Abstract
The AML-1/ETO fusion protein, created by the (8;21) translocation in M2-type acute myelogenous leukemia (AML), is a dominant repressive form of AML-1. This effect is due to the ability of the ETO portion of the protein to recruit co-repressors to promoters of AML-1 target genes. The t(11;17)(q21;q23)-associated acute promyelocytic leukemia creates the promyelocytic leukemia zinc finger PLZFt/RARα fusion protein and, in a similar manner, inhibits RARα target gene expression and myeloid differentiation. PLZF is expressed in hematopoietic progenitors and functions as a growth suppressor by repressing cyclin A2 and other targets. ETO is a corepressor for PLZF and potentiates transcriptional repression by linking PLZF to a histone deacetylase-containing complex. In transiently transfected cells and in a cell line derived from a patient with t(8;21) leukemia, PLZF and AML-1/ETO formed a tight complex. In transient assays, AML-1/ETO blocked transcriptional repression by PLZF, even at substoichiometric levels relative to PLZF. This effect was dependent on the presence of the ETO zinc finger domain, which recruits corepressors, and could not be rescued by overexpression of co-repressors that normally enhance PLZF repression. AML-1/ETO also excluded PLZF from the nuclear matrix and reduced its ability to bind to its cognate DNA-binding site. Finally, ETO interacted with PLZF/RARα and enhanced its ability to repress through the RARE. These data show a link in the transcriptional pathways of M2 and M3 leukemia.
Introduction
AML-1/ETO is a chimeric transcription factor generated as a consequence of the (8;21) chromosomal translocation, present in approximately 40% to 50% of patients with M2-type acute myelogenous leukemia (AML).1 The proteins that are fused in this event are AML-1 (also known as CBFA2, PEP2αB, and Runx-1), a transcription factor required for the development of definitive hematopoiesis,2,3 and ETO (also known as MTG8), which binds the mSin3 and N-CoR corepressors and histone deacetylases.4-6 AML-1 binds the CBP coactivator, activates the transcription of hematopoietic genes, and interacts and synergizes with other transcription factors such as PU.1, MEF, and C/EBPα7-10 The AML-1/ETO fusion product can bind to AML-1 binding sites and mediate inappropriate repression of these targets.11-14 This dominant repressive effect was further illustrated by the fact that AML-1/ETO knock-in mice display a similar phenotype to the AML-1–null mouse.2,3,15,16 AML-1 was also shown to repress transcription from the p21 promoter in a mSin3A-dependent manner17 and to interact with the groucho corepressor.18 However, AML-1/ETO lacks the AML-1 interaction domains for these corepressors.
Repression by AML-1/ETO is mediated by the ETO moiety and particularly requires the zinc finger or MYND domain of ETO, which binds to corepressors such as N-CoR and SMRT.4-6 ETO itself does not bind specific DNA sequences, and we recently showed that ETO is itself a corepressor for the promyelocytic leukemia zinc finger (PLZF) protein.19 Like ETO, PLZF is expressed in early hematopoietic cells and binds to N-Cor, SMRT, Sin3a, and histone deacetylase (HDAC).20-24 PLZF represses genes involved in cell proliferation and inhibits the growth of myeloid and other cells.25,26 PLZF is also disrupted by the t(11;17)(q21;q23) in retinoid-resistant acute promyelocytic leukemia (APL).27 This generates the PLZF/RARα fusion product, which inappropriately represses the transcription of RARα target genes by the recruitment of corepressors and HDACs (reviewed in28). Thus, in both M2 and M3 AML, a transcriptional repression complex is inappropriately targeted to promoters of genes involved in the normal process of blood cell maturation and differentiation.
Because ETO is a corepressor for PLZF, and both ETO and PLZF are expressed in the early stages of myeloid differentiation, we hypothesized that AML-1/ETO might interact with PLZF in t(8;21) leukemic cells. Similarly, because the PLZF/RARα protein retains the ETO-binding domain of PLZF (between amino acids 200 and 300),19 we hypothesized that ETO might be a corepressor for this APL fusion protein. We found that AML-1/ETO could bind to PLZF and severely impair its ability to repress transcription, whereas ETO enhanced transcriptional repression mediated by PLZF/RARα. We concluded that, in addition to its known repressor function, AML-1/ETO could function as an antirepressor. This may contribute to the leukemic transformation of myeloid progenitor cells by releasing PLZF-mediated inhibition of proliferation. Reciprocally, ETO may contribute to the retinoid-resistant phenotype of PLZF/RARα leukemia by augmenting the function of the corepressor complex attracted to RAR target genes.
Materials and methods
DNA constructs and cell lines
PLZF was expressed using both pSG5 (Stratagene, La Jolla, CA) or pCDNA3.1 + myc/his (Invitrogen, Carlsbad, CA) expression vectors. The GAL4-BTB/POZ (containing PLZF amino acids 1-137) and GAL4-RD2 (containing PLZF amino acids 200-300) mammalian expression vectors were described previously.19,29 ETO, AML-1/ETO full length, AML-1/ETOΔHHR , and AML-1/ETOΔHHR/MYNDconstructs14 were expressed from the pCMV5 vector. PML/RARα, PLZF/RARα, and RARα were expressed in the context of the pSG5 vector (Stratagene).30 N-CoR and SMRT were expressed from the pCMX vector and were a gift from Dr M. A. Lazar (University of Pennsylvania, Philadelphia, PA). GAL4-KRIP1 was a gift of Dr Joseph Bonaventre (Massachusetts General Hospital, Charlestown, MA). The highly transfectable human embryonic kidney cell line, 293T (gift of the late Dr Eugenia Spanapoulou), was used for co-immunoprecipitations, electrophoretic mobility shift assay (EMSA), and transcriptional assays. SKNO-1 cells (gift of Dr Y. Honma, Saitama Cancer Center, Saitama, Japan)31 were used to detect endogenous interactions between PLZF and AML-1/ETO.
Immunoprecipitations and immunoblotting
293T cells were plated at a density of 2 × 105cells per well and transfected with the indicated plasmids at a final concentration of 1.5 μg DNA per well of a 6-well plate. Transfections were performed with the Superfect reagent as directed (Qiagen, Valencia, CA). The cells were harvested 48 hours after transfection for immunoblotting. For analysis of endogenous proteins, SKNO-1 cells were cultured in RPMI 1640 with 10% fetal calf serum. Approximately 1 × 106 cells were lysed for each immunoprecipitation experiment. Twenty microliters of protein A Sepharose beads (Roche Molecular Biochemicals, Indianapolis, IN) was washed twice in 0.2 mol/L sodium borate (pH 9.0). The following antibodies were used for immunoprecipitation: PLZF mouse monoclonal antibody (IgG2aisotype),30 ETO C-terminal rabbit polyclonal antibody (Oncogene Research, Cambridge, MA), AML-1 runt domain rabbit polyclonal antibody (Oncogene Research), RARα polyclonal antibody (Oncogene Research), isotype control preimmune mouse monoclonal IgG2a(Jackson Immuno-Research, West Grove, PA), and control preimmune rabbit polyclonal IgG (Jackson Immuno-Research). The antibodies were added to the beads at a concentration of 1 mg/mL and mixed for 1 hour at room temperature. The beads were washed again at pH 9.0 at room temperature in sodium borate, followed by washing in 0.2 mol/L triethanolamine (pH 8.5) and freshly measured dimethyl pimelimidate at a final concentration of 20 mmol/L and were mixed for 1 hour at room temperature. The beads were resuspended in 0.2 mol/L ethanolamine for 5 minutes before they were transferred to phosphate-buffered saline (unless otherwise stated, all chemicals were obtained from Sigma, St Louis, MO). 293T cells and SKNO-1 cells were harvested with phosphate-buffered saline at 4°C and exposed to lysis buffer (150 mmol/L NaCl, 20 mmol/L Tris-Cl, Tween-20, pH 7.4, plus protease inhibitors) on ice for 15 minutes. This suspension was centrifuged at 3200g for 10 minutes at 4°C. The supernatant was precleared by exposure to beads bound to a nonspecific rabbit IgG (Zymed, San Francisco, CA) and mixed for approximately 1 hour at 4°C. The beads were then pelleted, and the lysates were exposed to specific antibodies of interest bound to protein A Sepharose or their isotype controls and were mixed on a rotator overnight. Pellets were washed in fresh cold lysis buffer 6 times, the last 3 times with NP-40 instead of Tween-20 in the lysis buffer. Precipitated proteins were released from the beads by boiling in Laemmelli buffer and then by electrophoresis through a 12% sodium dodecyl sulfate–polyacrylamide gel and transfer to an Immobilon P membrane (Millipore, Bedford, MA). Immunoblotting was performed using 1:1000 monoclonal PLZF (100 μg/mL) or 1:500 of ETO, AML, or RARα primary antibodies followed by 1:3000 horseradish peroxidase conjugated antimouse or antirabbit secondary antibody (Roche Molecular Biochemicals). Autoradiography was preformed using the ECL chemiluminescence kit (Amersham Pharmacia, Buckinghamshire, UK). Direct immunoblotting was performed using 100 μg protein from lysates from SKNO-1 cells or 20 μL lysates from transfected 293T cells.
Cell fractionation and electrophoretic mobility shift assay
Plates of 1 × 106 293T cells were transfected using Superfect (Qiagen) and 1.5 μg expression vectors for each of AML/ETO, wild-type PLZF, and control vectors. After 48 hours, the cells were harvested, nuclear extracts were prepared as described,32 and protein concentration of extracts were assayed using the DC Assay System (Bio-Rad, Hercules, CA). EMSA was performed with an [α-32P] dCTP-labeled double-stranded oligonucleotide containing a high-affinity binding site for PLZF, as described previously.33 Complementary oligonucleotides (2.5 pmol) were annealed in TE with 100 mmol/L NaCl, and the overhanging ends were filled in using Klenow and [α-32P] dCTP. For 20 minutes on ice, 10 fmol probe was incubated with 2 μg of each nuclear extract in 20 mmol/L HEPES, 1 mmol/L MgCl2, 10 μmol/L ZnCl2, 4% glycerol, and 100 μg/mL−1 bovine serum albumin. For supershift analysis, 100 ng PLZF mouse monoclonal antibody was pre-incubated with nuclear extracts for 20 minutes on ice before the probe was added. The DNA–protein complexes were separated by electrophoresis through a 4% nondenaturing, 0.5 × TBE acrylamide gel, dried, and visualized by exposure to Kodak XAR film (Eastman Kodak, Rochester, NY) and analyzed on a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). Nuclear matrix extracts were prepared as described.34
Immunofluorescence
293T cells were plated onto glass coverslips before transfection with PLZF and AML-1/ETO; 100 ng each of AML-1/ETO and PLZF was co-transfected, an amount that was carefully titrated to reproduce the natural staining pattern of each protein. After blocking in 10% donkey serum for 30 minutes, the cells were exposed to a 1:50 dilution of mouse monoclonal PLZF antibody for 1 hour. This was followed by exposure to a 1:100 dilution of rabbit polyclonal ETO antibodies or rabbit AML-1 (anti-RHD) polyclonal antibodies (Oncogene Research Products) for 1 hour. Samples were then treated for 30 minutes with donkey antimouse antibody conjugated to fluorescein isothiocyanate (FITC), goat antirabbit antibody conjugated to Texas red (Jackson Immuno-Research), or both. Vectashield mounting medium with DAPI (Vector Laboratories, Burlington, CA) was then applied, and the slides were examined with a Leica-TCS-SP (UV) confocal microscope (Leica, Heidelberg, Germany). To eliminate the possibility of cross-channel bleed-through, the samples were scanned separately in the nonoverlapping portion of the spectrum of each fluorescent marker. The resultant images were then overlaid to determine co-localization. This experiment was repeated 4 times, and multiple fields were imaged. As negative controls, the cells were incubated with secondary antibody alone or in the presence of irrelevant mouse antibody (DAKO, Carpinteria, CA) at a 1:50 dilution, irrelevant rabbit antibody (R&D, Minneapolis, MN) at a 1:100 dilution, or both, followed by incubation with the appropriate secondary antibodies.
Transcriptional assays
To determine the transcriptional effects of the GAL4-fusion constructs, we used a reporter containing 5 GAL4-binding sites linked 5′ to the herpes virus thymidine kinase (tk) promoter and the firefly luciferase gene.19 A reporter gene to assay the transcriptional effects of PLZF consisted of 4 copies of a high-affinity PLZF binding site found within the IL-3 receptor alpha chain (IL-3Rα) promoter linked 5′ to the tk-luciferase reporter.33 A tk-luciferase construct lacking specific binding sites was used as a negative control for the above reporters. 293T cells were plated at a density of 2 × 105 cells/well of 12-well or 4 × 105cells/well of 6-well tissue culture dishes and transfected using Superfect (Qiagen). In 12-well dish transfections, 100 ng reporter and 5 ng internal control (tk-renilla luciferase) were used, and in 6-well dishes 200 ng reporter and 10 ng internal control were used. The combinations and quantities of expression plasmids are noted in the figure legends. To determine the transcriptional effects of RARαPML/RARα, and PLZF/RARα, we used a RARE-tk-Luc reporter construct harboring a DR5 retinoid response element (gift of Dr Len Freedman, Memorial-Sloan Kettering Cancer Center, New York, NY). These experiments were performed in charcoal-stripped serum (Life Technologies, Rockville, MD). To assess transcriptional activity, dual luciferase assays were performed (Promega, Madison, WI), using an MLX Microtiter Plate Luminometer (Dynex Technologies, Chantilly, VA). All these experiments were performed in duplicate 3 to 6 times, with results averaged and normalized to the internal control. Immunoblotting, performed as above, confirmed the expression of proteins. To assess effects of the proteins on cell growth, 5 separate sets of 293T cell cultures at a density of 2.8 × 105/well (of a 6-well dish) were transfected with pCDNA, pCDNA + AML-1/ETO, PLZF or PLZF + AML-1/ETO as above. The cells were allowed to grow in parallel to the reporter assay cells, as described, and were harvested at identical times for counting.
Results
AML-1/ETO interacts with PLZF
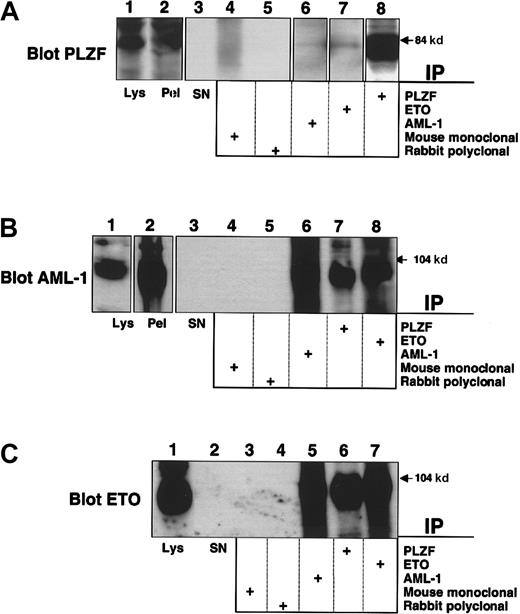
Because AML-1/ETO retains the domain (between ETO amino acids 220 and 330) required for interaction with PLZF,19 we transiently expressed both AML-1/ETO and PLZF in 293T cells to determine whether they formed a complex. Lysates from these cells were subjected to immunoprecipitation with antibodies against PLZF, the AML-1 runt domain, or the ETO C-terminal MYND domain. Lysates precipitated with AML-1 or ETO antibodies readily revealed the presence of PLZF (Figure 1A). In a reciprocal manner, immunoblots of lysates precipitated with PLZF antibody revealed the presence of AML-1/ETO, as detected by both the AML-1 and ETO antibodies (Figure 1B-C). 293T cells do not express AML-1 or ETO at detectable levels, as shown by the fact that immunoprecipitations performed with each antibody followed by immunoblotting with the same antibody did not show any endogenous protein. Thus, PLZF is interacting with the transfected chimeric protein. We then performed immunofluorescence studies in 293T cells to determine whether AML-1/ETO and PLZF colocalize in cell nuclei. PLZF normally localizes in a speckled nuclear pattern, and AML-1/ETO localizes in a combined speckled and diffuse nuclear pattern.30 35 Initially, we titrated the dose of each plasmid to achieve a staining pattern similar to that of the endogenous protein. Dual staining for both proteins with FITC and Texas red–conjugated secondary antibodies demonstrated that the speckled fraction of AML-1/ETO largely colocalizes with PLZF (Figure 1D). No staining was seen when nonspecific isotype antibodies were used as negative controls (data not shown). From these studies we concluded that PLZF and AML-1/ETO could associate in vivo in human cells.
AML-1/ETO interacts with PLZF.
293T cells were transfected with 750 ng each of pCMV-AML-1/ETO and pCDNA-PLZF expression vectors. The cells were lysed and subjected to immunoprecipitations and immunoblotting as indicated. (A) Immunoblot with PLZF mouse monoclonal antibody. (B) Immunoblot with ETO C-terminal rabbit polyclonal antibody. (C) Immunoblot with AML-1 runt domain rabbit polyclonal antibody. The lanes of all 3 panels correspond to similar experiments. Lane 1, direct immunoblot of cell lysate. Lanes 2-6, extracts immunoprecipitated with mouse control antibody, rabbit control antibody, PLZF antibody, AML-1 antibody, and ETO antibody, respectively. (D) Immunofluorescence staining of 293T cells co-transfected with 100 ng PLZF and AML-1/ETO expression vectors. DAPI, nuclear staining. ETO, AML-1/ETO located using ETO polyclonal antibodies and Texas red–conjugated secondary antibodies. PLZF was detected using monoclonal antibodies and FITC-conjugated secondary antibodies. Ov, overlay of images of PLZF and AML-1/ETO. Magnification, 400×.
AML-1/ETO interacts with PLZF.
293T cells were transfected with 750 ng each of pCMV-AML-1/ETO and pCDNA-PLZF expression vectors. The cells were lysed and subjected to immunoprecipitations and immunoblotting as indicated. (A) Immunoblot with PLZF mouse monoclonal antibody. (B) Immunoblot with ETO C-terminal rabbit polyclonal antibody. (C) Immunoblot with AML-1 runt domain rabbit polyclonal antibody. The lanes of all 3 panels correspond to similar experiments. Lane 1, direct immunoblot of cell lysate. Lanes 2-6, extracts immunoprecipitated with mouse control antibody, rabbit control antibody, PLZF antibody, AML-1 antibody, and ETO antibody, respectively. (D) Immunofluorescence staining of 293T cells co-transfected with 100 ng PLZF and AML-1/ETO expression vectors. DAPI, nuclear staining. ETO, AML-1/ETO located using ETO polyclonal antibodies and Texas red–conjugated secondary antibodies. PLZF was detected using monoclonal antibodies and FITC-conjugated secondary antibodies. Ov, overlay of images of PLZF and AML-1/ETO. Magnification, 400×.
Endogenous AML-1/ETO and PLZF interact in t(8;21) leukemia cells
PLZF is normally expressed in early myeloid cells and represses promoters of genes involved in both differentiation and cell proliferation, such as cyclin A2 and the IL-3Rα chain.25,33 Because the M2 leukemic phenotype corresponds to a myeloblastic morphology, we hypothesized that PLZF would be expressed in these cells. To answer this question we used the SKNO-1 cell line, derived from a young patient with t(8;21) M2 AML.31 These cells expressed PLZF at levels detectable by direct immunoblotting (Figure 2A, lane 1). We then subjected lysates from these cells to immunoprecipitation using the PLZF, AML-1, and ETO antibodies as above. AML-1/ETO was immunoprecipitated by antibodies to PLZF (Figure 2B-C). PLZF was also detectable in both the AML-1 and ETO immunoprecipitates (Figure 2A). Neither protein was present when the appropriate mouse and rabbit control antibodies were used for immunoprecipitation (Figure 2A-C). Thus, endogenous AML-1/ETO interacts with endogenous PLZF in t(8;21) leukemia cells.
Interaction of endogenous PLZF and AML-1/ETO in a t(8;21) M2 leukemia cell line.
SKNO-1 cells were lysed and subjected to immunoprecipitation–immunoblot analysis. (A) Immunoblot with PLZF mouse monoclonal antibody. Lane 1, direct immunoblot of SKNO-1 cell lysate. Lane 2, blot of immunopellet. Lane 3, lysate supernatant after immunoprecipitation. Lanes 4-8, extracts immunoprecipitated with mouse control antibody, rabbit control antibody, AML-1 antibody, ETO antibody, and PLZF antibody. (B) Immunoblot with AML-1 rabbit polyclonal antibody. Lane 1, direct immunoblot of SKNO-1 cell lysate. Lane 2, blot of immunopellet. Lane 3, lysate supernatant after immunoprecipitation. Lanes 4-8, extracts immunoprecipitated with mouse control antibody, rabbit control antibody, AML-1 antibody, PLZF antibody, and ETO antibody. (C) Immunoblot with ETO rabbit polyclonal antibody. Lane 1, direct immunoblot of SKNO-1 cell lysate. Lane 2, lysate supernatant after immunopelleting. Lanes 3-7, extracts immunoprecipitated with mouse control antibody, rabbit control antibody, AML-1 antibody, PLZF antibody, and ETO antibody.
Interaction of endogenous PLZF and AML-1/ETO in a t(8;21) M2 leukemia cell line.
SKNO-1 cells were lysed and subjected to immunoprecipitation–immunoblot analysis. (A) Immunoblot with PLZF mouse monoclonal antibody. Lane 1, direct immunoblot of SKNO-1 cell lysate. Lane 2, blot of immunopellet. Lane 3, lysate supernatant after immunoprecipitation. Lanes 4-8, extracts immunoprecipitated with mouse control antibody, rabbit control antibody, AML-1 antibody, ETO antibody, and PLZF antibody. (B) Immunoblot with AML-1 rabbit polyclonal antibody. Lane 1, direct immunoblot of SKNO-1 cell lysate. Lane 2, blot of immunopellet. Lane 3, lysate supernatant after immunoprecipitation. Lanes 4-8, extracts immunoprecipitated with mouse control antibody, rabbit control antibody, AML-1 antibody, PLZF antibody, and ETO antibody. (C) Immunoblot with ETO rabbit polyclonal antibody. Lane 1, direct immunoblot of SKNO-1 cell lysate. Lane 2, lysate supernatant after immunopelleting. Lanes 3-7, extracts immunoprecipitated with mouse control antibody, rabbit control antibody, AML-1 antibody, PLZF antibody, and ETO antibody.
AML-1/ETO antagonizes PLZF-mediated transcriptional repression
To determine the effect of AML-1/ETO on the transcriptional properties of PLZF, we transiently transfected both proteins along with a reporter containing PLZF binding sites from the IL-3Rα chain promoter. Repression of this reporter by PLZF is well established and is potentiated by ETO.19,33 We found that AML-1/ETO strikingly inhibited transcriptional repression by PLZF. This occurred when PLZF was expressed in the context of both pSG5 and pCDNA3.1+ expression vectors and was a specific effect of AML-1/ETO given that it did not occur in the presence of the empty pCMV expression vector (Figure 3A and data not shown). AML-1/ETO did not activate transcription from the reporter construct in the absence of PLZF, suggesting that the block of repression was related to the interaction of these 2 proteins (Figure 3B). AML-1/ETO blocks activation by AML-1 even when expressed at a lower level than AML-1.11,12 Similarly, AML-1/ETO antagonized repression by PLZF antagonism even when AML-1/ETO was cotransfected at an 8-fold lower concentration than PLZF. This effect was maintained over a broad range of doses of AML-1/ETO (Figure 3C). This was not caused by toxic effects because 293T cells transfected in conditions identical to those of the cells above grew to levels similar to those of mock-transfected cells and had similar levels of viability, as determined by trypan blue exclusion (Figure 3D and data not shown). Finally, the effect was specific for PLZF because transcriptional repression by the unrelated corepressor KRIP136 was not inhibited by the co-expression of AML-1/ETO (Figure 3E).
AML-1/ETO antagonizes PLZF-mediated transcriptional repression in a dominant-negative fashion.
Transient transfections and reporter assays were all performed in 293T cells. (A) 293T cells transfected with the IL-3Rα reporter construct. Lanes 1-8, (12-well dishes): 400 ng each of pCDNA expression vector, pCDNA-PLZF, pCMV expression vector, and pCMV-AML-1/ETO was transfected as indicated. Results are expressed as fold repression compared to the empty expression vector. (B) Transient transfections performed in conditions similar to those described for A. In this case the results are expressed as relative luciferase activity compared to vector alone. (C) 6-well dishes: 800 ng pCDNA or pCDNA-PLZF was cotransfected with increasing doses of AML-1/ETO as follows: lanes 2 and 8, 100 ng; lanes 3 and 9, 200 ng; lanes 4 and 10, 400 ng; lanes 5 and 11, 800 ng; lanes 6 and 12, 1200 ng AML-1/ETO, respectively. (D) 293T cells were plated at a density of 2.8 × 105 in 6-well dishes and transfected with 800 ng pCDNA, PLZF, or AML-1/ETO as indicated and identical amounts of reporter and internal controls, as in C. The cells were harvested simultaneously with those used for luciferase assays and were counted. (E) 293T cells transfected with the (GAL4)5-tk-Luc reporter. Lane 1, GAL41–147 (400 ng) transfected without AML-1/ETO. Lanes 2 and 3, GAL41–147 400 ng with AML-1/ETO 200 ng and 400 ng, respectively. Lane 4, GAL4-KRIP1 (400 ng). Lanes 5 and 6, GAL4-KRIP1 (400 ng) along with AML-1/ETO 200 ng and 400 ng, respectively. (C, D, E) Fold repression was compared to vector only. The results of all graphs reflect the average of multiple experiments ± SEM.
AML-1/ETO antagonizes PLZF-mediated transcriptional repression in a dominant-negative fashion.
Transient transfections and reporter assays were all performed in 293T cells. (A) 293T cells transfected with the IL-3Rα reporter construct. Lanes 1-8, (12-well dishes): 400 ng each of pCDNA expression vector, pCDNA-PLZF, pCMV expression vector, and pCMV-AML-1/ETO was transfected as indicated. Results are expressed as fold repression compared to the empty expression vector. (B) Transient transfections performed in conditions similar to those described for A. In this case the results are expressed as relative luciferase activity compared to vector alone. (C) 6-well dishes: 800 ng pCDNA or pCDNA-PLZF was cotransfected with increasing doses of AML-1/ETO as follows: lanes 2 and 8, 100 ng; lanes 3 and 9, 200 ng; lanes 4 and 10, 400 ng; lanes 5 and 11, 800 ng; lanes 6 and 12, 1200 ng AML-1/ETO, respectively. (D) 293T cells were plated at a density of 2.8 × 105 in 6-well dishes and transfected with 800 ng pCDNA, PLZF, or AML-1/ETO as indicated and identical amounts of reporter and internal controls, as in C. The cells were harvested simultaneously with those used for luciferase assays and were counted. (E) 293T cells transfected with the (GAL4)5-tk-Luc reporter. Lane 1, GAL41–147 (400 ng) transfected without AML-1/ETO. Lanes 2 and 3, GAL41–147 400 ng with AML-1/ETO 200 ng and 400 ng, respectively. Lane 4, GAL4-KRIP1 (400 ng). Lanes 5 and 6, GAL4-KRIP1 (400 ng) along with AML-1/ETO 200 ng and 400 ng, respectively. (C, D, E) Fold repression was compared to vector only. The results of all graphs reflect the average of multiple experiments ± SEM.
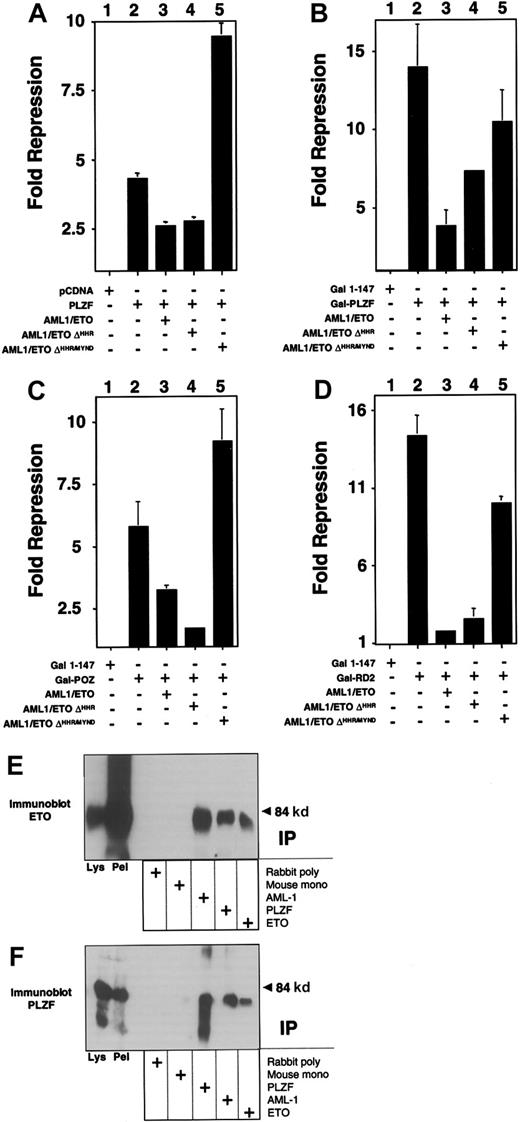
We next determined which of the 2 repression domains of PLZF could be inhibited by AML-1/ETO. For this purpose we fused amino acids 1 to 400 of PLZF to the GAL4 DNA-binding domain (GAL4-PLZF1–400) and cotransfected it with a reporter plasmid containing 5 GAL4-binding sites. This fragment includes both the N-terminal BTB/POZ repression domain (amino acids 1-130) and the second repression domain (RD2) between amino acids 200 to 300. AML-1/ETO was also able to inhibit repression by this construct, with a slightly lower efficiency than full-length PLZF, but also at a 1:8 ratio of AML-1/ETO to PLZF (Figure 4A and data not shown). The reporter was not activated by AML-1/ETO, indicating that the effect was related to the interaction between the 2 proteins (Figure 4B). We then analyzed this effect on the isolated repression domains of PLZF, each of which is required for PLZF function (see reference37 and see below). Wild-type ETO is able to enhance repression mediated by both domains, though it only binds to RD2.19 Both domains yielded powerful repression, with GAL4-RD2 repressing the luciferase reporter 70-fold and GAL4-BTB/POZ repressing it 14-fold (Figure 4C-D). AML-1/ETO inhibited the constructs in a fashion similar to that of full-length PLZF, suggesting that transcriptional disruption occurred through effects on both repression domains (Figure 4C-D).
AML-1/ETO antagonizes the transcriptional repression mediated by both repression domains of PLZF.
293T cells were transfected with the (GAL4)5-tk-Luc reporter. (A) Lanes 1 to 4 (12-well dishes), 400 ng GAL41–147, GAL4-PLZF, and AML-1/ETO as indicated. Results are expressed as fold repression compared to vector. (B) Transfections identical to those in A, with results shown as relative luciferase levels. (C) Lanes 1 to 4 (12-well dishes), 400 ng GAL41–147, GAL4-BTB/POZ, and AML-1/ETO as indicated. Lanes 5 to 14 (6-well dishes), 800 ng GAL41–147or GAL4-BTB/POZ was cotransfected with increasing doses of AML-1/ETO as follows: lanes 6 and 11, 100 ng; lanes 7 and 12, 200 ng; lanes 8 and 13, 400 ng; lanes 9 and 14, 800 ng, respectively. (D) Transfections with GAL4-RD2 with identical quantities as in the experiments described in C. (C,D) Fold repression compared to vector only. The results of all graphs reflect the average of multiple experiments ± SEM.
AML-1/ETO antagonizes the transcriptional repression mediated by both repression domains of PLZF.
293T cells were transfected with the (GAL4)5-tk-Luc reporter. (A) Lanes 1 to 4 (12-well dishes), 400 ng GAL41–147, GAL4-PLZF, and AML-1/ETO as indicated. Results are expressed as fold repression compared to vector. (B) Transfections identical to those in A, with results shown as relative luciferase levels. (C) Lanes 1 to 4 (12-well dishes), 400 ng GAL41–147, GAL4-BTB/POZ, and AML-1/ETO as indicated. Lanes 5 to 14 (6-well dishes), 800 ng GAL41–147or GAL4-BTB/POZ was cotransfected with increasing doses of AML-1/ETO as follows: lanes 6 and 11, 100 ng; lanes 7 and 12, 200 ng; lanes 8 and 13, 400 ng; lanes 9 and 14, 800 ng, respectively. (D) Transfections with GAL4-RD2 with identical quantities as in the experiments described in C. (C,D) Fold repression compared to vector only. The results of all graphs reflect the average of multiple experiments ± SEM.
The C-terminal corepressor binding domain of AML-1/ETO is required to block PLZF repression
Both the AML-1 and the ETO proteins can bind corepressors. However, AML-1 requires residues that are not present in the AML-1/ETO fusion protein to bind to the mSin3A and groucho corepressors.17,18 In contrast, AML-1/ETO contains all the domains required to associate with Sin3A, N-CoR, SMRT, and HDACs.4-6 Of these, the MYND domain is required to enhance repression by PLZF.19 We determined whether the same domain was required for the action of AML-1/ETO. PLZF and the isolated repression domains of PLZF were co-expressed along with AML-1/ETO or mutant AML-1/ETO constructs lacking either the HHR domain (AML-1/ETOΔHHR) or both the HHR and MYND domains (AML-1/ETOΔHHR/MYND). In all cases, the MYND corepressor-binding domain was absolutely required for AML-1/ETO to block transcriptional repression by full-length PLZF or the PLZF repression domains (Figure 5). We verified that AML-1/ETOΔHHR/MYND still associates with PLZF by coimmunoprecipitations in transiently transfected 293T cells (Figure 5).
The AML-1/ETO C-terminal corepressor binding domains are required to block PLZF repression.
Transient transfections and reporter assays performed in 293T cells in 12-well dishes. Results are all depicted as fold repression relative to the control plasmid basal expression level. (A) Experiments performed with the IL-3Rα chain reporter. Lane 1, pCDNA 400 ng. Lane 2, PLZF 400 ng. Lane 3, PLZF 400 ng plus AML-1/ETO 300 ng. Lane 4, PLZF 400 ng plus AML-1/ETOΔHHR 300 ng. Lane 5, PLZF 400 ng plus AML-1/ETOΔHHR/MYND 300 ng. (B-D) Experiments performed with the (GAL4)5-tk-Luc reporter and GAL4-PLZF in B, GAL4-BTB/POZ in C, and GAL4-RD2 in D. Doses are identical to those in A. Results of all graphs reflect the average of multiple experiments ± SEM. (E,F) 293T cells were transfected with 750 ng each of pCMV-AML-1/ETOΔHHR/MYND and pCDNA-PLZF expression vectors. Cells were lysed and subjected to immunoprecipitations and immunoblotting as indicated. (E) Immunoblot with ETO C-terminal rabbit polyclonal antibody. (F) Immunoblot with PLZF mouse monoclonal antibody. Lanes 1 and 2 of both panels correspond to immunoblots of cell lysates and precipitated pellet, respectively. Lanes 3 to 7 correspond to immunoprecipitates as indicated.
The AML-1/ETO C-terminal corepressor binding domains are required to block PLZF repression.
Transient transfections and reporter assays performed in 293T cells in 12-well dishes. Results are all depicted as fold repression relative to the control plasmid basal expression level. (A) Experiments performed with the IL-3Rα chain reporter. Lane 1, pCDNA 400 ng. Lane 2, PLZF 400 ng. Lane 3, PLZF 400 ng plus AML-1/ETO 300 ng. Lane 4, PLZF 400 ng plus AML-1/ETOΔHHR 300 ng. Lane 5, PLZF 400 ng plus AML-1/ETOΔHHR/MYND 300 ng. (B-D) Experiments performed with the (GAL4)5-tk-Luc reporter and GAL4-PLZF in B, GAL4-BTB/POZ in C, and GAL4-RD2 in D. Doses are identical to those in A. Results of all graphs reflect the average of multiple experiments ± SEM. (E,F) 293T cells were transfected with 750 ng each of pCMV-AML-1/ETOΔHHR/MYND and pCDNA-PLZF expression vectors. Cells were lysed and subjected to immunoprecipitations and immunoblotting as indicated. (E) Immunoblot with ETO C-terminal rabbit polyclonal antibody. (F) Immunoblot with PLZF mouse monoclonal antibody. Lanes 1 and 2 of both panels correspond to immunoblots of cell lysates and precipitated pellet, respectively. Lanes 3 to 7 correspond to immunoprecipitates as indicated.
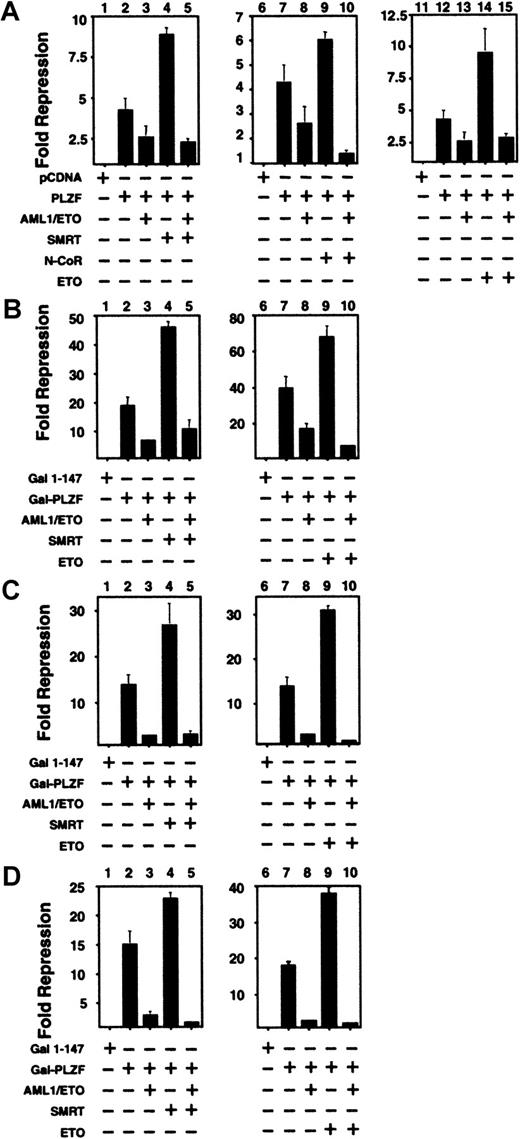
Corepressors do not rescue PLZF from inhibition by AML-1/ETO
The above results suggested that one possible mechanism of AML-1/ETO interference of PLZF action could be through competition for corepressors. To test this model, we cotransfected N-CoR, SMRT, or ETO along with PLZF and AML-1/ETO. When full-length PLZF was coexpressed along with N-CoR, SMRT, or ETO, transcriptional repression was enhanced (Figure 6A). However, when AML-1/ETO was added at substoichiometric levels to any of these PLZF/corepressor combinations, repression was still blocked. A similar phenomenon was observed when SMRT or ETO was transfected in combination with GAL4-PLZF1–400, GAL4-BTB/POZ, and GAL4-RD2. Thus, a simple competition for co-repressors is unlikely to fully explain the effects of AML-1/ETO on PLZF.
Co-repressors do not rescue PLZF from inhibition by AML-1/ETO.
293T cells were transiently transfected in 12-well dishes and subjected to luciferase reporter assays. All results are expressed as fold repression compared to the control plasmids. (A) Assays performed with the IL-3Rα-tk-Luc reporter. Lanes 1, 6, 11: pCDNA 400 ng. Lanes 2, 7, 12: PLZF 400 ng. Lanes 3, 8, 13: PLZF 400 ng plus AML-1/ETO 300 ng. Lane 4: PLZF 400 ng and SMRT 300 ng. Lane 5: PLZF 400 ng, SMRT 300 ng, and AML-1/ETO 300 ng. Lane 9: PLZF 400 ng and N-CoR 300 ng. Lane 10: PLZF 400 ng, N-CoR 300 ng, and AML-1/ETO 300 ng. Lane 14: PLZF 400 ng plus ETO 300 ng. Lane 15: PLZF 400 ng, ETO 300 ng, and AML-1/ETO 300 ng. Panels B, C and D: Assays performed with the (GAL4)5-tk-Luc reporter. (B) Lanes 1, 6: GAL41–147 400 ng. Lanes 2, 7: GAL4-PLZF 400 ng. Lanes 3, 8: GAL4-PLZF 400 ng and AML-1/ETO 300 ng. Lane 4: GAL4-PLZF 400 ng and SMRT 300 ng. Lane 5: GAL4-PLZF 400 ng, SMRT 300 ng, and AML-1/ETO 300 ng. Lane 9: GAL4-PLZF 400 ng and ETO 300 ng. Lane 10: GAL4-PLZF 400 ng, ETO 300 ng, and AML-1/ETO 300 ng. Panels C and D correspond to experiments performed with GAL4-BTB/POZ and GAL4-RD2, with doses similar to those in B. The results of all graphs reflect the average of multiple experiments ± SEM.
Co-repressors do not rescue PLZF from inhibition by AML-1/ETO.
293T cells were transiently transfected in 12-well dishes and subjected to luciferase reporter assays. All results are expressed as fold repression compared to the control plasmids. (A) Assays performed with the IL-3Rα-tk-Luc reporter. Lanes 1, 6, 11: pCDNA 400 ng. Lanes 2, 7, 12: PLZF 400 ng. Lanes 3, 8, 13: PLZF 400 ng plus AML-1/ETO 300 ng. Lane 4: PLZF 400 ng and SMRT 300 ng. Lane 5: PLZF 400 ng, SMRT 300 ng, and AML-1/ETO 300 ng. Lane 9: PLZF 400 ng and N-CoR 300 ng. Lane 10: PLZF 400 ng, N-CoR 300 ng, and AML-1/ETO 300 ng. Lane 14: PLZF 400 ng plus ETO 300 ng. Lane 15: PLZF 400 ng, ETO 300 ng, and AML-1/ETO 300 ng. Panels B, C and D: Assays performed with the (GAL4)5-tk-Luc reporter. (B) Lanes 1, 6: GAL41–147 400 ng. Lanes 2, 7: GAL4-PLZF 400 ng. Lanes 3, 8: GAL4-PLZF 400 ng and AML-1/ETO 300 ng. Lane 4: GAL4-PLZF 400 ng and SMRT 300 ng. Lane 5: GAL4-PLZF 400 ng, SMRT 300 ng, and AML-1/ETO 300 ng. Lane 9: GAL4-PLZF 400 ng and ETO 300 ng. Lane 10: GAL4-PLZF 400 ng, ETO 300 ng, and AML-1/ETO 300 ng. Panels C and D correspond to experiments performed with GAL4-BTB/POZ and GAL4-RD2, with doses similar to those in B. The results of all graphs reflect the average of multiple experiments ± SEM.
AML-1/ETO disrupts the nuclear matrix compartmentalization of PLZF and reduces its ability to bind to DNA
We next performed cell fractionation on 293T cells transiently transfected with the low levels of PLZF that reproduce the natural immunofluorescence pattern of the endogenous protein. Almost all the PLZF was present in the nuclear fraction (Figure7A). Intriguingly, a large amount of the protein was in the nuclear matrix fraction. This is consistent with the finding that PLZF can be found in nuclear bodies, which have been shown to be associated with the matrix38 39 (Figure 7A). When PLZF and AML-1/ETO were transfected together, there was almost no PLZF localization to the nuclear matrix, though the nuclear localization of PLZF was not affected (Figure 7A). Immunoblotting of whole cell extract showed that PLZF was expressed to similar levels in both the PLZF and the PLZF + AML-1/ETO cells (Figure 7A). Thus, sequestration from the nuclear matrix compartment might contribute to the effect of AML-1/ETO on transcriptional repression by PLZF.
AML-1/ETO disrupts nuclear matrix compartmentalization of PLZF and reduces its ability to bind to DNA.
(A) 200 ng vector, PLZF, AML-1/ETO, or PLZF plus AML-1/ETO was transfected in 293T cells. The cells were fractionated into cytoplasmic, nuclear, and nuclear matrix fractions. Immunoblotting with PLZF monoclonal antibodies was performed on each fraction and on the unfractionated whole cell lysates. (B) EMSA was performed on lysates from transfected 293T cells. Expression vectors in each transfection are indicated. The PLZF high-molecular–weight complex is indicated by the lower arrowhead, and the supershift generated by the addition of anti-PLZF monoclonal antibodies is indicated by the upper arrowhead. (C) Immunoblot of lysates from the cells used in the EMSA in B after normalization for protein concentration.
AML-1/ETO disrupts nuclear matrix compartmentalization of PLZF and reduces its ability to bind to DNA.
(A) 200 ng vector, PLZF, AML-1/ETO, or PLZF plus AML-1/ETO was transfected in 293T cells. The cells were fractionated into cytoplasmic, nuclear, and nuclear matrix fractions. Immunoblotting with PLZF monoclonal antibodies was performed on each fraction and on the unfractionated whole cell lysates. (B) EMSA was performed on lysates from transfected 293T cells. Expression vectors in each transfection are indicated. The PLZF high-molecular–weight complex is indicated by the lower arrowhead, and the supershift generated by the addition of anti-PLZF monoclonal antibodies is indicated by the upper arrowhead. (C) Immunoblot of lysates from the cells used in the EMSA in B after normalization for protein concentration.
We next analyzed whether AML-1/ETO could affect DNA binding by PLZF. We previously showed that PLZF forms a high-molecular–weight complex when allowed to bind with an oligonucleotide containing PLZF-binding sites from the IL-3Rα promoter, which can be further shifted by PLZF antibodies (Figure 7B).33 However, when PLZF and AML-1/ETO were coexpressed or expressed individually and then mixed, there was a 4-fold reduction in PLZF-DNA complexes detected by EMSA (Figure 7B). All samples were normalized for protein levels, and the expression level of PLZF was shown to be equivalent (Figure 7C). Thus, AML-1/ETO reduces the ability of PLZF to bind to its cognate DNA binding site. The combination of corepressor competition, sequestration from the nuclear matrix, reduced DNA binding, and possible aberrant complex formation may all contribute to the effect of AML-1/ETO on PLZF.
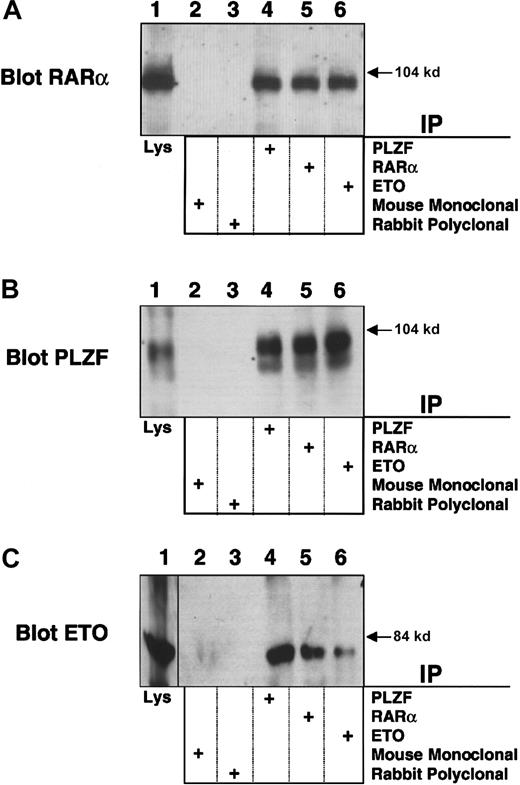
PLZF/RARα physically interacts with ETO
To further explore the mechanistic links between leukemias that target PLZF and ETO, we studied the PLZF/RARα product of the (11;17)(q23;q21) translocation from the retinoic acid (RA)-resistant variant of APL. PLZF/RARα recruits N-CoR, SMRT, and HDACs to RARα target genes and fails to release these proteins in response to retinoic acid.22-24 ETO, which is normally expressed in myeloblasts, binds the second repression domain of PLZF, retained in PLZF/RARα, and could participate in RA-independent PLZF/RARα transcriptional repression. To examine this possibility, we transiently expressed PLZF/RARα and ETO in 293T cells and performed immunoprecipitations with PLZF, RARα, or ETO rabbit polyclonal antibody. Immunoblots performed using the PLZF or RARα antibodies indicated that PLZF/RARα was present in all 3 of the precipitates (Figure 8A-B). Direct immunoblots of the cell lysates also confirmed expression of both proteins (Figure 8A-B). A reciprocal experiment was performed in which cell lysates were precipitated with the PLZF, RARα, or ETO antibodies and then immunoblotted for ETO. In this case ETO was observed in all 3 precipitates but not in precipitates obtained with mouse and rabbit control antibodies (Figure 8C). In contrast, we were unable to detect an interaction between ETO and the PML/RARα fusion protein of RA sensitive “classic” APL (data not shown).
PLZF/RARα interacts with ETO. PLZF/RARα and ETO (750 ng) were both transiently transfected in 293T cells grown in 6-well dishes.
Cells were lysed and subjected to immunoprecipitations and immunoblotting. (A) Immunoblot with RARα rabbit polyclonal antibody. (B) Immunoblot with PLZF mouse monoclonal antibody. (C) Immunoblot with ETO rabbit polyclonal antibody. The lanes of all 3 panels correspond to similar experiments. Lane 1, direct immunoblot of cell lysate. Lanes 2-6, extracts immunoprecipitated with mouse control antibody, rabbit control antibody, PLZF antibody, RARα antibody, and ETO antibody, respectively.
PLZF/RARα interacts with ETO. PLZF/RARα and ETO (750 ng) were both transiently transfected in 293T cells grown in 6-well dishes.
Cells were lysed and subjected to immunoprecipitations and immunoblotting. (A) Immunoblot with RARα rabbit polyclonal antibody. (B) Immunoblot with PLZF mouse monoclonal antibody. (C) Immunoblot with ETO rabbit polyclonal antibody. The lanes of all 3 panels correspond to similar experiments. Lane 1, direct immunoblot of cell lysate. Lanes 2-6, extracts immunoprecipitated with mouse control antibody, rabbit control antibody, PLZF antibody, RARα antibody, and ETO antibody, respectively.
ETO enhances PLZF/RARα-mediated repression of an RA response element
We used an RA response element (RARE) containing reporter to determine whether there was a functional interaction between ETO and PLZF/RARα and compared this to the effect of ETO on RARα and PML/RARα. 293T Cells were transiently transfected with empty vector, RARα, PML/RARα, or PLZF/RARα with or without ETO in charcoal-stripped serum. Consistent with previous reports, PML/RARα and PLZF/RARα had a modest repressive effect on their own on the RARE reporter in the absence of RA (Figure 9). However, the addition of ETO significantly enhanced repression of the RARE reporter by PLZF/RARα but did not enhance repression by RARα or PML/RARα. Thus, ETO is a corepressor for PLZF/RARα and for PLZF.
ETO enhances PLZF/RARα-mediated repression of a retinoic acid response element.
293T cells were transiently transfected with a DR5-RARE-tk-Luc reporter construct along with a tk-renilla internal control plasmid. Results are expressed as fold repression relative to the normalized basal expression of Luc by the pSG5 expression vector. Lane 1, 800 ng pSG5. Lane 2, 800 ng pSG5 and 600 ng ETO. Lane 3, 800 ng RARα. Lane 4, 800 ng RARα and 600 ng ETO. Lane 5, 800 ng PLZF/RARα. Lane 6, 800 ng PLZF/RARα and 600 ng ETO. Lane 7, 800 ng PML/RARα. Lane 8, 800 ng PML/RARα and 600 ng ETO.
ETO enhances PLZF/RARα-mediated repression of a retinoic acid response element.
293T cells were transiently transfected with a DR5-RARE-tk-Luc reporter construct along with a tk-renilla internal control plasmid. Results are expressed as fold repression relative to the normalized basal expression of Luc by the pSG5 expression vector. Lane 1, 800 ng pSG5. Lane 2, 800 ng pSG5 and 600 ng ETO. Lane 3, 800 ng RARα. Lane 4, 800 ng RARα and 600 ng ETO. Lane 5, 800 ng PLZF/RARα. Lane 6, 800 ng PLZF/RARα and 600 ng ETO. Lane 7, 800 ng PML/RARα. Lane 8, 800 ng PML/RARα and 600 ng ETO.
Discussion
The investigation of common pathways of leukemogenesis led us to study the interaction between proteins involved in M2 AML and retinoid-resistant APL. We previously reported that the ETO protein, involved in the (8;21) translocation of M2-AML is a corepressor for PLZF.19 This effect was dependent on the MYND corepressor/HDAC-binding domain of ETO and was blocked by HDAC inhibitors.19 We expected that AML-1/ETO would interact with PLZF because the PLZF-binding site located at ETO residues 220 to 330 is retained in the fusion protein. Consistent with this, the 2 proteins interacted when expressed in transfected cells and endogenously in a cell line derived from a patient with t(8;21) AML.
We modeled the interaction between PLZF and AML-1/ETO in transfected cells and found that the fusion protein strongly blocked repression by PLZF or the isolated repression domains of PLZF. This suggests a novel function for AML-1/ETO as de-repressor of a transcriptional repressor. AML-1/ETO functions include the blockade of AML-1 transcriptional activation in a dominant-negative fashion11,40 due to the inappropriate recruitment of corepressors to AML-1 target sequences,4-6 the active repression of basal transcription from the MDR-1 promoter independent of its dominant-negative effects on AML-1,14 the dominant-negative inhibition of transcriptional activation by C/EBPα and the Ets factors PU.1 and MEF,7-10 and the activation of transcription from an M-CSF-1 receptor promoter to a bcl-2 promoter construct.41 42 Our results suggest that such activation could be indirect because we have shown that AML-1/ETO can specifically inhibit the function of a repressor protein, PLZF.
The mechanism through which AML-1/ETO inhibits PLZF function is complex. The simplest explanation is that AML-1/ETO antagonized PLZF by competing for corepressors. Consistent with this explanation, AML-1/ETO requires the MYND corepressor/HDAC-binding motif to antagonize PLZF. However, the addition of corepressors at doses that enhance repression by PLZF alone could not enhance repression by PLZF in the presence of low levels of transfected AML-1/ETO. The fact that AML-1/ETO inhibits both repression domains of PLZF is also consistent with a model in which AML-1/ETO interferes with corepressor/PLZF function through noncompetitive mechanisms, possibly through an aberrant complex formation. This association might be stable and would preclude the formation of effective repression complexes, supported by the strong co-precipitation and co-localization of AML-1/ETO and PLZF when co-expressed in 293T cells and the co-precipitation of a significant amount of AML-1/ETO with PLZF in SKNO cells.
Part of the inhibition may be related to the ability of AML-1/ETO to almost completely exclude PLZF from the nuclear matrix compartment, an effect that correlates well with the suppression of PLZF function. It has been suggested that PLZF associates with the nuclear matrix.38 Proper nuclear matrix targeting may be essential for the function of certain transcriptional factors, such as AML-1.43 AML-1/ETO lacks the nuclear matrix signal sequence of AML-1 and localizes to a distinct matrix subcompartment.44 These results suggest a novel functional requirement for discrete nuclear matrix compartmentalization in PLZF repression, a possibility we are exploring further. Finally, PLZF binding to DNA was partially inhibited by AML-1/ETO. However, AML-1/ETO also blocked repression by GAL4-PLZF, GAL4-BTB/POZ, and GAL4-RD2, though it did not inhibit GAL4-KRIP1, suggesting that the effect on PLZF binding may be specific to PLZF. It should be noted that PLZF represses transcription through the action of HDACs, whereas the KRIP protein relies on a different mechanism, potentially through interaction with nonhistone, chromatin-associated proteins.45 Hence, it remains possible that AML-1/ETO may inhibit the function of other proteins related to PLZF in structure or function, such as FAZF/TZFP/ROG46-48 or LRF.49 In conclusion, we propose that AML-1/ETO inhibits PLZF through several mechanisms. These include the formation of an aberrant protein complex with nuclear corepressors and HDACs (unfavorable for transcriptional repression), sequestration of PLZF away from the nuclear matrix, and inhibition of the ability of PLZF to bind to DNA.
There may be significant biologic consequences to the ability of AML-1/ETO to inhibit transcriptional repression by PLZF and other proteins. PLZF is a powerful growth suppressor in myeloid and other cells.26 This may occur in part by the inhibition of cell cycle regulators such as cyclin A2, delaying entry into and transit through the S phase.25,26 In APL associated with t(11;17) (q21;q23), the RARα/PLZF protein activates rather than represses PLZF target genes, potentially contributing to leukemogenesis. AML-1/ETO may, in a similar manner, de-repress the expression of genes normally kept in check by PLZF. This is consistent with the observation that progenitor cells from an AML-1/ETO knock-in model exhibited increased self-renewal capacity, as did adult murine hematopoietic cells transduced with an AML-1/ETO retroviral vector.15 This also raises for the first time the possibility that PLZF could play a role in the pathophysiology of non-APL leukemia.
To examine whether ETO could play a central role in multiple forms of leukemia, we determined its ability to interact with the PLZF/RARα oncoprotein. PLZF/RARα contains the first 455 amino acids of PLZF and, therefore, includes the RD2-ETO binding domain. As anticipated, the 2 proteins could interact when coexpressed in mammalian cells. Unfortunately, we were unable to confirm this interaction with endogenous protein because no cell lines derived from these patients are currently available. However, because ETO is normally expressed in early myeloid cells,50 it is likely that leukemic APL blasts express ETO. Transcriptional repression by PLZF/RARα through the RARE, was enhanced by ETO, whereas ETO had no effect on transcription by wild-type RARα or the PML/RARα fusion of RA-sensitive APL. Thus, ETO may be a corepressor, specific for the retinoid-resistant PLZF/RARα protein. This may further explain the potent ability of PLZF/RARα to inhibit the expression of RARα target genes, block cellular differentiation,51 and induce a refractory form of leukemia.52 53
In conclusion, we report novel modes of action for AML-1/ETO on the PLZF growth suppressor and for the ETO corepressor on the PLZF/RARα fusion protein. This extends the paradigm of aberrant transcriptional repression as a central mechanism of leukemogenesis. Inappropriate repression of genes normally activated during myelopoiesis by crucial factors, such as the core binding factor complex or RARα, may contribute to the differentiation block of leukemia. De-repression of cell cycle regulators by the creation of dominant-active proteins such as RARα/PLZF or dominant-negative inhibitors of repression such as AML-1/ETO may contribute to abnormal growth control. These data also suggest that indirect genetic targets of oncoproteins affected by cross-talk through corepressor networks may be important in leukemogenesis.
Acknowledgments
We thank Drs Arthur Zelent and Samuel Waxman for continued support and Dr Kathy Borden for helpful advice.
Supported by National Institutes of Health grants R01 CA59936 (J.D.L.) and R01 CA64140 (S.W.H.) and by American Cancer Society Award DHP 160 (J.D.L.). J.D.L. is a Scholar of the Leukemia and Lymphoma Society. A.M. is supported by NIH grant K08 CA73762. Confocal laser scanning microscopy was performed at the MSSM-CLSM core facility, supported with funding from NIH shared instrumentation grant 1S10 RR0 9145-01 and National Science Foundation Major Research Instrumentation grant DBI-9724504.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jonathan D. Licht, Dept of Medicine, Derald H. Ruttenberg Cancer Center, Mount Sinai School of Medicine, Box 1130, 1 Gustave Levy Pl, New York, NY 10029; e-mail: Jonathan.licht@mssm.edu.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal