Abstract
An imbalance between cellular apoptosis and survival may be critical for the pathogenesis of lymphoma. Therefore, the gene expression pattern in lymph node preparations from patients with mantle cell lymphoma (MCL) was compared to the pattern in nonmalignant hyperplastic lymph nodes (HLs). Oligonucleotide microarray analysis was performed comparing 5 MCLs to 4 HLs using high-density microarrays. The expression data were analyzed using Genespring software. For confirmation, the expression of selected genes was analyzed by real-time polymerase chain reaction using the RNA extracted from 16 MCL and 12 HL samples. The focus was on 42 genes that were at least 3-fold down-regulated in MCL; in addition to the B-cell leukemia 2 (BCL2) system other apoptotic pathways were altered in MCL. The FAS-associated via death domain (FADD) gene that acts downstream of the FAS cascade as a key gene to induce apoptosis was more than 10-fold down-regulated in MCL. Furthermore, the death-associated protein 6(DAXX) gene, the caspase 2 (CASP2) gene, and the RIPK1 domain containing adapter with death domain(RAIDD) gene, which are key genes in other proapoptotic pathways, were also decreased in the MCL samples. The suggestion is made that in addition to the known overexpression of cyclin D1, which drives entry into the cell cycle, disturbances of pathways associated with apoptosis contribute to the development of MCL.
Introduction
Mantle cell lymphoma (MCL) is a subgroup of moderately aggressive B-cell lymphomas that seems incurable with available treatments. In addition to morphologic and immunologic features as proposed by the European Lymphoma Task Force (ELTF)1 and the Revised European American Lymphoma (REAL) classification,2 detection of the rearrangement and overexpression of the cyclin D1 (CCND1, BCL1/PRAD1) gene3 is used for characterization of MCL.4 CCND1 controls the progression from the G1 to the S phase of the cell cycle by binding to cyclin-dependent kinases (CDK4 and CDK6). This complex directly phosphorylates the retinoblastoma (RB) protein causing the release of the E2F family of transcription factors.5 BecauseCCND1 is not highly expressed in either normal lymphocytes or other low-grade non-Hodgkin lymphomas,6 overexpression of CCND1 in MCL suggests an important role for it in the pathogenesis of this lymphoma. CCND1 is associated with the development of several types of solid tumors7,8 and transfection experiments have shown that CCND1 may interact with oncogenes such as c-myc and ras.9 AlthoughCCND1 may be thought of as an oncogene, overexpression ofCCND1 alone seems not to be sufficient to promote malignant transformation.10 Additional abnormalities are required. We set out to find these additional abnormalities.
Microarray analysis11 has been recently developed as a highly efficient method of gene expression profiling.46 Recently, the microarray technique was used by several groups to subclassify malignant diseases including high-grade lymphoma.12-20
To expand the understanding of altered pathways in MCL, we performed oligonucleotide microarray analysis on well-characterized lymph node samples. All MCL samples used for our analyses had the morphologic and immunologic features of MCL and overexpressed CCND1. As a control, we investigated gene expression in nonmalignant hyperplastic lymph nodes (HLs). We used 2 different strategies for analysis of changes in gene expression in the tumor cells. First, we examined the expression of genes that are known to be involved in the pathogenesis of other hematologic malignancies and genes involved in cell cycle control and in signal transduction. Second, we focused on highly differentially expressed genes in MCL compared to the control normal HLs. In addition to the B-cell leukemia 2 (BCL2) pathway, we found that other apoptotic pathways were altered in MCL. The microarray results were confirmed by real-time polymerase chain reaction (PCR) and immunohistochemistry in a larger set of clinical samples.
Materials and methods
RNA isolation
Lymph nodes from 16 patients with MCL and 12 individuals with HLs due to nonmalignant causes were investigated. All morphologic diagnoses were confirmed by immunohistochemistry and CCND1 overexpression was detected in all of the MCL samples. The tissue sections were reviewed by 2 independent specialists. Total RNA was extracted from microdissected tumor cells from quick-frozen lymph nodes using the acid guanidium/phenol/chloroform method with minor modifications.21 In addition, RNA was purified by the RNeasy cleanup system (Qiagen, Valencia, CA) according to the manufacturer's protocol.
Oligonucleotidemicroarray
The detailed protocol for the sample preparation and microarray processing is available from Affymetrix (Santa Clara, CA). Briefly, at least 8 μg purified RNA was reverse transcribed by Superscript II reverse transcriptase (Life Technologies, Grand Island, NY) using T7-(dT)24 primer containing a T7 RNA polymerase promoter. After synthesis of the second complementary DNA (cDNA) strand, this product was used in an in vitro transcription reaction to generate biotinylated complementary RNA (cRNA).
Fifteen micrograms of fragmented cRNA was hybridized to a HuGeneFL microarray (Affymetrix) for 16 hours at 45°C with constant rotation at 60 rpm according to the Affymetrix protocol. This high-density oligonucleotide-based array targets 5600 human genes as selected from the National Center for Biotechnology Information (NCBI) Gene Bank database.47 Each microarray was used to assay a single sample. After hybridization, the microarray was washed and stained on an Affymetrix fluidics station and scanned with an argon ion confocal laser, with a 488-nm emission and detection at 570 nm. The fluorescence intensity was measured for each microarray and normalized by global scaling to 2500 and to the average fluorescence intensity for the entire microarray.22 The data were imported into a Microsoft Excel database.
Dataanalysis
Data analysis was performed with the Genespring software version 3.0 (Silicon Genetics, San Carlos, CA). To be able to calculate fold changes and not to lose genes that are not expressed in one sample, but switched on in the other, or vice versa, we set the lowest raw data value at an arbitrary “11.”23 To eliminate “fold-change” calls within the range of background noise, we required for significant changes that the genes to be classified as “up-regulated” had raw data values of at least 500 in the MCL samples, and to be called “down-regulated” had raw data values of at least 500 in the control.23 Within those parameters, genes were selected if they were either up- or down-regulated at least 3-fold in 4 of 5 cases of MCL studied by microarray.
Genelists
The gene expression profile of our clinical tumor samples includes: (1) changes potentially responsible for the development of MCL; (2) secondary gene expression alterations due to (1); (3) unrelated changes; and (4) false-positive/negative data. Nevertheless, the computational tools to better ascertain new insights about genes differentially expressed in MCL as compared to the control HLs are not yet at hand. Lacking a program that is able to discriminate reliably between these different possibilities and wishing to order the expression data into more meaningful pathways, we went through the public gene databases47 48 and developed a database containing information for 1149 genes that may be involved in the regulation of cell growth and cell signaling. We categorized these genes in 32 subgroups as given in Table 1and we created for each of the subgroups a separate Excel file containing the name of the gene, the GenBank accession number, and additional information if necessary.
Lists of genes involved in cell signaling pathways
| Subgroup . | No. of genes . |
|---|---|
| Adapter proteins | 19 |
| Caspases/caspase substrates | 12 |
| Cell-cycle proteins | 95 |
| Cell surface markers | 88 |
| Clusters of differentiation | 44 |
| Coactivators/HATs | 13 |
| Corepressors | 8 |
| Cytokines/cytokine receptors | 65 |
| Cytoskeletal proteins | 61 |
| Death domain adapters | 10 |
| DNA damage repair proteins | 32 |
| Enhancer binding proteins | 69 |
| Extracellular matrix proteins | 45 |
| Glutamate receptors | 4 |
| G-protein regulators | 17 |
| G-proteins | 33 |
| Growth factors/growth factor receptors | 98 |
| Histones | 15 |
| Ion channels | 49 |
| Kinases (Pl3, Ser-Thr) | 22 |
| MAPKs/SAPsK/JAKs | 64 |
| Mitochondrial proteins | 33 |
| NOSs | 2 |
| Nuclear receptors | 24 |
| PDZ adapters | 7 |
| Phosphatases | 56 |
| Signaling effectors | 65 |
| SMADs/STATs | 10 |
| Synaptic vesicles | 17 |
| TGF/TNF | 31 |
| Transcription factors | 23 |
| Tyrosine kinases | 18 |
| Subgroup . | No. of genes . |
|---|---|
| Adapter proteins | 19 |
| Caspases/caspase substrates | 12 |
| Cell-cycle proteins | 95 |
| Cell surface markers | 88 |
| Clusters of differentiation | 44 |
| Coactivators/HATs | 13 |
| Corepressors | 8 |
| Cytokines/cytokine receptors | 65 |
| Cytoskeletal proteins | 61 |
| Death domain adapters | 10 |
| DNA damage repair proteins | 32 |
| Enhancer binding proteins | 69 |
| Extracellular matrix proteins | 45 |
| Glutamate receptors | 4 |
| G-protein regulators | 17 |
| G-proteins | 33 |
| Growth factors/growth factor receptors | 98 |
| Histones | 15 |
| Ion channels | 49 |
| Kinases (Pl3, Ser-Thr) | 22 |
| MAPKs/SAPsK/JAKs | 64 |
| Mitochondrial proteins | 33 |
| NOSs | 2 |
| Nuclear receptors | 24 |
| PDZ adapters | 7 |
| Phosphatases | 56 |
| Signaling effectors | 65 |
| SMADs/STATs | 10 |
| Synaptic vesicles | 17 |
| TGF/TNF | 31 |
| Transcription factors | 23 |
| Tyrosine kinases | 18 |
By screening the public available gene databases (http://www4.ncbi.nlm.nih.gov/ and http://www4.ncbi.nlm.nih.gov/omim/), 1149 genes that may be involved in the regulation of cellular growth and cell signaling were selected and categorized. For each of the subgroups, a separate Excel file was created containing the name of the gene, the GenBank accession number and additional information.
By combining these gene lists with the genes that were selected due to changes of expression by comparing MCL to the control, we identified pathways that were altered in our tumor samples.
Real-time PCR
Total RNA (2 μg) was processed directly to cDNA by reverse transcription with Superscript II (Life Technologies) according to the manufacturer's protocol in a total volume of 100 μL. All PCR primers and TaqMan probes (Table 2) were designed using software PRIMER349 using published sequence data from the NCBI database. Primers were synthesized by Life Technologies. TaqMan probes were purchased from Applied Biosystems (Foster City, CA) and were labeled with the reporter dye FAM in the 5′ end and the quencher dye TAMRA in the 3′ end. In control experiments with 12 replicates, no false positives were detected and the variance between each of the replicates was within 5%. Amplification reactions contained 5 μL cDNA, 12.5 μL of the Universal Taqman 2× PCR mastermix (Applied Biosystems), and 2.5 μL of each of the specific primers and the probe. Primer and TaqMan probe concentrations in the final volume of 25 μL were 300 nM and 100 nM, respectively. All reactions were performed in triplicate in an iCycler iQ system (Biorad, Hercules, CA) and the thermal cycling conditions were: 2 minutes at 50°C, 10 minutes at 95°C, followed by 45 cycles of 95°C for 15 seconds, and 60°C for 1 minute.
Primers and probes used for real-time polymerase chain reaction
| Gene . | Primer 1 (5′-3′) . | Primer 2 (5′-3′) . | Probe (5′-FAM-3′-TAMRA) . |
|---|---|---|---|
| CASP8 | AAGCAAACCTCGGGGATACT | TCCATTCCTGTCCCTAATGC | AGCACGGGAGAAAGTGCCCAAA |
| CASP9 | GTTGAGACCCTGGACGACAT | CCTTTCACCGAAACAGCATT | CAGTCCCTCCTGCTTAGGGTCGC |
| RAIDD | TGGAGGGACTGGTTCTTCAG | ATCCAGCAGGAGCATTGTTT | TGCTCAAACCACAGGCCTCCG |
| DAXX | GACGGACATTTCCTCTTCCA | CGCCTCCATTGAAGGAAGTA | CAATCAGAGGAGCCCTTCACCACTG |
| DP2 | AACCATTTGGCTGCTGATTC | GATTCTGACATTCCTGAGCAGAA | CAAGTGGATTGGCCTGCCTACCA |
| FADD | GTGCGGGAGTCACTGAGAAT | GTCAGCCACCAGGTTCATCT | AACGCAACAGTGGCCCACCTG |
| FAS | TCCAAGGGATTGGAATTGAG | ACCTGGAGGACAGGGCTTAT | AGGCCTGCATCATGATGGCCA |
| ICSBP1 | GCTGATCAAGGAGCCTTCTG | ACCAGTCTGGAAGGAGCTGA | AAAAGGAGCCCTTCCCCGCC |
| PDCD1 | TGTCTTTCCTAGCGGAATGG | CCAAGAGCAGTGTCCATCCT | TCGGAGTGCCCAGCCACTGA |
| STAF50 | TCATTTGCTGGGTTTGTGAA | GCTACCTGCAGCTTTTCCTG | CCAAGGTCACCAAACATTCCGC |
| TRAIL | TCACAGTGCTCCTGCAGTCT | GCAATGCCACTTTTGGAGTA | CCAACGAGCTGAAGCAGATGCA |
| BAC | TCCCTGGAGAAGAGCTACGA | AGGAAGGAAGGCTGGAAGAG | CAATGAGCGGTTCCGCTGCC |
| Gene . | Primer 1 (5′-3′) . | Primer 2 (5′-3′) . | Probe (5′-FAM-3′-TAMRA) . |
|---|---|---|---|
| CASP8 | AAGCAAACCTCGGGGATACT | TCCATTCCTGTCCCTAATGC | AGCACGGGAGAAAGTGCCCAAA |
| CASP9 | GTTGAGACCCTGGACGACAT | CCTTTCACCGAAACAGCATT | CAGTCCCTCCTGCTTAGGGTCGC |
| RAIDD | TGGAGGGACTGGTTCTTCAG | ATCCAGCAGGAGCATTGTTT | TGCTCAAACCACAGGCCTCCG |
| DAXX | GACGGACATTTCCTCTTCCA | CGCCTCCATTGAAGGAAGTA | CAATCAGAGGAGCCCTTCACCACTG |
| DP2 | AACCATTTGGCTGCTGATTC | GATTCTGACATTCCTGAGCAGAA | CAAGTGGATTGGCCTGCCTACCA |
| FADD | GTGCGGGAGTCACTGAGAAT | GTCAGCCACCAGGTTCATCT | AACGCAACAGTGGCCCACCTG |
| FAS | TCCAAGGGATTGGAATTGAG | ACCTGGAGGACAGGGCTTAT | AGGCCTGCATCATGATGGCCA |
| ICSBP1 | GCTGATCAAGGAGCCTTCTG | ACCAGTCTGGAAGGAGCTGA | AAAAGGAGCCCTTCCCCGCC |
| PDCD1 | TGTCTTTCCTAGCGGAATGG | CCAAGAGCAGTGTCCATCCT | TCGGAGTGCCCAGCCACTGA |
| STAF50 | TCATTTGCTGGGTTTGTGAA | GCTACCTGCAGCTTTTCCTG | CCAAGGTCACCAAACATTCCGC |
| TRAIL | TCACAGTGCTCCTGCAGTCT | GCAATGCCACTTTTGGAGTA | CCAACGAGCTGAAGCAGATGCA |
| BAC | TCCCTGGAGAAGAGCTACGA | AGGAAGGAAGGCTGGAAGAG | CAATGAGCGGTTCCGCTGCC |
Primers and TaqMan probes were designed using software PRIMER3 (http://www.genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) using sequence data from the NCBI database.
We and others used the expression of β-actin (BAC) in malignant hematopoietic cells to normalize the expression data of each of the genes.24 25 BAC was used as an active and endogenous reference to correct for differences in the amount of total RNA added to a reaction and to compensate for different levels of inhibition during reverse transcription of RNA and during PCR.
Immunohistochemistry
Immunohistochemical analysis was performed on cryostat sections from snap-frozen tissue blocks using modifications of immunoperoxidase techniques as previously described.26 CCND1 and BCL2 antibodies were purchased from Dako (Carpinteria, CA). Antibody detection was performed with the Dako labeled streptavidin-biotin system (LSAB).
Results
Distribution of gene expression in MCL
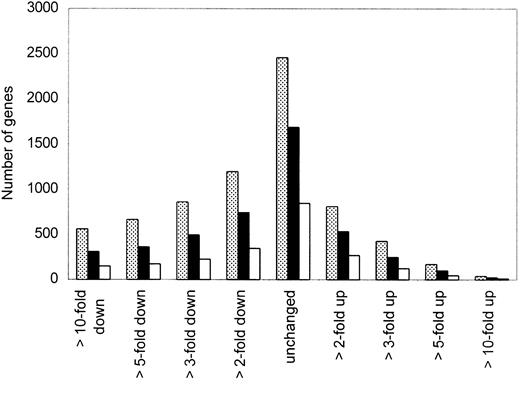
To select the most interesting genes from a total of 5600 genes analyzed by oligonucleotide microarray in our clinical samples, we performed several restrictions as described in “Materials and methods.” In this study, we focused on 4731 genes that had raw data of more than 500 according to our background cutoff. Microarray data were available from 5 MCL and 4 HL samples; fold changes of gene expression were based on the comparison between each of the MCL with the control HLs. Figure 1 provides a snapshot overview of the number of genes that were unchanged or either increased or decreased at least 2-, 3-, 5-, or 10-fold in either 3, 4, or 5 of the 5 samples compared to the expression in HLs. We focused on genes that were more than 3-fold up- or down-regulated in at least 4 of the 5 MCL samples.
Distribution of gene expression in MCL samples.
Overview of the number of genes that were unchanged or either up- or down-regulated at least 2-, 3-, 5-, or 10-fold in 3 (░), 4 (▪), or 5 (■) of the 5 MCL samples compared to expression in HL samples. We focused on genes that were more than 3-fold up- or down-regulated in at least 4 of 5 MCL samples.
Distribution of gene expression in MCL samples.
Overview of the number of genes that were unchanged or either up- or down-regulated at least 2-, 3-, 5-, or 10-fold in 3 (░), 4 (▪), or 5 (■) of the 5 MCL samples compared to expression in HL samples. We focused on genes that were more than 3-fold up- or down-regulated in at least 4 of 5 MCL samples.
Most differentially regulated genes in MCL
The comparison of the genes that were 3-fold or greater up-regulated in at least 4 of the 5 MCL with the list that we culled from the public databases containing a total of 1149 genes (Table 1) involved in cell growth and signaling resulted in a list of 92 highly differentially expressed genes. Fifty of them were up-regulated in MCL (Table 3) and 42 of them were down-regulated (Table 4). Many of the up-regulated genes were transcription factors (eg, AP2, ATF1, C/EBPγ, JUN, MYB, MYC), cell cycle–related genes (eg,CCND1, CDK4, DP2, E2F5), and growth factors and their receptors (eg, IL-1R, IL-3, IL-8, IL-13, IL-18).
Genes whose expression is up-regulated in mantle cell lymphoma compared to hyperplastic lymph nodes as analyzed by oligonucleotide microarray
| Gene . | Access . | Map . | Description . | Change . |
|---|---|---|---|---|
| Transcription factors | ||||
| AF10 | U13948 | 10p12 | ALL1 fused gene from chromosome 10 | |
| AP2α | M61156 | 6p22.3 | Transcription factor AP2α | > 10 |
| ATF1 | X55544 | 12q13 | Activating transcription factor | > 10 |
| C/EBPγ | U20240 | CCAAT/enhancer binding protein γ | > 10 | |
| DP2 | L40386 | E2F dimerization partner DP2 | > 10 | |
| E2F5 | U15642 | E2F transcription factor 5 | > 10 | |
| JUN | U65928 | 1p32 | V-jun avian sarcoma virus oncogene homolog | 8 |
| MYBL2 | X13293 | 20q13.1 | V-myb avian myeloblastosis viral oncogene | |
| MYC | M13929 | 8q24.12 | Avian myelocytomatosis viral oncogene | > 10 |
| PBX2 | X59842 | 6p21.3 | Pre-B-cell transcription factor 2 | > 10 |
| TAF2 | X84003 | TBP associated factor, RNA polymerase 2 | > 10 | |
| TAF2D | X95525 | 10q24 | 100-kd subunit of Pol II | > 10 |
| TBP | X54993 | 6q27 | TATA box binding protein | 6 |
| YES1 | M15990 | 18p11.31 | Yamaguchi sarcoma virual oncogene | > 10 |
| Cell-cycle–related genes | ||||
| CCND1 | X59798 | 11q13 | Cyclin D1 | > 10 |
| CCNG2 | U47414 | 4 | Cyclin G2 | > 10 |
| CDC8 | L16991 | Deoxythymidylate kinase | > 10 | |
| CDK4 | U37022 | 12q14 | Cyclin-dependent kinase 4 | > 10 |
| CKS1 | X54941 | 8q21 | CDC28 protein kinase 1 | > 10 |
| MDM2 | U33203 | 12q14.3 | Homolog of mouse double minute 2 | > 10 |
| RB1 | L49209 | 13q14.1 | Retinoblastoma, exon 4 | > 10 |
| RBBP2 | S66431 | 12p11 | Retinoblastoma-binding protein 2 | > 10 |
| RBBP5 | X85134 | 1q32 | Retinoblastoma-binding protein 5 | > 10 |
| SKP2 | U33761 | 5p13 | S-phase kinase-associated protein 2 | > 10 |
| Growth factors/receptors | ||||
| ICSBP1 | M91196 | Interferon consensus sequence binding protein 1 | > 10 | |
| IFNA16 | M27318 | 9q22 | Interferon alpha 16 | 6 |
| IFNA201 | K01900 | 9q22 | Interferon alpha-201 | > 10 |
| IL-1R1 | M27492 | 2q12 | Interleukin-1 receptor, type 1 | > 10 |
| IL-3 | M20137 | 5q23 | Interleukin-3 | > 10 |
| IL-8 | M28130 | 4q12 | Interleukin-8 | > 10 |
| IL-13 | U31120 | 5q31 | Interleukin-13 | > 10 |
| IL-13RA1 | Y10659 | Chr X | Interleukin-13 receptor, alpha 1 | > 10 |
| IL-18 | D49950 | 11q22.2 | Interleukin-18 | > 10 |
| ILF3 | U10324 | Interleukin enhancer binding factor 3 | > 10 | |
| LIFR | X61615 | 5p13 | Leukemia inhibitory factor receptor | 4 |
| Signal proteins and others | ||||
| BCL2 | U15173 | Oncogene B-cell leukemia 2 | > 10 | |
| BLK | S76617 | 8p23 | B lymphoid tyrosine kinase | 6 |
| FLI1 | M98833 | 11q24.1 | Friend leukemia virus integration 1 | 4 |
| H1F0 | X03473 | 22q13.1 | H1 histone family, member 0 | > 10 |
| H2BFQ | M60571 | 1q21 | H2B histone family, member Q | > 10 |
| NCK1 | X17576 | 3q21 | Melanoma NCK protein | > 10 |
| NRAS | X02751 | 1p13 | Neuroblastoma RAS viral oncogene homolog | > 10 |
| OGR1 | U48405 | 14q31 | Ovarian cancer G protein-coupled receptor 1 | 6 |
| PRKCB1 | X06318 | 16p12 | Protein kinase C, beta 1 | 8 |
| PRKM8 | U35005 | Protein kinase mitogen-activated 8 (JNK1) | > 10 | |
| RAB2 | M28213 | RAS-associated protein 2 | 8 | |
| RAB9 | U44103 | RAS-associated protein 9 | > 10 | |
| REL | X75042 | 2p13 | V-rel avian reticuloendotheliosis viral oncogene | 6 |
| RXRG | U38480 | 1q22 | Retinoic X receptor gamma | > 10 |
| STAF50 | X82200 | Interferon inducible gene | 8 |
| Gene . | Access . | Map . | Description . | Change . |
|---|---|---|---|---|
| Transcription factors | ||||
| AF10 | U13948 | 10p12 | ALL1 fused gene from chromosome 10 | |
| AP2α | M61156 | 6p22.3 | Transcription factor AP2α | > 10 |
| ATF1 | X55544 | 12q13 | Activating transcription factor | > 10 |
| C/EBPγ | U20240 | CCAAT/enhancer binding protein γ | > 10 | |
| DP2 | L40386 | E2F dimerization partner DP2 | > 10 | |
| E2F5 | U15642 | E2F transcription factor 5 | > 10 | |
| JUN | U65928 | 1p32 | V-jun avian sarcoma virus oncogene homolog | 8 |
| MYBL2 | X13293 | 20q13.1 | V-myb avian myeloblastosis viral oncogene | |
| MYC | M13929 | 8q24.12 | Avian myelocytomatosis viral oncogene | > 10 |
| PBX2 | X59842 | 6p21.3 | Pre-B-cell transcription factor 2 | > 10 |
| TAF2 | X84003 | TBP associated factor, RNA polymerase 2 | > 10 | |
| TAF2D | X95525 | 10q24 | 100-kd subunit of Pol II | > 10 |
| TBP | X54993 | 6q27 | TATA box binding protein | 6 |
| YES1 | M15990 | 18p11.31 | Yamaguchi sarcoma virual oncogene | > 10 |
| Cell-cycle–related genes | ||||
| CCND1 | X59798 | 11q13 | Cyclin D1 | > 10 |
| CCNG2 | U47414 | 4 | Cyclin G2 | > 10 |
| CDC8 | L16991 | Deoxythymidylate kinase | > 10 | |
| CDK4 | U37022 | 12q14 | Cyclin-dependent kinase 4 | > 10 |
| CKS1 | X54941 | 8q21 | CDC28 protein kinase 1 | > 10 |
| MDM2 | U33203 | 12q14.3 | Homolog of mouse double minute 2 | > 10 |
| RB1 | L49209 | 13q14.1 | Retinoblastoma, exon 4 | > 10 |
| RBBP2 | S66431 | 12p11 | Retinoblastoma-binding protein 2 | > 10 |
| RBBP5 | X85134 | 1q32 | Retinoblastoma-binding protein 5 | > 10 |
| SKP2 | U33761 | 5p13 | S-phase kinase-associated protein 2 | > 10 |
| Growth factors/receptors | ||||
| ICSBP1 | M91196 | Interferon consensus sequence binding protein 1 | > 10 | |
| IFNA16 | M27318 | 9q22 | Interferon alpha 16 | 6 |
| IFNA201 | K01900 | 9q22 | Interferon alpha-201 | > 10 |
| IL-1R1 | M27492 | 2q12 | Interleukin-1 receptor, type 1 | > 10 |
| IL-3 | M20137 | 5q23 | Interleukin-3 | > 10 |
| IL-8 | M28130 | 4q12 | Interleukin-8 | > 10 |
| IL-13 | U31120 | 5q31 | Interleukin-13 | > 10 |
| IL-13RA1 | Y10659 | Chr X | Interleukin-13 receptor, alpha 1 | > 10 |
| IL-18 | D49950 | 11q22.2 | Interleukin-18 | > 10 |
| ILF3 | U10324 | Interleukin enhancer binding factor 3 | > 10 | |
| LIFR | X61615 | 5p13 | Leukemia inhibitory factor receptor | 4 |
| Signal proteins and others | ||||
| BCL2 | U15173 | Oncogene B-cell leukemia 2 | > 10 | |
| BLK | S76617 | 8p23 | B lymphoid tyrosine kinase | 6 |
| FLI1 | M98833 | 11q24.1 | Friend leukemia virus integration 1 | 4 |
| H1F0 | X03473 | 22q13.1 | H1 histone family, member 0 | > 10 |
| H2BFQ | M60571 | 1q21 | H2B histone family, member Q | > 10 |
| NCK1 | X17576 | 3q21 | Melanoma NCK protein | > 10 |
| NRAS | X02751 | 1p13 | Neuroblastoma RAS viral oncogene homolog | > 10 |
| OGR1 | U48405 | 14q31 | Ovarian cancer G protein-coupled receptor 1 | 6 |
| PRKCB1 | X06318 | 16p12 | Protein kinase C, beta 1 | 8 |
| PRKM8 | U35005 | Protein kinase mitogen-activated 8 (JNK1) | > 10 | |
| RAB2 | M28213 | RAS-associated protein 2 | 8 | |
| RAB9 | U44103 | RAS-associated protein 9 | > 10 | |
| REL | X75042 | 2p13 | V-rel avian reticuloendotheliosis viral oncogene | 6 |
| RXRG | U38480 | 1q22 | Retinoic X receptor gamma | > 10 |
| STAF50 | X82200 | Interferon inducible gene | 8 |
These genes were selected because their expression was up-regulated ≥ 3-fold in at least 4 of 5 mantle cell lymphoma samples as compared to the nonmalignant hyperplastic lymph nodes.
Access indicates GenBank accession number.
Genes whose expression is down-regulated in mantle cell lymphoma compared to hyperplastic lymph nodes as analyzed by oligonucleotide microarray
| Gene . | Access . | Map . | Description . | Change . |
|---|---|---|---|---|
| Apoptosis-related genes | ||||
| CASP9 | U60521 | Caspase 9, apoptosis-related cysteine protease | > 10 | |
| CASP10 | U60519 | 2q33-q34 | Caspase 10, apoptosis-related cysteine protease | 8 |
| CHARAS1 | J00277 | C-Ha-ras1 p21 proto-oncogene | > 10 | |
| RAIDD | U79115 | 12q21.33 | CASP2 and RIPK1 domain-containing adaptor with death domain | > 10 |
| CYC1 | J0444 | 8q24.3 | Cytochrome C1 | > 10 |
| FADD | X84709 | 11q13.3 | Fas (TNFRSF6)-associated via death domain | > 10 |
| DAXX | AF006041 | 6p21.3 | Death-associated protein 6 | 3 |
| PDCD1 | U64863 | 2q37.3 | Programmed cell death 1 | > 10 |
| RIN1 | L36463 | Ras interactor | 3 | |
| Retinoid acid–related genes | ||||
| RARA | X06614 | 17q12 | Retinoic acid receptor, alpha | 8 |
| RARG | M38258 | 12q13 | Retinoic acid receptor, gamma | > 10 |
| RI58 | U34605 | Retinoic acid and interferon–inducible 58K protein | > 10 | |
| RORA | U04898 | 15q21 | RAR-related orphan receptor A | > 10 |
| Kinases | ||||
| PRKAA2 | U06454 | 1p31 | Protein kinase, AMP-activated, alpha 2 | 3 |
| PRKG1 | Y07512 | 10q11.2 | Protein kinase, cGMP-dependent, type I | 8 |
| PRKM7 | U29725 | 17p11.2 | Protein kinase mitogen-activated 7 (MAP kinase) | > 10 |
| Growth factors/receptors | ||||
| IFI17 | J04164 | Interferon-induced protein 17 | 3 | |
| IFNAR1 | J03171 | 21q22.1 | Interferon (alpha, beta, and omega) receptor 1 | 10 |
| IL-1RL2 | U49065 | Interleukin-1 receptor-like 2 | 10 | |
| IL-6R | M20566 | 1q21 | Interleukin-6 receptor | 10 |
| IL-9R | L39064 | Xq28 | Interleukin-9 receptor | 4 |
| IL-11RA | U32324 | 9p13 | Interleukin-11 receptor, alpha | > 10 |
| IL-12RB2 | U64198 | 1p31.3 | Interleukin-12 receptor, beta 2 | 10 |
| IL-15RA | U31628 | 10p15 | Interleukin-15 receptor, alpha | > 10 |
| IL-18R1 | U43672 | 2q12 | Interleukin-18 receptor 1 | 3 |
| TGFBR3 | L07594 | 1p33-p32 | Transforming growth factor, beta receptor III | 8 |
| TNFRSF12 | U83598 | 1p36.2 | TNF receptor superfamily, member 12 | 4 |
| TNFRSF1A | M58286 | 12p13.2 | TNF receptor superfamily, member 1A | > 10 |
| Cell-cycle–related genes | ||||
| CCNA1 | U66838 | 13q12.3 | Cyclin A1 | 5 |
| CDC34 | L22005 | 19p13.3 | Cell division cycle 34 | > 10 |
| MAD1L1 | U33822 | 7p22 | MAD1 (mitotic arrest deficient, yeast, homolog) | 3 |
| MAD2L1 | U65410 | 4q27 | MAD2 (mitotic arrest deficient, yeast, homolog) | 4 |
| RBBP1 | S57153 | Retinoblastoma-binding protein 1 | 10 | |
| Others | ||||
| APR | D90070 | Adult T-cell leukemia–derived PMA-responsive | 3 | |
| H1F4 | M60748 | 6p22.2 | H1 histone family, member 4 | 6 |
| KIR2DL4 | X99479 | 19q13.4 | Killer cell immunoglobulin-like receptor | 10 |
| KLRC1 | X54867 | 12p13 | Killer cell lectin-like receptor subfamily C | > 10 |
| MAGEB1 | X82539 | Xp21.3 | Melanoma antigen, family B, 1 | > 10 |
| PPP2R5B | L42374 | 11q12 | Protein phosphatase 2, regulatory subunit B | > 10 |
| TAF2A | D90359 | Xq13.1 | TATA box binding protein associated factor | 10 |
| TAL1 | M63589 | 1p32 | T-cell acute lymphocytic leukemia 1 | 3 |
| XPA | D14533 | 9q22.3 | Xeroderma pigmentosum, complementation A | > 10 |
| Gene . | Access . | Map . | Description . | Change . |
|---|---|---|---|---|
| Apoptosis-related genes | ||||
| CASP9 | U60521 | Caspase 9, apoptosis-related cysteine protease | > 10 | |
| CASP10 | U60519 | 2q33-q34 | Caspase 10, apoptosis-related cysteine protease | 8 |
| CHARAS1 | J00277 | C-Ha-ras1 p21 proto-oncogene | > 10 | |
| RAIDD | U79115 | 12q21.33 | CASP2 and RIPK1 domain-containing adaptor with death domain | > 10 |
| CYC1 | J0444 | 8q24.3 | Cytochrome C1 | > 10 |
| FADD | X84709 | 11q13.3 | Fas (TNFRSF6)-associated via death domain | > 10 |
| DAXX | AF006041 | 6p21.3 | Death-associated protein 6 | 3 |
| PDCD1 | U64863 | 2q37.3 | Programmed cell death 1 | > 10 |
| RIN1 | L36463 | Ras interactor | 3 | |
| Retinoid acid–related genes | ||||
| RARA | X06614 | 17q12 | Retinoic acid receptor, alpha | 8 |
| RARG | M38258 | 12q13 | Retinoic acid receptor, gamma | > 10 |
| RI58 | U34605 | Retinoic acid and interferon–inducible 58K protein | > 10 | |
| RORA | U04898 | 15q21 | RAR-related orphan receptor A | > 10 |
| Kinases | ||||
| PRKAA2 | U06454 | 1p31 | Protein kinase, AMP-activated, alpha 2 | 3 |
| PRKG1 | Y07512 | 10q11.2 | Protein kinase, cGMP-dependent, type I | 8 |
| PRKM7 | U29725 | 17p11.2 | Protein kinase mitogen-activated 7 (MAP kinase) | > 10 |
| Growth factors/receptors | ||||
| IFI17 | J04164 | Interferon-induced protein 17 | 3 | |
| IFNAR1 | J03171 | 21q22.1 | Interferon (alpha, beta, and omega) receptor 1 | 10 |
| IL-1RL2 | U49065 | Interleukin-1 receptor-like 2 | 10 | |
| IL-6R | M20566 | 1q21 | Interleukin-6 receptor | 10 |
| IL-9R | L39064 | Xq28 | Interleukin-9 receptor | 4 |
| IL-11RA | U32324 | 9p13 | Interleukin-11 receptor, alpha | > 10 |
| IL-12RB2 | U64198 | 1p31.3 | Interleukin-12 receptor, beta 2 | 10 |
| IL-15RA | U31628 | 10p15 | Interleukin-15 receptor, alpha | > 10 |
| IL-18R1 | U43672 | 2q12 | Interleukin-18 receptor 1 | 3 |
| TGFBR3 | L07594 | 1p33-p32 | Transforming growth factor, beta receptor III | 8 |
| TNFRSF12 | U83598 | 1p36.2 | TNF receptor superfamily, member 12 | 4 |
| TNFRSF1A | M58286 | 12p13.2 | TNF receptor superfamily, member 1A | > 10 |
| Cell-cycle–related genes | ||||
| CCNA1 | U66838 | 13q12.3 | Cyclin A1 | 5 |
| CDC34 | L22005 | 19p13.3 | Cell division cycle 34 | > 10 |
| MAD1L1 | U33822 | 7p22 | MAD1 (mitotic arrest deficient, yeast, homolog) | 3 |
| MAD2L1 | U65410 | 4q27 | MAD2 (mitotic arrest deficient, yeast, homolog) | 4 |
| RBBP1 | S57153 | Retinoblastoma-binding protein 1 | 10 | |
| Others | ||||
| APR | D90070 | Adult T-cell leukemia–derived PMA-responsive | 3 | |
| H1F4 | M60748 | 6p22.2 | H1 histone family, member 4 | 6 |
| KIR2DL4 | X99479 | 19q13.4 | Killer cell immunoglobulin-like receptor | 10 |
| KLRC1 | X54867 | 12p13 | Killer cell lectin-like receptor subfamily C | > 10 |
| MAGEB1 | X82539 | Xp21.3 | Melanoma antigen, family B, 1 | > 10 |
| PPP2R5B | L42374 | 11q12 | Protein phosphatase 2, regulatory subunit B | > 10 |
| TAF2A | D90359 | Xq13.1 | TATA box binding protein associated factor | 10 |
| TAL1 | M63589 | 1p32 | T-cell acute lymphocytic leukemia 1 | 3 |
| XPA | D14533 | 9q22.3 | Xeroderma pigmentosum, complementation A | > 10 |
These genes were selected because their expression was down-regulated ≥ 3-fold in at least 4 of 5 mantle cell lymphoma samples as compared to the nonmalignant hyperplastic lymph nodes.
Access indicates GenBank accession number.
Apoptosis-related genes in MCL
We found genes that are involved in common apoptotic pathways abnormally expressed in MCL. BCL2 and cytochrome C1(CYC1), which were previously shown to be up- and down-regulated in several subtypes of low-grade lymphoma,27 were similarly altered in our MCL samples (Tables 3 and 4). Furthermore, we discovered an altered pathway in MCL that is known to be associated with apoptosis, but that has not been described to be abnormal in lymphoma cells. The cell surface receptor FAS (Apo-1, CD95) has been well characterized in apoptosis. One of the downstream interacting proteins is FAS-associated via death domain (FADD). We found that this gene was markedly down-regulated in all of the MCL samples as compared to the HL samples. In addition, several other apoptosis-related genes, such as caspase 2 (CASP2)and RIPK1 domain containing adapter with death domain(RAIDD), death-associated protein 6 (DAXX), and programmed cell death 1 (PDCD1, PD1), were substantially down-regulated in the tumor samples (Table 4).
Gene expression of MCL confirmed and extended by real-time PCR
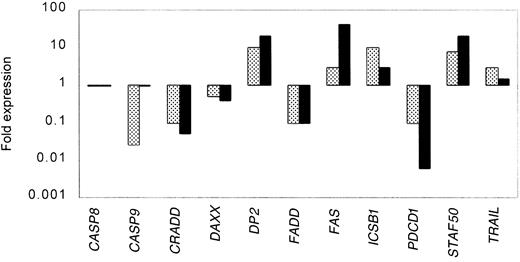
To confirm the expression data from the oligonucleotide microarray studies and to extend the analysis to a larger set of clinical samples, we analyzed by real-time PCR the expression of a total of 11 genes in 16 samples from MCL and 12 samples from HLs. Table5 shows the results of each of the genes analyzed in each of the samples. All the assays were done in triplicate. The variance between the triplicates was less than 5%. The results were normalized to the expression of BAC in each of the samples. Figure 2 summarizes the expression data measured by oligonucleotide microarray compared to real-time PCR for the selected genes. We found with one exception (caspase 9) that for both the up-regulated and down-regulated genes, both of the methods detected similar patterns of expression.
Results from real-time polymerase chain reaction analysis of selected genes
| Sample . | CASP8 . | CASP9 . | RAIDD . | DAXX . | DP2 . | FADD . | FAS . | ICSBP1 . | PDCD1 . | STAF50 . | TRAIL . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 12.4 | 10.2 | 11.0 | 6.8 | 12.2 | 9.8 | 8.1 | 10.6 | 16.3 | 8.7 | 8.0 |
| B | 11.5 | 9.7 | 10.0 | 7.1 | 11.6 | 9.5 | 8.9 | 11.7 | 13.1 | 9.7 | 6.2 |
| C | 12.4 | 10.1 | 11.0 | 8.5 | 12.7 | 10.6 | 8.6 | 13.1 | 15.1 | 9.9 | 9.7 |
| D | 11.6 | 11.6 | 11.3 | 8.2 | 17.9 | 11.2 | 5.5 | 7.2 | 12.2 | 4.7 | 8.5 |
| E | 10.4 | 9.6 | 11.7 | 7.1 | 12.0 | 10.2 | 10.8 | 12.9 | 14.8 | 9.0 | 8.4 |
| F | 10.3 | 9.9 | 12.2 | 8.1 | 13.5 | 10.2 | 6.3 | 13.4 | 16.2 | 10.3 | 9.8 |
| G | 10.9 | 9.9 | 12.1 | 8.7 | 12.9 | 9.7 | 5.5 | 11.7 | 14.9 | 8.3 | 8.2 |
| H | 9.9 | 10.5 | 11.3 | 8.2 | 11.2 | 9.7 | 5.9 | 12.1 | 15.4 | 8.2 | 8.7 |
| I | 9.9 | 10.1 | 12.0 | 9.3 | 12.1 | 10.0 | 6.7 | 11.8 | 12.5 | 9.3 | 7.7 |
| K | 10.4 | 10.2 | 10.5 | 7.7 | 11.7 | 10.2 | 8.6 | 11.8 | 11.1 | 8.6 | 7.9 |
| L | 12.3 | 9.1 | 11.9 | 9.0 | 13.5 | 10.5 | 6.0 | 11.6 | 15.5 | 9.3 | 10.0 |
| M | 8.8 | 10.6 | 11.0 | 8.3 | 12.1 | 9.4 | 6.8 | 8.4 | 12.9 | 7.8 | 7.8 |
| N | 10.1 | 9.7 | 10.5 | 7.5 | 13.4 | 8.1 | 6.6 | 12.1 | 13.8 | 7.8 | 6.6 |
| O | 12.1 | 10.6 | 12.2 | 8.6 | 14.0 | 10.0 | 4.5 | 12.4 | 14.2 | 11.4 | 9.2 |
| P | 10.2 | 10.0 | 11.1 | 7.4 | 12.9 | 10.0 | 6.1 | 13.2 | 14.5 | 10.4 | 9.5 |
| Q | 13.4 | 11.9 | 10.1 | 9.4 | 15.8 | 8.9 | 1.9 | 12.9 | 11.3 | 8.8 | 10.1 |
| Mean (SD) | 11.0 (1.2) | 10.2 (0.7) | 11.2 (0.7) | 8.1 (0.8) | 13.1 (1.7) | 9.9 (0.7) | 5.7 (2.1) | 11.7 (1.7) | 14.0 (1.6) | 8.9 (1.5) | 8.5 (1.2) |
| 1245T | 8.3 | 10.1 | 5.7 | 6.2 | 19.1 | 4.9 | 11.9 | 11.3 | 3.2 | 14.4 | 7.6 |
| 1278T | 17.7 | 13.8 | 7.5 | 8.9 | 19.2 | 4.3 | 8.2 | 14.2 | 7.8 | 10.8 | 8.7 |
| 1408T | 8.7 | 11.3 | 6.1 | 6.7 | 19.3 | 4.3 | 10.8 | 12.4 | 4.3 | 15.3 | 9.3 |
| 1440T | 10.4 | 12.3 | 7.3 | 8.9 | 20.0 | 5.8 | 10.3 | 12.0 | 6.5 | 13.3 | 10.4 |
| 1935T | 14.3 | 13.6 | 1.4 | 3.4 | 15.2 | 0.0 | 6.2 | 12.4 | 0.4 | 10.4 | 10.6 |
| 1956T | 13.5 | 9.6 | 9.6 | 6.6 | 17.6 | 8.8 | 12.4 | 15.5 | 10.9 | 12.1 | 9.3 |
| 2850T | 3.2 | 1.7 | 2.9 | 3.5 | 13.4 | 1.1 | 7.7 | 7.8 | 1.1 | 13.5 | 9.1 |
| 3853T | 11.8 | 9.5 | 9.8 | 8.3 | 15.6 | 10.4 | 12.0 | 15.4 | 11.1 | 12.6 | 8.9 |
| 4010T | 12.7 | 10.0 | 9.6 | 8.0 | 14.6 | 10.8 | 13.5 | 15.8 | 14.5 | 13.1 | 7.2 |
| 4893T | 11.1 | 9.8 | 7.3 | 7.1 | 18.0 | 7.7 | 13.0 | 14.1 | 7.4 | 14.1 | 7.1 |
| 4896T | 10.7 | 10.6 | 5.7 | 6.5 | 17.6 | 5.5 | 10.6 | 16.6 | 5.5 | 14.4 | 10.0 |
| 5341T | 11.5 | 12.0 | 9.5 | 7.3 | 20.0 | 9.2 | 16.0 | 13.8 | 8.1 | 16.1 | 11.0 |
| Mean (SD) | 11.2 (3.6) | 10.4 (3.1) | 6.9 (2.7) | 6.8 (1.8) | 17.5 (2.2) | 6.1 (3.4) | 11.1 (2.7) | 13.4 (2.4) | 6.7 (4.2) | 13.3 (1.7) | 9.1 (1.3) |
| Fold | unchg | unchg | 18 (down) | 3 (down) | 21 (up) | 14 (down) | 42 (up) | 3 (up) | 157 (down) | 20 (up) | 2 (up) |
| Sample . | CASP8 . | CASP9 . | RAIDD . | DAXX . | DP2 . | FADD . | FAS . | ICSBP1 . | PDCD1 . | STAF50 . | TRAIL . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 12.4 | 10.2 | 11.0 | 6.8 | 12.2 | 9.8 | 8.1 | 10.6 | 16.3 | 8.7 | 8.0 |
| B | 11.5 | 9.7 | 10.0 | 7.1 | 11.6 | 9.5 | 8.9 | 11.7 | 13.1 | 9.7 | 6.2 |
| C | 12.4 | 10.1 | 11.0 | 8.5 | 12.7 | 10.6 | 8.6 | 13.1 | 15.1 | 9.9 | 9.7 |
| D | 11.6 | 11.6 | 11.3 | 8.2 | 17.9 | 11.2 | 5.5 | 7.2 | 12.2 | 4.7 | 8.5 |
| E | 10.4 | 9.6 | 11.7 | 7.1 | 12.0 | 10.2 | 10.8 | 12.9 | 14.8 | 9.0 | 8.4 |
| F | 10.3 | 9.9 | 12.2 | 8.1 | 13.5 | 10.2 | 6.3 | 13.4 | 16.2 | 10.3 | 9.8 |
| G | 10.9 | 9.9 | 12.1 | 8.7 | 12.9 | 9.7 | 5.5 | 11.7 | 14.9 | 8.3 | 8.2 |
| H | 9.9 | 10.5 | 11.3 | 8.2 | 11.2 | 9.7 | 5.9 | 12.1 | 15.4 | 8.2 | 8.7 |
| I | 9.9 | 10.1 | 12.0 | 9.3 | 12.1 | 10.0 | 6.7 | 11.8 | 12.5 | 9.3 | 7.7 |
| K | 10.4 | 10.2 | 10.5 | 7.7 | 11.7 | 10.2 | 8.6 | 11.8 | 11.1 | 8.6 | 7.9 |
| L | 12.3 | 9.1 | 11.9 | 9.0 | 13.5 | 10.5 | 6.0 | 11.6 | 15.5 | 9.3 | 10.0 |
| M | 8.8 | 10.6 | 11.0 | 8.3 | 12.1 | 9.4 | 6.8 | 8.4 | 12.9 | 7.8 | 7.8 |
| N | 10.1 | 9.7 | 10.5 | 7.5 | 13.4 | 8.1 | 6.6 | 12.1 | 13.8 | 7.8 | 6.6 |
| O | 12.1 | 10.6 | 12.2 | 8.6 | 14.0 | 10.0 | 4.5 | 12.4 | 14.2 | 11.4 | 9.2 |
| P | 10.2 | 10.0 | 11.1 | 7.4 | 12.9 | 10.0 | 6.1 | 13.2 | 14.5 | 10.4 | 9.5 |
| Q | 13.4 | 11.9 | 10.1 | 9.4 | 15.8 | 8.9 | 1.9 | 12.9 | 11.3 | 8.8 | 10.1 |
| Mean (SD) | 11.0 (1.2) | 10.2 (0.7) | 11.2 (0.7) | 8.1 (0.8) | 13.1 (1.7) | 9.9 (0.7) | 5.7 (2.1) | 11.7 (1.7) | 14.0 (1.6) | 8.9 (1.5) | 8.5 (1.2) |
| 1245T | 8.3 | 10.1 | 5.7 | 6.2 | 19.1 | 4.9 | 11.9 | 11.3 | 3.2 | 14.4 | 7.6 |
| 1278T | 17.7 | 13.8 | 7.5 | 8.9 | 19.2 | 4.3 | 8.2 | 14.2 | 7.8 | 10.8 | 8.7 |
| 1408T | 8.7 | 11.3 | 6.1 | 6.7 | 19.3 | 4.3 | 10.8 | 12.4 | 4.3 | 15.3 | 9.3 |
| 1440T | 10.4 | 12.3 | 7.3 | 8.9 | 20.0 | 5.8 | 10.3 | 12.0 | 6.5 | 13.3 | 10.4 |
| 1935T | 14.3 | 13.6 | 1.4 | 3.4 | 15.2 | 0.0 | 6.2 | 12.4 | 0.4 | 10.4 | 10.6 |
| 1956T | 13.5 | 9.6 | 9.6 | 6.6 | 17.6 | 8.8 | 12.4 | 15.5 | 10.9 | 12.1 | 9.3 |
| 2850T | 3.2 | 1.7 | 2.9 | 3.5 | 13.4 | 1.1 | 7.7 | 7.8 | 1.1 | 13.5 | 9.1 |
| 3853T | 11.8 | 9.5 | 9.8 | 8.3 | 15.6 | 10.4 | 12.0 | 15.4 | 11.1 | 12.6 | 8.9 |
| 4010T | 12.7 | 10.0 | 9.6 | 8.0 | 14.6 | 10.8 | 13.5 | 15.8 | 14.5 | 13.1 | 7.2 |
| 4893T | 11.1 | 9.8 | 7.3 | 7.1 | 18.0 | 7.7 | 13.0 | 14.1 | 7.4 | 14.1 | 7.1 |
| 4896T | 10.7 | 10.6 | 5.7 | 6.5 | 17.6 | 5.5 | 10.6 | 16.6 | 5.5 | 14.4 | 10.0 |
| 5341T | 11.5 | 12.0 | 9.5 | 7.3 | 20.0 | 9.2 | 16.0 | 13.8 | 8.1 | 16.1 | 11.0 |
| Mean (SD) | 11.2 (3.6) | 10.4 (3.1) | 6.9 (2.7) | 6.8 (1.8) | 17.5 (2.2) | 6.1 (3.4) | 11.1 (2.7) | 13.4 (2.4) | 6.7 (4.2) | 13.3 (1.7) | 9.1 (1.3) |
| Fold | unchg | unchg | 18 (down) | 3 (down) | 21 (up) | 14 (down) | 42 (up) | 3 (up) | 157 (down) | 20 (up) | 2 (up) |
The value of the threshold cycle after normalization to β-actin is shown for each gene. Each experiment was done in triplicate. The variance between the triplicates was < 5%.
A to Q indicates each of the MCL samples; 1245T to 5341T, each of the HL samples; Fold, fold increase or decrease in expression in mantle cell lymphoma samples compared to the control hyperplastic lymph nodes; unchg, unchanged.
Correlation between gene expression measured by microarray and real-time PCR for selected genes.
Expression data from the oligonucleotide microarray analyses (░) were confirmed for selected genes in a larger set of clinical samples from MCL by real-time PCR (▪). All the real-time PCR experiments were done in triplicate. The variance between the triplicates was less than 5%, and the results were normalized to the expression of β-actin in each of the samples.
Correlation between gene expression measured by microarray and real-time PCR for selected genes.
Expression data from the oligonucleotide microarray analyses (░) were confirmed for selected genes in a larger set of clinical samples from MCL by real-time PCR (▪). All the real-time PCR experiments were done in triplicate. The variance between the triplicates was less than 5%, and the results were normalized to the expression of β-actin in each of the samples.
Immunohistochemistry
CCND1 and BCL2 are proteins that are important in the regulation of cell cycle and apoptosis. We found the genes coding for these proteins overexpressed in MCL by microarray analysis and confirmed our expression data for these genes as detected by immunohistochemical staining on cryostat sections from snap-frozen lymph nodes. We showed that these proteins were markedly up-regulated in the cytoplasm of the MCL cells as compared to the HL control cells (data not shown).
Discussion
Microarray technology provides a powerful new tool to asses the expression of a large number of genes in a single experiment. The initial use of array technology in medical science was to characterize global gene expression profiles during physiologic changes or disease progression.19,28The aim of those studies was to accelerate the identification of diagnostic and prognostic markers and to identify novel tumor-specific markers for certain malignant diseases.12,13 20
Recently, experience has shown that microarray analyses can provide sufficient data to detect genes or gene patterns that are associated with alterations of specific cellular pathways or signal cascades. For example, several new genes regulated downstream of p53 have been identified and are important in the p53-induced cell cycle arrest.29 The development of software specific for the analysis of gene expression data provides the tools not only to analyze single genes or gene clusters with diagnostic and prognostic importance, but also offers the possibility to correlate expression data of unknown genes with expression patterns for genes known to be involved in regulation of the biology of the cell. Knowledge of the genes involved in specific pathways facilitates the discovery of those coding for interactive proteins giving insights into their functional importance. An example of this use of microarray data is the detection of several redox and mitochondrial elements that control either resistance or sensitivity to apoptosis.22
Here, we show that this technique can be used to analyze clinical tumor samples for alterations of specific cellular signal pathways. To date, the analysis of specific pathways was restricted to cell lines or animal models where sequential data after treatment with chemicals or radiation or induction of immunologic response were obtained to characterize new genes involved in common pathways or to describe new pathways. We analyzed clinical lymph node samples from individuals with MCL and compared their gene expression to those of highly proliferative nonmalignant lymph nodes. These control samples were used as our first screen to exclude gene expression changes resulting from cellular proliferation in general.
To ensure that the gene expression that we measured by microarray assay was not affected by degradation of the RNA extracted from the frozen lymph node samples, we used denaturating gel electrophoresis to evaluate the quality of the RNA. Furthermore, expression of well-known housekeeping genes28 including BAC andGAPDH was found to be nearly equal in all our MCL and HL samples (raw data 250 000, variance between the samples <10%). As a third quality control, CCND1 was found to be overexpressed in each of our MCL samples, but not in the HL samples, as measured by microarray analysis and immunohistochemistry.
Analysis of our microarray data identified a group of 50 genes that were significantly up-regulated in 4 of 5 MCL samples, suggesting that these genes may play an important role in the development of this type of lymphoma. These genes will be studied in greater detail in future studies. Various transcription factors (Table 3) including the E2F transcription factor 5 (E2F5), its dimerization partner DP2, and the CDC28 protein kinase 1 (CKS1) were substantially up-regulated. We confirmed the overexpression of selected genes by real-time PCR (Table5) in our set of MCL samples. To our knowledge, these genes have not been reported to be aberrantly expressed in any type of lymphomas.
Because studies have shown that the overexpression of CCND1in MCL does not correlate with the proliferative activity of these tumor cells,30 other regulatory elements might cooperate with CCND1 in deregulating cell cycle control, but little is known about which proteins these may be. We found other up-regulated genes that are involved in cell cycle control including CDK4, MDM2, and SKP2. These genes were at least 10-fold higher expressed in MCL in comparison to the control HLs. AlsoE2F5, DP2 and their targets, CMYC andCDC8, were overexpressed (Table 3) in our MCL samples. These genes are downstream of CCND1/CDK4.
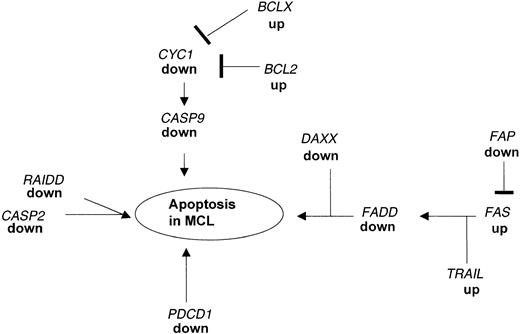
An imbalance between cellular apoptosis and survival may be critical for the pathogenesis of lymphoma. Programmed cell death is essential for the generation of the complex structural and functional organization of various organs and systems in the living organism. Most of the cells undergoing programmed cell death require transcriptional activation of genes that are essential for cell death. BCL2 is overexpressed in many low-grade lymphomas27 leading to the hypothesis that high levels of this protein inhibit the CYC1 system resulting in the inhibition of the caspase 9 (CASP9)-mediated cell death in these tumors. Besides the increased expression ofBCL2 and BCLX (Table 3), we found in our MCL samples that the downstream target genes, CYC1 andCASP9, were markedly decreased (Table 4, Figure3).
Apoptotic pathways disrupted in MCL as detected by microarray analysis and confirmed by real-time PCR.
Expression of several genes acting in concert to regulate the balance between proliferation and programmed cell death are altered in MCL as compared to nonmalignant HLs. The down-regulation of FADD causes a blockade of the FAS-FADD apoptotic pathway leading to secondary changes in expression of proapoptotic genes. In addition, PDCD1 and RAIDD, which can directly induce apoptosis, are markedly down-regulated in MCL.
Apoptotic pathways disrupted in MCL as detected by microarray analysis and confirmed by real-time PCR.
Expression of several genes acting in concert to regulate the balance between proliferation and programmed cell death are altered in MCL as compared to nonmalignant HLs. The down-regulation of FADD causes a blockade of the FAS-FADD apoptotic pathway leading to secondary changes in expression of proapoptotic genes. In addition, PDCD1 and RAIDD, which can directly induce apoptosis, are markedly down-regulated in MCL.
In addition, our analysis identified several other pathways associated with apoptosis that are substantially down-regulated in MCL. One of the key genes involved in apoptosis is FADD, which is a downstream target for the FAS pathway and which can directly induce apotosis.31,32 FAS cannot induce apoptosis in FADD-deficient embryonic fibroblasts.33 These findings indicate that the FAS-FADD system is important for the endogenous regulation of the balance between survival and death of human cells34 and that the lack of expression of FADD in MCL may tilt the balance providing these tumor cells with a growth advantage. The dissection of this apoptotic pathway was shown in human leukemia and lymphoma cell lines but so far no alterations of genes that are involved in the FAS-FADD system in primary non-Hodgkin lymphoma cells are reported.35-37 As shown in Figure 3, low expression ofFADD is associated with changes in the expression of genes in the FAS cascade including TRAIL and FAP. In addition, we found the gene coding for DAXX down-regulated in MCL; this protein enhances the FAS-mediated apoptosis38 by binding to the cytosolic domain of FAS.39 Changes in expression of these genes as shown by the microarray data were confirmed by real-time PCR.
The disruption of the gene encoding for the negative immunoregulatory receptor PDCD1 in BALB/c mice causes several autoimmune diseases such as a lupuslike disease40 and an autoimmune cardiomyopathy.41 PDCD1 was, therefore, thought to be an important factor for the prevention of these diseases. PDCD1 is highly expressed in activated human T and B lymphocytes42 as well as in pro-B cells in the bone marrow.43 The engagement of PDCD1 with its ligand PD-L1 leads to the inhibition of anti-IgM–stimulated B cells41 indicating the importance of PDCD1 as a negative immunoregulatory protein. Furthermore, PDCD1 was found to be activated in lymphatic cell lines undergoing programmed cell death,44 suggesting a role of this protein in apoptosis. Expression of this gene was low in our MCL samples as determined by microarray analysis and real-time PCR. We propose that the low expression of PDCD1 may contribute to the accumulation of abnormal B cells in MCL.
We identified a third key gene, RAIDD, which is associated with apoptosis and is down-regulated in expression in MCL (Table 4). RAIDD can function as an adaptor molecule in recruiting CASP2 to the tumor necrosis factor receptor 1 (TNFR1) signaling complex. The overexpression of RAIDD in mammalian cells induces apoptosis45 and the lack of expression of RAIDD may contribute to altered apoptosis in the MCL cells (Figure 3).
In conclusion, we show in this study that microarray data can markedly enhance the search for the molecular pathogenesis of tumors. We detected by oligonucleotide microarray and confirmed by quantitative PCR as well as by immunohistochemistry that apoptotic pathways are altered in MCL. The lack of appropriate regulation of apoptosis may be an additional step in the tumorogenesis of this B-cell malignancy.
We thank Drs Jonathan Braun and Mike Teitell for their helpful discussions and the continuous encouragement by Neil Ruzic. We thank the VASDHS gene chip core lab and the UCSD Cancer Center microarray facility for the technical assistance with the microarray assay.
Division of Hematology and Oncology, Cedars Sinai Medical Center, University of California Los Angeles, School of Medicine; Department of Pathology, University of California Los Angeles, School of Medicine; Research Service, VASDHS and Division of Hematology/Oncology and Cancer Center, University of California San Diego, School of Medicine, La Jolla; and Department of Pathology, Brigham and Women′s Hospital, Boston, MA.
Submitted January 9, 2001; accepted March 15, 2001.
Supported in part by National Institutes of Health grants, Neil Ruzic Fund, Ko-So Foundation, C. and H. Koeffler Fund and Lymphoma Foundation of America. W.K.H. is a recipient of a scholarship (HO2207/1-1) from the Deutsche Forschungsgemeinschaft. S.d.V. was supported by the UCLA STAR program as a Hematology/Oncology fellow and as an Advanced Research fellow. H.P.K. is a member of the Jonsson Comprehensive Cancer Center and holds the endowed Mark Goodson Chair of Oncology Research at Cedars Sinai Medical Center/UCLA School of Medicine.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Wolf-K. Hofmann, Division of Hematology and Oncology, Cedars Sinai Medical Center, UCLA School of Medicine, 8700 Beverly Blvd, Suite BM-1, Rm 109, Los Angeles, CA 90048; e-mail: w.k.hofmann@em.uni-frankfurt.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal