Abstract
Mammalian CBFB encodes a transcription factor (CBFβ) that in combination with CBFα2 binds to specific DNA sequences and regulates expression of a number of hematopoietic genes.CBFB is associated with human leukemias through a chromosome 16 inversion and is essential for definitive hematopoiesis during mouse embryo development. We have isolated a zebrafishcbfb complementary DNA (cDNA) clone from a zebrafish kidney cDNA library. This cbfb is highly homologous to human and mouseCBFB/Cbfb genes at both the DNA and protein level. In biochemical analyses, cbfβ binds to human CBFα2 and enhances its DNA binding. During zebrafish development, cbfb is expressed in the lateral plate mesoderm at tail bud stage and in the intermediate cell mass (ICM, the location of embryonic hematopoiesis) between the 21- to 26-somite stages. The cbfb is also expressed in Rohon-Beard cells, cranial nerve ganglia, hindbrain, retina, branchial arches, jaw, and fin buds. Expression ofcbfb is decreased or absent in the ICM and Rohon-Beard cells in some hematopoietic mutants and is unaffected in others. We have also analyzed the expression of scl andgata-1 in the same hematopoietic mutants to ascertain the relative order of these transcription factors to cbfb in zebrafish hematopoiesis. Our results indicate that cbfb is expressed in early hematopoietic progenitors and that its expression pattern in the hematopoietic mutants is similar to that ofscl.
Introduction
Hematopoiesis is a complex and tightly regulated process with concordant expression of genes involved in differentiation, proliferation, and apoptosis.1-4 Recent studies have indicated the importance of a series of transcription factors during hematopoiesis. These transcription factors regulate expression of lineage-specific genes, and many of them are indispensable for embryonic and adult hematopoiesis.5Moreover, some of these transcription factors, when mutated through chromosomal translocations, contribute to the pathogenesis of hematologic malignancies.6
CBFB (also called PEBP2B) encodes a transcription factor (CBFβ) that plays important roles in hematopoiesis and in leukemia.7,8 CBFβ associates with a family of runt-domain–containing proteins, AML1-3/CBFα1-3/PEBP2αA-C/RUNX1-3 (referred to as CBFα proteins below), to form a DNA-binding heterodimer.9,10 CBFβ does not bind to DNA directly but enhances the DNA binding affinity of CBFα proteins. Mouse knockout models demonstrated that Cbfbis required for definitive hematopoiesis but not for primitive hematopoiesis during embryogenesis.11-13 Mouse embryos with mutations in both copies of the Cbfb gene lack definitive hematopoiesis in the liver, whereas yolk sac primitive hematopoiesis is relatively intact. The blockage of definitive hematopoiesis by Cbfb appears to be cell autonomous becauseCbfb−/− fetal liver and yolk sac cells fail to generate hematopoietic colonies in vitro. In addition,Cbfb−/− embryos develop central nervous system hemorrhage that is likely the reason for midgestation lethality of those embryos. CBFB is frequently mutated in acute myeloid leukemias, especially the M4Eo subtype, through a chromosome 16 inversion, inv(16)(p13q22), which generates a fusion gene betweenCBFB and MYH11 (a gene coding for the smooth muscle myosin heavy chain).14 This CBFB-MYH11fusion gene has been shown to dominantly suppress CBF functions and predispose mice to acute myeloid leukemia.15 16
Two CBFB homologs, called Brother (Bro) andBig brother (Bgb), exist inDrosophila.17 Similar to mammalian CBFβ proteins, Bro and Bgb interact with runt and lozenge, the Drosophila homologs of mammalian CBFα proteins, and are required for proper functions of runt and lozenge for sex determination and eye development.17 18 TheBro and Bgb genes presumably arose through a gene duplication, because they share extensive sequence homology to each other and are closely linked physically—about 8 kilobases (kb) apart in tandem orientation (Bgb is upstream) on the left arm of chromosome 3.
GATA-1 and SCL are 2 other transcription factors that play important roles in hematopoiesis. The zinc finger protein GATA-1 has been shown to be a central regulator of erythroid differentiation.19The cognate binding sequence (A/T)GATA(A/G) is present in the regulatory sequences of many erythroid-specific genes, and murine knockout experiments demonstrated that GATA-1 is specifically required for the maturation of proerythroblasts of both primitive and definitive hematopoiesis.20 On the other hand, SCL is a transcription factor with a basic helix-loop-helix motif and is expressed in stem cells or early progenitors as well as in erythroid cells, mast cells, and megakaryocytes.21 In murine knockout models, removal of Scl results in blockage of all lineages in both embryonic (primitive and definitive) and adult hematopoiesis.22 In vitro differentiation with embryonic stem cells confirmed thatScl is required for hematopoietic differentiation at the multipotent stem cell level.23 In addition, recent studies in zebrafish suggest that scl is expressed in the so-called “hemangioblasts,” which are capable of both hematopoietic and vascular differentiation.24
Animal model studies reported so far suggest that SCL is probably the earliest transcriptional regulator of hematopoiesis, whereas GATA-1 is a late transcription factor that determines specific lineages. CBFβ functions at the stem cell level in definitive hematopoiesis, becauseCbfb knockout mice have intact primitive hematopoiesis but all lineages of definitive hematopoiesis are blocked. However, the exact roles of these proteins during hematopoiesis and their relationship to each other remain unresolved.
Zebrafish is a vertebrate model organism that has attracted significant attention in recent years because of its many advantageous features for genetic and developmental studies.25,26 Zebrafish is particularly useful for studies of embryonic hematopoiesis because of the easy access and analysis of transparent external developing embryos. Moreover, the availability of many hematopoietic mutant fish lines,27 28 and the potential of generating additional mutants through random mutagenesis, makes it possible to dissect hematopoietic pathways with unprecedented depth and detail.
Here we report the cloning of a zebrafish cbfb gene. The zebrafish cbfb gene is more similar to mammalianCBFB/Cbfb genes than to Drosophila Bro/Bgb genes at both DNA and protein levels. The encoded zebrafish cbfβ protein binds to human CBFα2 and enhances its DNA binding as efficiently as human CBFβ. The expression ofcbfb during fish embryo development was analyzed, both in wild-type embryos and in hematopoietic mutants, showing thatcbfb is expressed in early hematopoietic cells during zebrafish embryogenesis.
Material and methods
Zebrafish strains and maintenance
Zebrafish were raised and handled as described,29under an approved National Institutes of Health animal use protocol. Fish of the wild-type strain ekw30 were used for the production of wild-type embryos for in situ hybridization and RNA isolation. Three mutants were used in the study: cloche, vampire, and m683. Cloche andvampire have been described27,31;m683 is a recessive lethal and fully penetrant bloodless mutation (with a similar phenotype to vampire) that was isolated during the Boston large-scale mutagenesis effort32but not characterized at that time (B.M.W., unpublished results, 1996).
Cloning and sequencing of zebrafish cbfb cDNA and promoter
Random-primed and oligo-dT–primed zebrafish kidney phage complementary DNA (cDNA) libraries24 were screened with the32P-labeled 250-base pair (bp)SalI/EcoRI fragment from CK9, a humanCBFB cDNA clone.33 One positive clone was identified from the random primed library. The λ clone was excised using the Stratagene Rapid Excision Kit (Stratagene, La Jolla, CA) and sequenced. Genomic clones were obtained by hybridization of the zebrafish cbfb cDNA clone to the zebrafish PAC filters (The Resource Center/Primary Database of the German Human Genome Project, Berlin, Germany). Two positive PAC clones were isolated, BUSMP706O13197Q3 (PAC 197) and BUSMP706I09152Q3 (PAC 152). Sequence for the 5′ flanking region was obtained by sequencing PAC 197 using a primer (5′-TCCTGAAGAACTCCTCGTTC-3′) designed from exon 1 of the zebrafish cbfb cDNA clone.
Linkage group assignment
The T51 radiation–reduced zebrafish/hamster hybrid panel (Research Genetics, Huntsville, AL) was used to map cbfb by polymerase chain reaction (PCR).34 PCR was performed with an annealing temperature of 60°C and 31 cycles of amplification using the forward primer ATAAGAATGCGGCCGCTAACTATGGGAAGTGGGAACATATCCAACC and the reverse primer GGACTAGTCCAGAAGAGTGAAGCGTTGCCTTG. The PCR results were sent to Robert Geisler at MPI fuer Entwicklungsbiologie, Tuebingen, Germany, for linkage group assignment.
In situ hybridization
Zebrafish embryos were dechorionated with protease (P8811, Sigma, St Louis, MO) prior to fixation. The probe for hybridization was generated by digestion of the cbfb cDNA withEcoRI and incorporation of digoxigenin-UTP by in vitro transcription with T7 polymerase using DIG RNA Labeling Mix (Boehringer Mannheim, Indianapolis, IN). In situ hybridization was performed according to standard procedure29 at 55°C with the following modifications: Block solution was 2% blocking reagent (Boehringer Mannheim) with 5% lamb serum (Life Technologies, Rockville, MD) in 0.1 mol/L maleic acid and 150 mmol/L NaCl, pH 7.5, and signal was detected with BM Purple AP Substrate (Boehringer Mannheim). Zebrafish gata-1 and scl probes have been described.24 35
For hematopoietic mutants, embryos produced by crosses between heterozygous males and females were used for in situ hybridization. At least 20 embryos were scored in each hybridization, and the results were consistent with approximately 25% of the embryos being homozygous mutants.
Gel electrophoretic mobility shift assay
The 18-bp fragment containing the high-affinity (HA) core site33 was prepared by annealing 2 oligonucleotides (5′-GGATATTTGCGGTTAGCA-3′ and 5′-TGCTAACCGCAAATAT-3′) such that 2 Gs were left protruding at the 5′ end of the sense strand in the double helix. The HA-containing oligo was32P-labeled by filling in the ends in the presence of [α-32P] dCTP and Klenow enzyme. The labeled DNA was purified with the G25 spin column (5Prime → 3Prime, Inc, Boulder, CO) and used for gel shift analysis. Proteins used in the assays were in vitro–translated in the absence of any radioisotope using the Promega in vitro translation kit. The DNA binding reaction typically consisted of approximately 5 ng (10 000 cpm) of radiolabeled DNA fragment, 0.5 to 1 μg of poly dI-dC, and 5 to 10 μL of in vitro–translated proteins in 1 × binding buffer.16 36The binding reaction was incubated for 30 minutes at room temperature and then electrophoresed in a 6% polyacrylamide gel. The gel was subsequently dried and autoradiographed.
Immunoprecipitation
CK933 and cbfb plasmids were translated in vitro with 35S-labeled methionine using the Promega in vitro translation kit (Promega, Madison, WI); pCbfa216 was translated in vitro without radioactive methionine. Immunoprecipitation was carried out as described previously.16 Briefly, protein A Sepharose-plus (Pierce, Rockford, IL) was washed with phosphate-buffered saline (PBS) and then blocked with 2% bovine serum albumin in phosphate-buffered saline. A total of 30 μL of the blocked protein A was incubated with antibody α3043 for 2 hours at 4°C on a rotator and then washed twice with Triton buffer. The protein A beads were incubated overnight at 4°C with 35S-labeled human CBFβ and zebrafish cbfβ in the presence or absence of unlabeled in vitro–translated CBFα2. The Sepharose beads were collected by centrifugation and washed with Triton buffer 4 times. The beads were resuspended and boiled for 5 minutes in 25 μL of 2 × sodium dodecyl sulfate buffer. A total of 10 μL of this buffer containing the eluted proteins was analyzed by denaturing polyacrylamide gel electrophoresis and autoradiography.
Immunochemical staining of fish embryos
Antibody staining was performed as described37subsequent to in situ hybridization. Briefly, after in situ hybridization, embryos were washed several times and incubated for more than 1 hour in blocking solution (5% goat serum in PBT [PBS with 0.1% Tween-20]). Embryos were then incubated with a mouse anti–HNK-1 antibody (anti–HNK-1/N-CAM; Sigma) overnight at 4°C. Washes with PBT were performed at room temperature for 3 times at 5 minutes each and 3 times at 30 minutes each. Secondary antibody staining was carried out using biotinylated antimouse IgM and the Vectastain avidin/biotin/horseradish peroxidase ABC Elite System (Vector Laboratories, Burlingame, CA). Embryos stained with antibody were kept in 70% glycerol.
Results
Isolation of zebrafish cbfb cDNA
A random-primed zebrafish kidney cDNA library24 was screened by filter hybridization with a human CBFB cDNA probe CK9 under low-stringency conditions. One positive clone was identified from the library, which contains a 2.7-kb insert that was fully sequenced. Sequence analysis revealed an open-reading frame encoding a protein of 187 amino acids, the same size as that encoded by the longest alternatively spliced transcript of human CBFB. Sequence comparison analysis indicates that the deduced protein sequence is very similar to those of the human and mouse CBFβ proteins (87% identity) and, to a lesser degree, theDrosophila Brother and Big Brother proteins (around 47% identity) (Figure 1). Therefore, this cDNA contains a zebrafish cbfb gene.
Protein sequence comparison.
Sequences of zebrafish (zcbfb), human (hCBFb), mouse (mCbfb) CBFβ proteins, and the Drosophila Bro and Bgb proteins are compared using the ClustalW program. Amino acid identities are in black boxes, and conserved amino acid changes are in shaded areas. The GenBank accession number for human CBFβ is AF294326, for mouse Cbfβ is Q08024, for Drosophila Bro is U22176, and for Bgb isU22177. Additional N-terminal sequences of Bro and Bgb are from Golling et al.17
Protein sequence comparison.
Sequences of zebrafish (zcbfb), human (hCBFb), mouse (mCbfb) CBFβ proteins, and the Drosophila Bro and Bgb proteins are compared using the ClustalW program. Amino acid identities are in black boxes, and conserved amino acid changes are in shaded areas. The GenBank accession number for human CBFβ is AF294326, for mouse Cbfβ is Q08024, for Drosophila Bro is U22176, and for Bgb isU22177. Additional N-terminal sequences of Bro and Bgb are from Golling et al.17
cDNA sequence analysis
Of the total 187 amino acid positions of the cbfβ protein sequence, 163 are identical to those of the longest splicing variant of the human and the mouse CBFβ proteins. Among the 24 amino acids that are different between human and zebrafish, 10 are conserved changes. Most of these 24 amino acids are clustered in 3 regions of the protein: 8 between amino acids 72 and 92, 3 between amino acids 163 and 166, and 5 between amino acids 178 and 187. Interestingly, amino acids 72 to 92 encompass a loop between β-sheets 3 and 4,38 where there is virtually no sequence homology among different species (Figure 1). Amino acids 163 to 166 and 178 to 187 are in the C-terminal region of the protein that is not required for binding with CBFα proteins.10 39 Therefore, these amino acid differences are not expected to affect the ability of the zebrafish cbfβ protein to bind with CBFα. The overall sequence similarities between the zebrafish protein and the Drosophila Brother and Big brother proteins are much reduced. The amino acid sequence similarities between the Drosophila proteins and those of the vertebrates are mostly clustered in amino acids 1 to 68 and 87 to 140 (Figure 1), suggesting that those regions of the protein are crucial for its function.
At the DNA level, cbfb is also highly similar to the humanCBFB gene (76% identical) and to the mouse Cbfbgene (78% identical) in the coding region. The isolated cDNA clone also contains about 330 bp 5′- and 1.8 kb 3′-untranslated region (UTR) sequences. The 5′- and 3′-UTR sequences do not contain obvious homology to the human 5′- and 3′-UTR sequences, although both human and zebrafish 3′-UTRs are very long and AT-rich (67% AT for the human gene and 63% for the zebrafish gene).
Additional 5′ upstream sequence of the cbfb gene was obtained by sequencing a PAC clone that was isolated by screening a zebrafish PAC library with the cbfb cDNA probe. The PAC clone (PAC 197) was sequenced for over 1 kb by primer-walking starting from exon 1. In total, 836 bp 5′ from the beginning of the cDNA was obtained by sequencing this PAC (data not shown). A cluster of tetranucleotide repeats, including (CCAT)12, (CTAT)8, (CTAT)17, and (CTGT)18, was discovered between −836 and −517 bp from the beginning of the cDNA. Comparison of the 5′ flanking sequences in different species did not reveal significant sequence similarity between zebrafishcbfb and the human and the mouse CBFB/Cbfb genes (data not shown).
Chromosome mapping
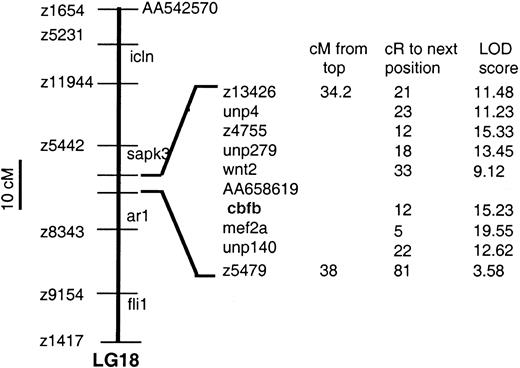
The T51 zebrafish hybrid mapping panel34 was typed by PCR designed to amplify the 5′-UTR of cbfb. Analysis of the PCR data indicates that cbfb is located between 34.2 and 38 cM from the top of linkage group 18, with a significant LOD score of 15.23 (a LOD score > 6 indicates significant linkage). LOD scores and relative positions of genes in the region are shown in Figure2. The flanking genes for cbfbare wnt2 and mef2a (12 and 5 cR fromcbfb, respectively). The human CBFB gene is located on chromosome 16,14 whereas the human homologs ofwnt2 and mef2a are located on chromosomes 740 and 15,41 respectively. Therefore, no apparent synteny exists between this region of the zebrafish genome and a corresponding region of the human genome.
Map location of
cbfb in zebrafish linkage group 18. Thecbfb is located between wnt2 and mef2ausing a radiation hybrid panel.
Map location of
cbfb in zebrafish linkage group 18. Thecbfb is located between wnt2 and mef2ausing a radiation hybrid panel.
Zebrafish cbfβ protein associates with mammalian CBFα2
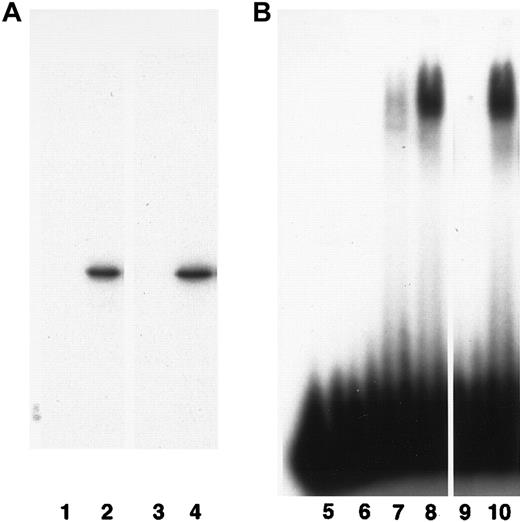
Mammalian CBFβ and Drosophila Bro and Bgb proteins are able to bind mammalian CBFα proteins and enhance their DNA binding affinity.9,10 17 To test if the zebrafish cbfβ protein has a similar function, we produced zebrafish cbfβ protein by in vitro translation from the cbfb cDNA clone and used it in immunoprecipitation and electrophoretic mobility shift assays. In these assays cbfβ was able to associate with CBFα2 and enhance its DNA binding affinity as efficiently as the human CBFβ protein (Figure3). Therefore, zebrafish cbfβ protein functions similarly to human CBFβ with respect to association with CBFα.
Biochemical properties of cbfβ protein.
(A) The cbfβ associates with human CBFα2. 35S-labeled in vitro–translated human CBFβ (lanes 1 and 2) and zebrafish cbfβ (lanes 3 and 4) proteins were immunoprecipitated by an anti-CBFα2 antibody in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of the CBFα2 protein and separated by electrophoresis. (B) The cbfβ enhances DNA binding by CBFα2. A 32P-labeled oligonucleotide (HA) containing the recognition site for CBF was incubated with the in vitro–translated proteins and separated by electrophoresis. H2O control, lane 5; in vitro translation lysate control, lane 6; human CBFα2 alone, lane 7; human CBFα2 plus CBFβ, lane 8; zebrafish cbfβ alone, lane 9; human CBFα2 plus zebrafish cbfβ, lane 10.
Biochemical properties of cbfβ protein.
(A) The cbfβ associates with human CBFα2. 35S-labeled in vitro–translated human CBFβ (lanes 1 and 2) and zebrafish cbfβ (lanes 3 and 4) proteins were immunoprecipitated by an anti-CBFα2 antibody in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of the CBFα2 protein and separated by electrophoresis. (B) The cbfβ enhances DNA binding by CBFα2. A 32P-labeled oligonucleotide (HA) containing the recognition site for CBF was incubated with the in vitro–translated proteins and separated by electrophoresis. H2O control, lane 5; in vitro translation lysate control, lane 6; human CBFα2 alone, lane 7; human CBFα2 plus CBFβ, lane 8; zebrafish cbfβ alone, lane 9; human CBFα2 plus zebrafish cbfβ, lane 10.
Expression pattern in wild-type fish embryos
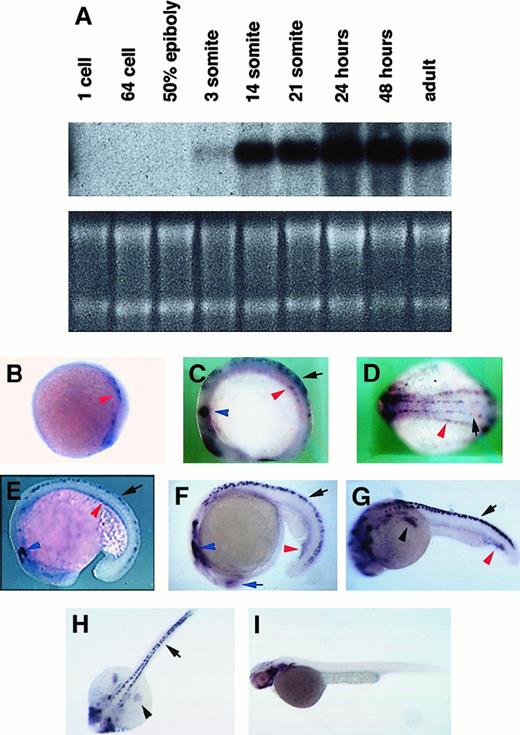
Expression of cbfb during zebrafish embryogenesis was analyzed by Northern blot hybridization and RNA in situ hybridization. By Northern blot hybridization, cbfb expression was observed starting at the 3-somite stage, continued through at least 2 days postfertilization, and was also detected in adults (Figure4A). The size of the detected transcript is 5 kb, indicating that our cDNA is not complete and suggesting thatcbfb contains additional 5′ or 3′ flanking sequences, similar to the human CBFB gene14 (Karla Henning and P.P.L., unpublished results, 2000).
Expression of
cbfb in zebrafish embryos. (A) Northern blot hybridization with 32P-labeled cbfb cDNA. RNA quantities in each lane of the Northern blot are shown by ethidium bromide staining of the gel. (B-I) In situ hybridization with digoxigenin-labeled cbfb antisense RNA probe on embryos of 10 hpf (B), 10-somite/14 hpf (C,D for lateral and dorsal views), 18-somite/18 hpf (E), 21-somite/19.5 hpf (F), 24 hpf (G,H for lateral and dorsal views), and 2 days postfertilization (I). Expression ofcbfb in Rohon-Beard cells are labeled with black arrows, in lateral plate mesoderm and ICM with red arrowheads, in fin buds with black arrowheads, in trigeminal ganglion with blue arrowheads, and in the retina with blue arrows. Embryos are all oriented with anterior to the left and posterior to the right. For the lateral views, the dorsal side of the embryos is in the upper part of the picture and the ventral is lower. Embryos in the subsequent figures are all oriented the same way.
Expression of
cbfb in zebrafish embryos. (A) Northern blot hybridization with 32P-labeled cbfb cDNA. RNA quantities in each lane of the Northern blot are shown by ethidium bromide staining of the gel. (B-I) In situ hybridization with digoxigenin-labeled cbfb antisense RNA probe on embryos of 10 hpf (B), 10-somite/14 hpf (C,D for lateral and dorsal views), 18-somite/18 hpf (E), 21-somite/19.5 hpf (F), 24 hpf (G,H for lateral and dorsal views), and 2 days postfertilization (I). Expression ofcbfb in Rohon-Beard cells are labeled with black arrows, in lateral plate mesoderm and ICM with red arrowheads, in fin buds with black arrowheads, in trigeminal ganglion with blue arrowheads, and in the retina with blue arrows. Embryos are all oriented with anterior to the left and posterior to the right. For the lateral views, the dorsal side of the embryos is in the upper part of the picture and the ventral is lower. Embryos in the subsequent figures are all oriented the same way.
By in situ hybridization, cbfb expression was observed in embryos of 10 hours postfertilization (hpf), as 2 stripes along the lateral plate mesoderm in both the anterior and posterior ends of the embryo body (Figure 4B, red arrowhead). By the 10-somite stage, 2 pairs of symmetrically distributed cbfb expression stripes were observed along the anterior-posterior axis of the embryo (Figure 4C-D). The inside pair of expression stripes is in the Rohon-Beard cells (black arrow; see below for more discussion), whereas the outside pair of expression stripes is in the lateral plate mesoderm (red arrowhead).
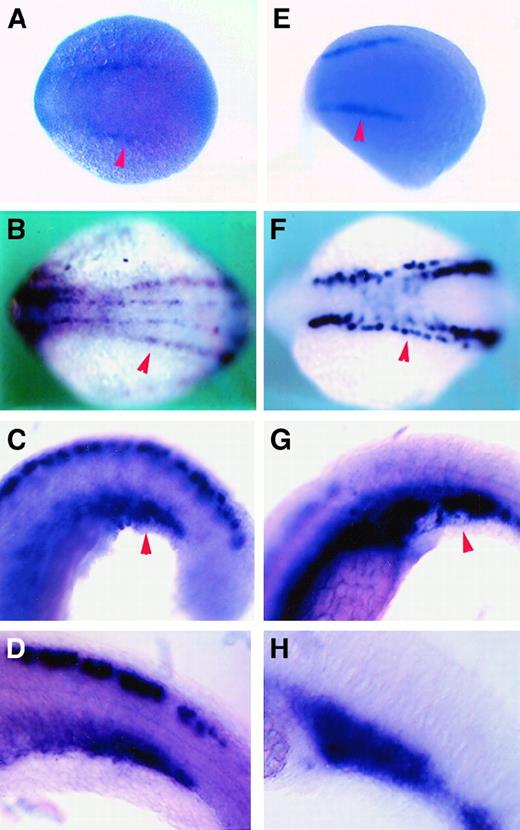
This expression of cbfb in the lateral plate mesoderm is very similar to the expression pattern of scl at these early stages of development, and these mesoderm stripes have been suggested to contain precursors of hematopoietic cells24 (Figure5A,B,E,F, red arrowheads).
Expression pattern comparison between
cbfb and scl by RNA in situ hybridization. (A-D) The cbfb antisense RNA probe; (E-H) scl antisense RNA probe. (A,E) Embryos at 10 hpf; (B,F) 10-somite/14 hpf embryos; (C,G) 21-somite/19.5 hpf embryos; and (D,H) 24-somite/21 hpf embryos. Red arrowheads point to lateral plate mesoderm in panels A, B, E, and F; ICM in panels C and G.
Expression pattern comparison between
cbfb and scl by RNA in situ hybridization. (A-D) The cbfb antisense RNA probe; (E-H) scl antisense RNA probe. (A,E) Embryos at 10 hpf; (B,F) 10-somite/14 hpf embryos; (C,G) 21-somite/19.5 hpf embryos; and (D,H) 24-somite/21 hpf embryos. Red arrowheads point to lateral plate mesoderm in panels A, B, E, and F; ICM in panels C and G.
By 21 to 24 hours, cbfb expression was detected in the intermediate cell mass (ICM), a structure formed when the lateral plate mesoderm from each side of the embryo converges to the midline, where hematopoiesis takes place during this period of embryo development42 (Figure 4E-G, red arrowheads). Expression ofcbfb was stronger in the posterior region of the ICM and was detected in a subset of large, round cells that also expressedscl (Figure 5C,D,G,H). The morphology of the cells suggests that they are hematopoietic.
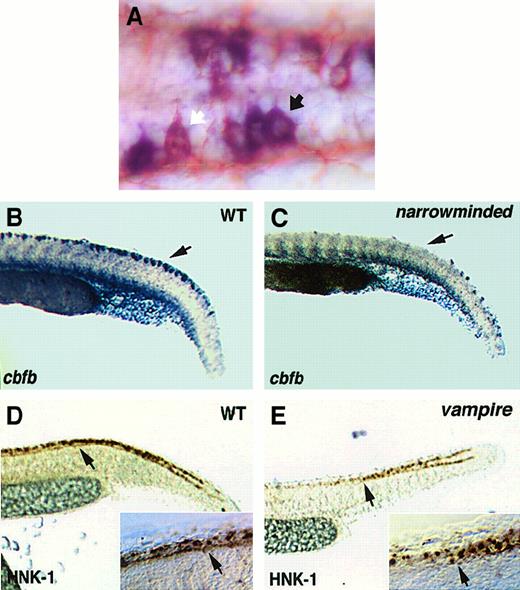
From 10-somite stage to 24 hpf, nervous system expression of cbfbwas observed in the retina (Figure 4F, blue arrow), 3 pairs of cranial nerve ganglia (Figure 4C,E,F, blue arrowheads), and a subset of cells within bilateral stripes of the neural tube, extending from the hindbrain to the tail (Figure 4C-H, black arrows). These bilateral stripes were shown to be Rohon-Beard sensory neurons by immunostaining for HNK-1, a neuronal marker, and examining the morphology of the stained cells (Figure 6A). The cbfbis expressed at higher levels in some Rohon-Beard sensory neurons than others (Figure 6A). Expression of cbfb in the Rohon-Beard cells was decreased in narrowminded (Figure6B,C), a mutant with reduced number of early neural crest cells and Rohon-Beard cells.43 In addition, cbfbexpression was observed in the pectoral fin buds by 24 hours (Figure4G,H, black arrowheads).
Analysis of Rohon-Beard cells.
(A) A higher-magnification view of a 24-hpf embryo hybridized with thecbfb antisense RNA probe and immunostained with an anti–HNK-1 antibody. The white arrow indicates a Rohon-Beard cell with lower level of cbfb expression, whereas the black arrow indicates a Rohon-Beard cell with higher level of cbfb expression. (B,C) Expression of cbfb in Rohon-Beard cells of wild-type andnarrowminded embryos, respectively. Black arrows point to the location of Rohon-Beard cells. (D,E) HNK-1 staining of Rohon-Beard cells in wild-type and vampire embryos, respectively. The arrows point to the interruption of Rohon-Beard cells at the level of the end of yolk sac extension. The insets at the lower right corner of panels D and E show close-up views of the Rohon-Beard cells as large neurons with horizontal axons, which do not extend further anteriorly after the place indicated by the arrowhead in the vampiremutant embryo.
Analysis of Rohon-Beard cells.
(A) A higher-magnification view of a 24-hpf embryo hybridized with thecbfb antisense RNA probe and immunostained with an anti–HNK-1 antibody. The white arrow indicates a Rohon-Beard cell with lower level of cbfb expression, whereas the black arrow indicates a Rohon-Beard cell with higher level of cbfb expression. (B,C) Expression of cbfb in Rohon-Beard cells of wild-type andnarrowminded embryos, respectively. Black arrows point to the location of Rohon-Beard cells. (D,E) HNK-1 staining of Rohon-Beard cells in wild-type and vampire embryos, respectively. The arrows point to the interruption of Rohon-Beard cells at the level of the end of yolk sac extension. The insets at the lower right corner of panels D and E show close-up views of the Rohon-Beard cells as large neurons with horizontal axons, which do not extend further anteriorly after the place indicated by the arrowhead in the vampiremutant embryo.
By 2 days postfertilization (Figure 4I), the expression ofcbfb in the Rohon-Beard cells has disappeared, and neuronal expression can be observed in the retina, forebrain, and hindbrain. Thecbfb is also expressed in branchial arches and the jaw at this time of development.
Expression pattern in hematopoietic mutants
To verify the expression of cbfb during zebrafish hematopoiesis, the expression pattern of cbfb in several blood mutants was analyzed by in situ hybridization using embryos at 24 hpf.
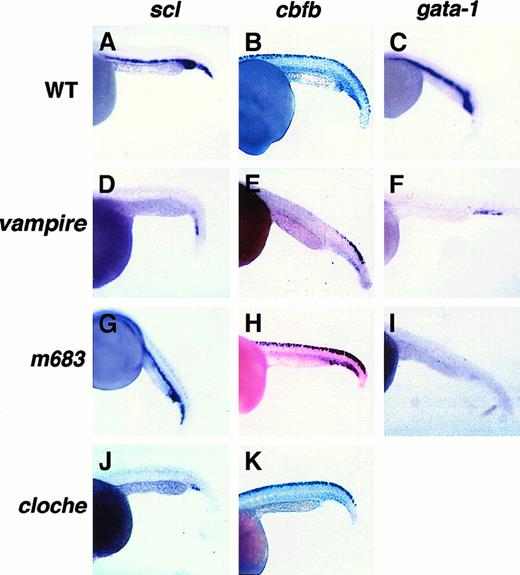
Three mutants were used in the study: cloche (m378), vampire (m62), and m683. Cloche is a recessive lethal mutation with defects in both hematopoiesis and vasculogenesis.31Vampire and m683are recessive lethal mutations that result in no or very few blood cells during embryo development, whereas blood vessels and other tissues formed normally27 (B.M.W., unpublished results, 1998). Recent studies suggest that the clochemutation results in an early blockage at the hemangioblast level.24 44 Even though there have been no published in-depth studies on mutants vampire and m683, their early bloodless phenotype suggests defects at the stem/progenitor cell level.
Expression of both scl and gata-1 was lost in the ICM of the vampire mutant embryos except for a few cells in the posterior ICM (Figure 7D,F). In them683 mutant embryos, scl expression was retained whereas gata-1 expression was lost (Figure 7G,I). We also confirmed the previously published findings in the clochemutant that expression of both scl and gata-1 is absent from the anterior and trunk ICM, whereas a few scattered cells persist in the posterior ICM (Figure 7J and data not shown).
Expression of
scl, cbfb, and gata-1 in zebrafish hematopoietic mutants. Representative whole-mount RNA in situ hybridization results with digoxigenin-labeled antisense RNA probes for scl, cbfb, and gata-1 on 24-hpf embryos of the indicated genotypes are shown in the corresponding panels.
Expression of
scl, cbfb, and gata-1 in zebrafish hematopoietic mutants. Representative whole-mount RNA in situ hybridization results with digoxigenin-labeled antisense RNA probes for scl, cbfb, and gata-1 on 24-hpf embryos of the indicated genotypes are shown in the corresponding panels.
Similar to scl, expression of cbfb was significantly decreased in the ICM of the cloche andvampire mutant embryos, except for a few scattered cells in the posterior ICM (Figure 7E,K), but was unaffected in m683(Figure 7H). Expression of cbfb in other tissues was not affected in the cloche mutant embryos (Figure 7K). On the other hand, cbfb expression in the trunk Rohon-Beard cells was lost in the vampire mutant embryos (Figure 7E). To determine if this loss of cbfb expression is due to change in transcriptional regulation or loss of Rohon-Beard cells,vampire embryos were stained with the neuronal marker HNK-1. As shown in Figure 6D-E, the number of Rohon-Beard cells was decreased in the trunk region of the vampire mutant embryo, whereas their number remained the same in the caudal region. This pattern is consistent with that of cbfb expression in this mutant, suggesting that trunk Rohon-Beard cells are selectively lost in this mutant.
Discussion
In this study we isolated the zebrafish cbfb gene and analyzed its expression pattern during embryogenesis, both in wild-type and in several hematopoietic mutants. Sequence analysis revealed a very high level of sequence identity between cbfβ and mammalian CBFβ proteins. Importantly, the sequence comparison analysis confirmed the existence of domains in the proteins that are highly conserved in diverse species of the animal kingdom. Functional similarity between zebrafish cbfβ and CBFβ/Bro proteins in other species was demonstrated by the ability of zebrafish cbfβ to bind CBFα2 and increase its binding affinity for target DNA sequence in immunoprecipitation and gel shift assays.
Embryonic expression of the mammalian CBFB/Cbfb genes has not been studied in detail. In the only reported study ofCbfb expression during mouse embryonic development,Cbfb was found to be expressed in the telencephalon, cranial nerve ganglia, and the dorsal root ganglia in 11.5 days postcoitum embryos.11 In addition, faint expression in the fetal liver was observed at 11.5 days postcoitum. Expression ofCbfb during other stages of embryonic development was not analyzed. In this study we examined the expression of cbfbduring zebrafish embryonic development at several stages. We found thatcbfb is expressed in the organs of zebrafish that are anatomically or developmentally similar to the murine organs that express Cbfb. These include the brain, cranial nerve ganglia, and the Rohon-Beard cells (which are related to neural crest cells that gave rise to both dorsal root and cranial nerve ganglia). Expression of cbfb was also found in the fin buds, retina, branchial arches, and the jaw, suggesting potential roles ofcbfb in the development of those tissues.
More importantly, expression of cbfb in hematopoietic tissues was observed clearly, starting in the lateral plate mesoderm, where hematopoietic precursors are formed, to the ICM, where embryonic hematopoiesis takes place. The observation that such cbfbexpression in ICM was lost or decreased in known hematopoietic mutants confirmed that cbfb was expressed in the hematopoietic cells. In addition, cbfb might be expressed in endothelial cells in the ICM region, based on the distribution ofcbfb-expressing cells in ICM.
The expression of cbfb in Rohon-Beard cells is very prominent in embryos at 24 hpf. Rohon-Beard cells are sensory neurons that are present only during embryogenesis and young adult life. Not present in mammalian species, Rohon-Beard cells are closely related developmentally to neural crest cells in higher vertebrates.43 In this regard, it is interesting thatxaml, a Xenopus homolog of the humanCBFA2 gene, is also expressed in the Rohon-Beard cells.45,46 This would suggest that CBF genes play some role in the development of Rohon-Beard cells. Dorsal root and cranial nerve ganglia (both neural crest cell derivatives) are also sites of murine Cbfb expression and sites of frequent hemorrhage inCbfb−/− embryos.11 Earlier studies suggested that lack of CBF function led to necrosis in the dorsal root and cranial nerve ganglia, which may result in damage to blood vessel walls.11 Recently published results also indicate that CBF proteins may play important roles in angiogenesis during embryo development.47
An interesting finding in this study is the concordant loss ofcbfb expression in ICM and Rohon-Beard cells in the hematopoietic mutant vampire. The loss of cbfbexpression in Rohon-Beard cells in mutant vampire is surprising because Rohon-Beard cells were not known to play any role during hematopoiesis. One explanation for our observation is that thevampire gene controls cbfb expression in both ICM and the Rohon-Beard cells. Another explanation is that thevampire gene controls development and growth/survival of both cell types. Our data showing that trunk Rohon-Beard cells are lost in the vampire mutant support the second explanation.
Studying gene expression in zebrafish hematopoietic mutants can potentially subdivide those mutants even though they share the same bloodless phenotype. The mutations in cloche andvampire likely affect genes required at early stages of hematopoiesis, because expression of all 3 genes studied here are abolished or decreased. On the other hand, the mutation inm683 likely affects a lineage-specific gene becausescl and cbfb expression is unaffected in this mutant, whereas gata-1 expression is missing.
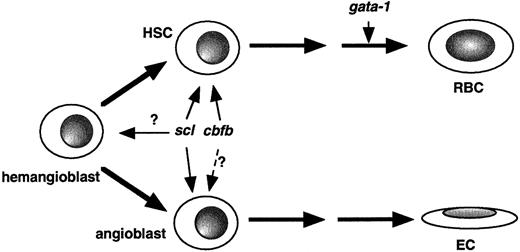
Based on the findings in this paper, we can propose thatcbf genes are involved in the early stages of hematopoiesis, at a similar level as that for scl (Figure8). Studying additional hematopoietic mutants may help separate scl and cbfb. The expression of cbfb in the lateral plate mesoderm in early fish embryos and the expression pattern of cbfb in the ICM also suggest that cbfb might be expressed in hemangioblasts and endothelial cells as well. This needs to be further explored by studying cbfb expression in angiogenesis mutants.
Model of
cbfb function in embryonic hematopoiesis. Based on results described in this study and published earlier,cbfb can be placed at the hematopoietic stem cell (HSC) level, similar to scl, whereas gata-1 is involved in later steps during hematopoiesis. Previous studies and expression pattern ofcbfb reported here also suggest that cbfb is expressed in hemangioblasts and might play a role in angiogenesis. RBC indicates red blood cell; EC, endothelial cell.
Model of
cbfb function in embryonic hematopoiesis. Based on results described in this study and published earlier,cbfb can be placed at the hematopoietic stem cell (HSC) level, similar to scl, whereas gata-1 is involved in later steps during hematopoiesis. Previous studies and expression pattern ofcbfb reported here also suggest that cbfb is expressed in hemangioblasts and might play a role in angiogenesis. RBC indicates red blood cell; EC, endothelial cell.
In conclusion, we have identified the zebrafish cbfbgene that shares very high sequence identity with the mammalianCBFB/Cbfb genes. The zebrafish cbfb gene is expressed in hematopoietic tissues during zebrafish embryonic development as well as in Rohon-Beard cells and several other tissues. The results of gene expression patterns in hematopoietic mutants help to dissect the mechanisms of embryonic hematopoiesis further and reveal unexpected connections between hematopoiesis and neuronal development.
Acknowledgments
We thank Karla Henning for the sequence of the humanCBFB promoter region; Susan Lyons, Bixiong Xu, and other lab members for helpful discussions and technical assistance; Pachiappan Manickam for the zebrafish RNA blot; and Alan Davidson for helpful discussions and for sharing scl expression data. We thank David Bodine for critical reading of the manuscript and Robert Geisler for radiation hybrid mapping data analysis.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Pu Paul Liu, NHGRI, NIH, 49 Convent Dr, Rm 3A18, Bethesda, MD 20892; e-mail: pliu@nhgri.nih.gov.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal