesting to see that the percentage of GFP-positive cells in the NOD/SCID mice is lower than the percentage of vector-positive colonies in both primary and secondary recipients. Similar results were also reported by Miyoshi and colleagues18 for their primary recipients. A part of this difference could be explained by low-level expression in a portion of the differentiated hematopoietic cells, but it is also possible that a higher proportion of progenitors in the bone marrow are vector positive than the proportion of vector-positive hematopoietic cells of all types for reasons that are poorly understood. The latter phenomenon has been reported in clinical studies where the gene-marking efficiency of bone marrow progenitors has been higher than the marking of differentiated cells in the peripheral blood.34
The immunophenotype of the engrafted cells was analyzed by FACS. The results showed that all animals analyzed in this way had both lymphoid and myeloid human cells. Cells of the B-cell lineage (CD19+) were the most abundant and a surprisingly high proportion of CD34+ cells was detected. Many cells with a myeloid cell surface phenotype were also detected. GFP+cells were seen in all these fractions detected with antibodies toward antigens characterizing different lineages or stages of maturation. CD34+, CD19+, CD38+, CD33+, and CD14+ cells were all found to be positive and the proportion of positive cells was rather similar in all these fractions, even in the secondary NOD/SCID mice after serial transplantation. These results suggest that we are transducing secondary SRC that can serially engraft in NOD/SCID mice and differentiate into both myeloid and lymphoid progeny cells in the secondary recipient.
To further support the notion that we are transducing very primitive SRC that can engraft NOD/SCID mice and develop into both lymphoid and myeloid lineages, we performed limiting dilution experiments where reducing numbers of transduced CD34+ cells were transplanted into primary recipients. The results clearly show that there is a direct relationship between the number of recipients that engraft human cells and the dose of cells transplanted. Furthermore, there is also a relationship between the number of cells transplanted and the number of animals expressing the GFP transgene. In one recipient (mouse 7.23), which received only 35 000 cells, the engraftment was relatively low (3.8%) as expected but 91% of the human cells were expressing the transgene. All the CFU-GM colonies from this animal were visually green and almost all myeloid and lymphoid cells expressed the GFP as determined by FACS. The most likely interpretation of these results is that one transduced SRC has generated the myeloid and lymphoid progeny that is expressing the transgene. It is possible that 2 or more transduced SRC that have all been transduced are responsible for repopulating this recipient with human hematopoietic cells, but this possibility is less likely. The secondary transplantation experiments in Table 3 show also that only a fraction of the secondary recipient animals are expressing the transgene. Collectively these data indicate that very primitive repopulating cells that are able to differentiate into both myeloid and lymphoid progeny cells have been successfully transduced.
It is important to know whether the lentiviral vector has integrated into chromosome(s) of the primitive hematopoietic candidate stem cells, which are the key target cells in our study. Lentiviral transduction using vectors with the GFP gene has been measured by FACS analysis or by PCR analysis where internal primers amplify the transgene alone in DNA from hematopoietic colonies. This prompted us to determine whether the primary CFU-GM colonies and the secondary and tertiary ones from the primary and secondary recipients contained integrated vector copies. We used ligation-mediated PCR to detect vector-chromosomal junctional fragments in colonies that were visually green. We have tested a significant number of lentivirally transduced embryonic stem cell colonies by this method and have compared results from this assay and from Southern blot analysis of vector integration sites using BamHI to cut the DNA. The results show correlation between the 2 assays except in circumstances where the Southern blot indicates large BamHI fragments that are vector positive.33 Therefore, the method is much less sensitive than ordinary PCR used to detect the presence of theGFP gene. Vector-genomic DNA junctions have been sequenced in CFU-GM colonies that are derived from NOD/SCID mice and confirm integration of the vector proviral DNA in the progeny of SRC. Unfortunately, the assay is not sensitive enough to allow clonal analysis.
The expression level of the GFP gene generated by the PGK promoter in the transplanted recipients was in the range of 30 to 1000 fluorescent units in the primary mice with a mean fluorescent intensity of 230, but somewhat lower levels were seen in the secondary recipients on average. These results may indicate that some silencing of lentiviral transgene expression occurs on secondary transplantation as has been described for oncoretroviral vectors,35 but there are also secondary animals that express similar levels as their respective primary donors. The expression levels in primary and secondary recipients are high enough to be easily detected using vectors with the GFP gene, but the levels achieved may be inadequate for many therapeutic genes. Therefore, there is a need to optimize the design of lentiviral vectors to achieve higher expression levels in hematopoietic cells. Studies in cell lines have demonstrated that the woodchuck hepatitis virus posttranscriptional regulatory element increases expression levels in many human cell lines by a factor of 3 to 4.36 Similarly, the effects of deletions in the HIV LTR aimed at improving safety of vector design (self-inactivating [SIN] vectors) have to be studied with regard to expression levels to ask whether these deletions have a beneficial or a deleterious effect in hematopoietic cells.37 In the future, lentiviral vectors with various regulatory elements need to be tested in the NOD/SCID model to generate a more optimal vector design for expression of therapeutic genes in human gene therapy protocols.
Acknowledgments
We are indebted to Lilian Wittman for expert technical assistance with animal experiments. We would also like to thank Kristina Sundgren and Eva Gynnstam for taking care of the animals, Sverken Segrén for assistance and advice with FACS analysis, Dr Johan Richter for help with the control oncoretroviral vectors, Dr Xiaolong Fan for advice on cell cycle analysis, members of the Stem Cell Laboratory, Lund for their general support, Drs John Dick and Connie Eaves for invaluable help and advice in establishing the NOD/SCID assay, and Dr Saemundur Gudmundsson for help in obtaining a continuous supply of cord blood cells. The generous supply of growth factors by Amgen and Novartis is gratefully acknowledged.
Supported by grants from the Swedish Cancer Society, Swedish Children's Cancer Foundation, Swedish Medical Research Council, and John and Augusta Persson Foundation (to S.K. and S.E.J.) and from the Swiss National Foundation and the Gabriella Giorgi-Cavaglieri Foundation (to D.T.).
N.-B.W. and C.F. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Stefan Karlsson, Department of Molecular Medicine and Gene Therapy, Lund University, Sölvegatan 17, 223 62 Lund, Sweden; e-mail: Stefan.Karlsson@molmed.lu.se.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal