Abstract
Multiple myeloma (MM) is identified by unique immunoglobulin heavy chain (IgH) variable diversity joining region gene rearrangements, termed clonotypic, and an M protein termed the “clinical” isotype. Transcripts encoding clonotypic pre and postswitch IgH isotypes were identified in MM peripheral blood mononuclear cells (PBMCs), bone marrow (BM), and mobilized blood. For 29 patients, 38 BM, 17 mobilized blood, and 334 sequential PBMC samples were analyzed at diagnosis, before and after transplantation for 2 to 107 months. The clinical clonotypic isotype was readily detectable and persisted throughout treatment. Eighty-two percent of BM and 38% of PBMC samples also expressed nonclinical clonotypic isotypes. Clonotypic immunoglobulin M (IgM) was detectable in 68% of BM and 25% of PBMC samples. Nonclinical clonotypic isotypes were detected in 41% of mobilized blood samples, but clonotypic IgM was detected in only 12%. Patients with persistent clonotypic IgM expression had adverse prognostic features at diagnosis (lower hemoglobin, higher β2-microglobulin) and higher numbers of BM plasma cells compared with patients with infrequent/absent clonotypic IgM. Patients with persistent clonotypic IgM expression had significantly poorer survival than patients with infrequent IgM expression (P < .0001). In a multivariate analysis, persistent clonotypic IgM expression in the blood correlated independently with poor survival (P = .01). In nonobese diabetic severe combined immunodeficiency mice, xenografted MM cells expressed clinical and nonclinical postswitch clonotypic isotypes. MM expressing clonotypic IgM engrafted both primary and secondary mice, indicating their persistence within the murine BM. This study demonstrates that MM clonotypic cells expressing preswitch transcripts are tied to disease burden and outcomes. Because MM pathology involves postswitch plasma cells, this raises the possibility that IgH isotype switching in MM may accompany worsening disease.
Introduction
Multiple myeloma (MM) is an incurable cancer affecting the B lineage. The ultimate neoplastic plasma cell is located in the bone marrow (BM) and produces monoclonal immunoglobulin of a fixed isotype (referred to here as the clinical isotype) found in the serum and sometimes in urine of MM patients. MM may arise in a multistep process as mutations leading to neoplastic transformation accumulate.1 The immunoglobulin heavy chain (IgH) gene rearrangement provides a unique molecular signature, termed clonotypic, to identify members of the myeloma clone. Populations of circulating clonotypic B cells have been postulated to include MM progenitors.2-9 However, the maturation stage of the MM progenitor cell remains controversial.
Rearrangement of IgH variable diversity joining region (VDJ) signals commitment to the B lineage. Selection of V D and J genes in progenitor development and the subsequent hypermutation pattern of the V region becomes the clonotypic signature of B cells and their malignant derivatives.4,10-14 B-lineage cells undergo further programmed genetic instability when isotype class switching occurs to join the rearranged VDJ to postswitch heavy chain genes (IgG isotypes, IgA, and IgE) from an IgM/IgD preswitch heavy chain gene. Chromosomal translocations that occur during immunoglobulin class switching may be important in MM.1 15-20 Clinically, myeloma is a disease of postswitch B-lineage cells, most frequently of the clinical IgG or IgA isotype. For this study, clonotypic isotypes other than the MM clinical isotype are termed nonclinical isotypes.
Analysis of pre and postswitch clonal isotype expression indicates differentiation stages within the MM clone. Molecular analysis of peripheral blood mononuclear cells (PBMCs) in myeloma demonstrated the presence of circulating B cells that are clonally related to BM plasma cells.2,8,14,21-23 Single cell analysis showed that on average 66% of circulating MM B cells are clonotypic, exhibiting strict intraclonal homogeneity.14 Molecular studies of MM B cells have revealed isotype switching, implying the potential for differentiation within the myeloma clone.7,8,24 Palumbo et al25 reported results suggesting that, for some patients, MM may have a preswitch origin. Billadeau et al7demonstrated pre and postswitch clonal isotypes in BM B cells, whereas among the plasma cells only the clonotypic clinical isotype was detected. Significantly, for BM-localized B cells, even preswitch members of the myeloma clone appear to lack intraclonal diversity.4 7 This finding implies that the transformation event in myeloma must have occurred after germinal center maturation and that the myeloma clone has escaped hypermutator mechanisms and antigen-driven selection.
B-lineage cells belonging to the myeloma clone from blood, BM, and granulocyte colony-stimulating factor (G-CSF)–mobilized blood were identified by their clonotypic IgH VDJ and characterized for immunoglobulin isotype expression. For 29 patients, PBMCs and BM cells were analyzed sequentially from 2 months to 9 years, before and after transplantation. The engraftment potential of pre and postswitch clonotypic cells was confirmed by analysis of xenografted human myeloma.26 We find a highly significant correlation between the persistent expression of preswitch transcripts and worsened survival, as well as between persistent expression of preswitch transcripts and more advanced disease at the time of diagnosis, suggesting that preswitch clonotypic cells and isotype switching may be significant components of disease progression.
Patients, materials, and methods
Patients
For the clinical study, 29 patients were studied, including 27 patients expressing an M protein and 2 patients with light chain MM. A summary of patient characteristics is listed in Table1. Five MM patients provided PBMCs or, in one case, cells from a pleural effusion, at the time of aggressive disease progression, for xenografting into nonobese diabetic severe combined immunodeficiency (NOD SCID) mice.
Patient characteristics
| Mean age, y (range) | 56 (41-77) |
| Sex | |
| Male | 21 |
| Female | 8 |
| Durie-Salmon stage | |
| IA | 1 |
| IIA | 4 |
| IIIA | 21 |
| IIIB | 3 |
| Clinical isotype | |
| IgG | 19 |
| IgA | 7 |
| IgD | 1 |
| Light chain | 2 |
| Therapy* | |
| Conventional | 12 |
| High dose | 17 |
| Mean age, y (range) | 56 (41-77) |
| Sex | |
| Male | 21 |
| Female | 8 |
| Durie-Salmon stage | |
| IA | 1 |
| IIA | 4 |
| IIIA | 21 |
| IIIB | 3 |
| Clinical isotype | |
| IgG | 19 |
| IgA | 7 |
| IgD | 1 |
| Light chain | 2 |
| Therapy* | |
| Conventional | 12 |
| High dose | 17 |
Conventional therapy refers to standard-dose chemotherapy and/or corticosteroid therapy without growth factor or stem cell support. High dose refers to high-dose melphalan with autologous stem cell support.
BM was available from all patients listed in Table 1, to determine the clonotypic sequence. Sufficient BM remained for further analysis in 26 patients, including BM from the time of diagnosis in 15 patients. BM samples were generally obtained when a sample was needed for clinical decision making, during periods of active disease, or prior to high-dose therapy with autologous stem cell support. Multiple samples were obtained from 10 patients, for a total of 38 BM samples analyzed.
Of the patients listed in Table 1, 17 of 29 provided samples from blood mobilized with chemotherapy and G-CSF. Mobilized blood samples were obtained during periods of maximal disease regression, following 3 to 4 cycles of conventional chemotherapy. These patients were then treated with high-dose melphalan and autologous peripheral blood stem cell rescue. Twelve patients were ineligible for high-dose therapy on clinical grounds and were treated with chemotherapy and/or corticosteroids. All patients with lytic bone disease received bisphosphonates.
All but 3 patients in Table 1 were followed from the time of diagnosis. All 29 were followed until death (n = 15) or the last clinic visit (n = 14). Follow-up times were significant (median follow-up time, 26 months; range, 2-107 months). No patient still alive was followed for less than 15 months from the time of diagnosis, with 13 of 14 living patients followed for at least 2 years. Each patient provided peripheral blood for study at multiple points (range, 1-23 samples per patient) during the course of follow-up; a total of 334 blood samples were analyzed. The median sampling interval was 70 days; in all but 5 patients the sampling interval was 4 months or less. Blood samples were available during both active and quiescent phases of clinical disease; approximately one third of samples were obtained at times of no active therapy.
Total RNA was extracted from cell pellets of fresh PBMCs or BM cells and subjected to reverse transcriptase–polymerase chain reaction (RT-PCR) as previously described,12 using high-fidelity Platinum TAQ DNA polymerase (GIBCO/BRL, Burlington, ON, Canada). Vh family leader-specific primers were the following: Vh2 leader, CTTTGCTCCACGCTCCTG; Vh3 leader, GA(A/G)TT(G/T)GGGCTGAGCTGG; Vh4 leader, ATGAAACACCTGTGGTTCTTC; and Vh5 leader, TGGGGTCAACCGCCATC.
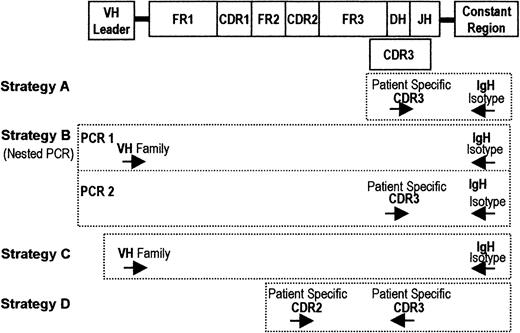
Sequences for the primers encoding the constant heavy chain regions were from Billadeau et al.7 Primers used to derive patient-specific sequences were from Aubin et al.27 For all 38 patients, the clonotypic IgH sequence was identified and validated as previously described.14 Specific IgH VDJ primers were selected in the CDR2 and CDR3 regions. We used 3 strategies to amplify the RT-PCR product. Figure1 shows the primers for each strategy. Strategy A was used to analyze clonotypic transcripts detectable in a single-stage PCR reaction, as a presumptive measure of relative frequency, involving 35 cycles of PCR with CDR3 and constant chain primers. Strategy B used nested PCR to determine whether any clonotypic transcripts were detectable within a given isotype class. This process involved 35 cycles of PCR with primers to Vh leader and a given constant chain, followed by reamplification of 2% (vol/vol) of the primary PCR product in 25 cycles of PCR with patient-specific CDR3 and nested constant chain primers. Strategy C was used to estimate the relative frequency of clonotypic transcripts within each of the 4 isotype classes (IgM, IgD, IgG, and IgA), involving 35 cycles of PCR with primers to the Vh leader and a constant chain, to amplify only the relevant isotype in each reaction, independent of its VDJ rearrangement. This amplification step was followed by subcloning and sequencing as described below. For all strategies PCR products of appropriate sizes were detected in 2% agarose electrophoresis containing ethidium bromide. For all samples, in the nested PCR experiments, aliquots of complementary DNA (cDNA) were amplified by using primers to histone to confirm the integrity of the RNA; all samples were positive for histone transcripts. For every sample, control lanes lacking cDNA were always run for each primer set and were always negative. All samples were also subjected to a single-stage RT-PCR, using patient-specific CDR2/CDR3 primers (strategy D). All BM, all mobilized blood samples, and 312 of 334 PBMC samples showed amplification of this patient-specific product in a single-stage RT-PCR. As previously described,14 all CDR2 and both 3′ and 5′ CDR3 primers were tested on RNA from several unrelated patients and have been shown to be rigorously specific for the IgH VDJ of the patient from which they were derived. We observed that, although specific 3′ CDR3 primers were readily designed, it was quite difficult to design patient-specific CDR3 primers in the 5′ direction, requiring multiple attempts.
RT-PCR strategies used in the detection of clonotypic IgH transcripts.
At the top of the figure is a schematic diagram of the structure of the immunoglobulin heavy chain gene, including the Vh leader, complementarity determining regions (CDRs), and constant chains (μ, δ, γ, α, and ε). Primers specific to unique CDR2 and CDR3 sequences in the MM clone were generated. All strategies were performed using cDNA derived from patient or mouse samples. Strategy A involved 35 cycles of PCR with CDR3 and constant chain (μ, δ, γ, and α) primers. Strategy B involved 35 cycles of PCR with primers to Vh leader and a given constant chain, followed by reamplification with 25 cycles of PCR using patient-specific CDR3 and nested constant chain primers. Strategy C involved 35 cycles of PCR with primers to the Vh leader and a constant chain to amplify only the relevant isotype in each reaction, independent of its VDJ rearrangement. All samples were also subjected to a single-stage RT-PCR using patient-specific CDR2/CDR3 primers (strategy D).
RT-PCR strategies used in the detection of clonotypic IgH transcripts.
At the top of the figure is a schematic diagram of the structure of the immunoglobulin heavy chain gene, including the Vh leader, complementarity determining regions (CDRs), and constant chains (μ, δ, γ, α, and ε). Primers specific to unique CDR2 and CDR3 sequences in the MM clone were generated. All strategies were performed using cDNA derived from patient or mouse samples. Strategy A involved 35 cycles of PCR with CDR3 and constant chain (μ, δ, γ, and α) primers. Strategy B involved 35 cycles of PCR with primers to Vh leader and a given constant chain, followed by reamplification with 25 cycles of PCR using patient-specific CDR3 and nested constant chain primers. Strategy C involved 35 cycles of PCR with primers to the Vh leader and a constant chain to amplify only the relevant isotype in each reaction, independent of its VDJ rearrangement. All samples were also subjected to a single-stage RT-PCR using patient-specific CDR2/CDR3 primers (strategy D).
Cloning and sequencing
PCR products were cloned using the INVITROGEN (Carlsbad, CA) TA TOPO cloning system accordingly to the manufacturer's directions. The transformants were analyzed by direct-lysis PCR with a standard M13 primer set. PCR products were sequenced by using a fluorescent di-deoxyterminator sequencing kit (PerkinElmer, Mississauga, ON, Canada), and analyzed on an ABI310 capillary genetic analyzer (PerkinElmer).
Analysis of the DNA sequences
The primary analysis was performed with a V-Base Internet-based program as previously described14to establish the type of variable region and the pattern of somatic hypermutations against the germline. The secondary analysis was done by alignment of the variable regions against the plasma cell-derived sequence for each patient, using the AutoAssembler (Perkin Elmer) program.
Human myeloma xenografts
NOD/LtSz-SCID (NOD SCID) mice at 6 to 8 weeks of age were irradiated (300-340 Gy) and injected by the intracardiac route26 with peripheral cells from the 5 indicated patients. Cells were isolated from the tumor masses, femoral BM, and spleen. RNA was purified as described above and analyzed for clinical and nonclinical clonotypic transcripts.26 For secondary transplantations, BM was harvested from a xenografted mouse that had been injected 3 months previously with human peripheral cells and injected into a second recipient. At 3 months, tissues were harvested from secondary recipients for PCR analysis.
Statistics
Student t test was used to compare the mean values of continuous variables between 2 groups. Correlations between 2 continuous variables of interest were assessed using the Pearson correlation coefficient. Correlations involving noncontinuous variables were assessed using Spearman (nonparametric) correlation analysis. Correlations between variables of interest and survival were assessed using Cox univariate regression analysis. Variables found to correlate with survival on univariate analysis were combined in a model using stepwise Cox multivariate regression analysis. Statistical significance for any test or correlation was defined as a 2-sided P value of .05 or less. Statistics were calculated using SAS software (SAS Institute, Cary, NC) and GraphPad Prism software (GraphPad Software, San Diego, CA).
Results
BM contains clonotypic cells expressing nonclinical isotypes, including IgM
Thirty-eight samples of BM from 26 of the 29 patients studied, including 2 samples from light chain MM patients, were tested for clinical and nonclinical clonotypic isotype expression (Table2, Figure2, Figure3, and Figure4). As expected, clonotypic transcripts of the clinical isotype were present in the BM in all samples from MM patients expressing an M protein and were detectable by both single-stage and nested RT-PCR (strategies A and B), suggesting that, in relative terms, the clinical clonotypic isotype was frequent. Single-stage RT-PCR revealed multiple nonclinical clonotypic isotypes in the BM of 15 of 26 patients, or 20 of 38 samples. Clonotypic IgM was seen in the BM on single-stage RT-PCR in 6 of 26 patients, or 8 of 38 samples. On nested RT-PCR, multiple clonotypic isotypes were seen in the BM in 22 of 26 patients, or 31 of 38 samples. Clonotypic IgM was seen in the BM on nested RT-PCR in 18 of 26 patients, or 26 of 38 samples. Each of the 2 light chain MM patients provided a single BM sample. Multiple postswitch clonotypic isotypes were present in the BM in both patients. Clonotypic IgM and IgD were present in 1 of 2 light chain patients.
Persistent presence of nonclinical and/or clinical clonotypic isotypes in bone marrow, mobilized blood, and peripheral blood mononuclear cells
| . | Bone marrow . | Mobilized blood . | PBMC* . |
|---|---|---|---|
| Single-stage RT-PCR† | |||
| No. of samples | 38 | 17 | 334 |
| Clonotypic (%)‡ | 38 (100) | 17 (100) | 295 (88) |
| Clinical (%)2-153 | 38 (100) | 17 (100) | 295 (88) |
| Nonclinical (%)2-155 | 20 (53) | 2 (12) | 70 (21) |
| IgM (%)2-154 | 8 (21) | 0 | 26 (8) |
| Nested RT-PCR† | |||
| No. of samples | 38 | 17 | 334 |
| Clonotypic (%)‡ | 38 (100) | 17 (100) | 312 (93) |
| Clinical (%)2-153 | 38 (100) | 17 (100) | 299 (90) |
| Nonclinical (%)2-155 | 31 (82) | 7 (41) | 127 (38) |
| IgM (%)2-154 | 26 (68) | 2 (12) | 83 (25) |
| . | Bone marrow . | Mobilized blood . | PBMC* . |
|---|---|---|---|
| Single-stage RT-PCR† | |||
| No. of samples | 38 | 17 | 334 |
| Clonotypic (%)‡ | 38 (100) | 17 (100) | 295 (88) |
| Clinical (%)2-153 | 38 (100) | 17 (100) | 295 (88) |
| Nonclinical (%)2-155 | 20 (53) | 2 (12) | 70 (21) |
| IgM (%)2-154 | 8 (21) | 0 | 26 (8) |
| Nested RT-PCR† | |||
| No. of samples | 38 | 17 | 334 |
| Clonotypic (%)‡ | 38 (100) | 17 (100) | 312 (93) |
| Clinical (%)2-153 | 38 (100) | 17 (100) | 299 (90) |
| Nonclinical (%)2-155 | 31 (82) | 7 (41) | 127 (38) |
| IgM (%)2-154 | 26 (68) | 2 (12) | 83 (25) |
PBMC indicates peripheral blood mononuclear cell; RT-PCR, reverse transcriptase polymerase chain reaction; IgM, immunoglobulin M, MM, multiple myeloma.
The PBMC samples were obtained from 29 patients, including 26 who provided BM and 17 who provided mobilized blood.
Single stage RT-PCR was strategy A and nested RT-PCR was strategy B (Figure 1).
The number of samples containing any clonotypic transcripts.
The number of samples containing clonotypic transcripts of the clinical isotype (or of at least one isotype, in the case of light chain MM patients).
The number of samples containing clonotypic transcripts of nonclinical isotypes (or of more than one isotype, in the case of light chain MM patients).
The number of samples containing clonotypic IgM transcripts.
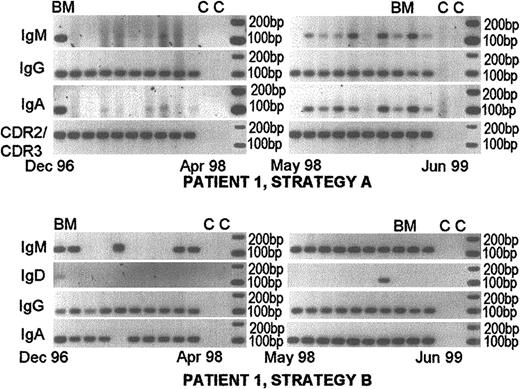
Persistent expression of nonclinical and/or clinical isotypes at diagnosis and throughout the course of disease.
Nonclinical isotypes are detectable in a single-stage RT-PCR (strategy A). Patient 1 was followed from diagnosis in December 1996 to June 1999. She was treated with melphalan and prednisone from diagnosis until April 1998. She was on dexamethasone therapy from April 1998 until her death in November 1999 from MM-related BM failure and sepsis. Her treatment was only of modest clinical benefit. Her clinical isotype was IgG. The top panels show RT-PCR results using strategy A and the bottom panels show results using strategy B. This patient is representative of those having nonclinical clonotypic isotype expression sufficiently frequent to be detected in a single-stage PCR reaction. Some samples that were negative with strategy A, became positive when tested using nested PCR (strategy B). All samples were positive for expression of the patient-specific VDJ gene rearrangement (strategy D). BM indicates bone marrow samples; c, control lanes lacking cDNA.
Persistent expression of nonclinical and/or clinical isotypes at diagnosis and throughout the course of disease.
Nonclinical isotypes are detectable in a single-stage RT-PCR (strategy A). Patient 1 was followed from diagnosis in December 1996 to June 1999. She was treated with melphalan and prednisone from diagnosis until April 1998. She was on dexamethasone therapy from April 1998 until her death in November 1999 from MM-related BM failure and sepsis. Her treatment was only of modest clinical benefit. Her clinical isotype was IgG. The top panels show RT-PCR results using strategy A and the bottom panels show results using strategy B. This patient is representative of those having nonclinical clonotypic isotype expression sufficiently frequent to be detected in a single-stage PCR reaction. Some samples that were negative with strategy A, became positive when tested using nested PCR (strategy B). All samples were positive for expression of the patient-specific VDJ gene rearrangement (strategy D). BM indicates bone marrow samples; c, control lanes lacking cDNA.
Persistent pretransplantation nonclinical clonotypic isotypes in blood, BM, and autografts, followed by a loss of nonclinical isotypes after transplantation and re-emergence of clonotypic IgM at the time of relapse.
Patient 2 was diagnosed in May 1997. She provided PBMC samples until January 1999. Her treatment consisted of 3 cycles of chemotherapy, followed by high-dose melphalan with autologous blood stem cell support in January 1998. Her disease was clinically stable until she relapsed in January 1999. She was not a candidate for further treatment, and she died in July 2000. Her clinical isotype was IgG. The clinical isotype was always detectable using strategy A. Nonclinical isotypes were only detected using strategy B. All samples were positive for strategy D. Mobilized blood had detectable clinical and nonclinical clonotypic isotypes, and the clinical isotype persisted postautologous transplantation. All samples to the right of the vertical line were taken after transplantation. M, mobilized blood autografts; R, relapse. Other abbreviations are as for Figure 2.
Persistent pretransplantation nonclinical clonotypic isotypes in blood, BM, and autografts, followed by a loss of nonclinical isotypes after transplantation and re-emergence of clonotypic IgM at the time of relapse.
Patient 2 was diagnosed in May 1997. She provided PBMC samples until January 1999. Her treatment consisted of 3 cycles of chemotherapy, followed by high-dose melphalan with autologous blood stem cell support in January 1998. Her disease was clinically stable until she relapsed in January 1999. She was not a candidate for further treatment, and she died in July 2000. Her clinical isotype was IgG. The clinical isotype was always detectable using strategy A. Nonclinical isotypes were only detected using strategy B. All samples were positive for strategy D. Mobilized blood had detectable clinical and nonclinical clonotypic isotypes, and the clinical isotype persisted postautologous transplantation. All samples to the right of the vertical line were taken after transplantation. M, mobilized blood autografts; R, relapse. Other abbreviations are as for Figure 2.
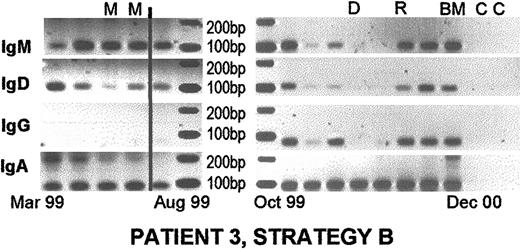
Persistent presence of nonclinical isotypes during treatment, in mobilized autografts, and after transplantation.
Patient 3 was diagnosed in February 1999 and followed until December 2000. He was treated with 4 cycles of combination chemotherapy, followed by high-dose melphalan and autologous blood stem cell support in July 1999. His disease was stable until May 2000, when his serum M protein began to rise. He was treated with dexamethasone from June 2000 until December 2000. He had a transient response to therapy, followed by progression in November 2000. Thalidomide was added in December 2000. The clinical isotype was IgA. Some samples were weakly positive for clinical isotype in strategy A (not shown) and strongly positive for rearranged IgH VDJ in strategy D (not shown), supporting the suggestion that strategy D is considerably more sensitive than strategy A. Nonclinical isotypes were weakly detectable using strategy A but were readily apparent using strategy B as shown. All samples to the right of the vertical line were postautologous transplantation. D, response to dexamethasone therapy. Other abbreviations are as for Figures 2 and 3.
Persistent presence of nonclinical isotypes during treatment, in mobilized autografts, and after transplantation.
Patient 3 was diagnosed in February 1999 and followed until December 2000. He was treated with 4 cycles of combination chemotherapy, followed by high-dose melphalan and autologous blood stem cell support in July 1999. His disease was stable until May 2000, when his serum M protein began to rise. He was treated with dexamethasone from June 2000 until December 2000. He had a transient response to therapy, followed by progression in November 2000. Thalidomide was added in December 2000. The clinical isotype was IgA. Some samples were weakly positive for clinical isotype in strategy A (not shown) and strongly positive for rearranged IgH VDJ in strategy D (not shown), supporting the suggestion that strategy D is considerably more sensitive than strategy A. Nonclinical isotypes were weakly detectable using strategy A but were readily apparent using strategy B as shown. All samples to the right of the vertical line were postautologous transplantation. D, response to dexamethasone therapy. Other abbreviations are as for Figures 2 and 3.
The 10 patients who provided BM at more than one point in time were reviewed for clinical correlation with the RT-PCR results. In 4 patients, there was clinical improvement (ie, lowering of serum M protein in response to therapy) in the interval between BM aspirates. In 3 of these 4 patients, there was an associated disappearance (on strategy B) or reduced abundance (ie, absent on strategy A but still present on strategy B) of nonclinical clonotypic isotypes that were initially detectable in the BM. In 5 patients, the multiple BM samples were obtained only at times of disease progression (ie, at diagnosis or at times of a rising M-protein level); in those patients, no change in the RT-PCR results was seen over time. In the remaining patient, there was disease stability at the time of the initial BM aspirate, followed by disease progression at the second BM aspirate; this finding was associated with the appearance of previously undetected nonclinical clonotypic transcripts in the second BM aspirate. These patterns suggest that the presence of multiple nonclinical clonotypic isotypes in the BM is associated with progressive disease, whereas their disappearance is associated with disease regression.
G-CSF–mobilized blood collections contain both clinical and nonclinical clonotypic transcripts, but clonotypic IgM transcripts are rarely detected
Seventeen of the patients studied underwent stem cell mobilization for autologous transplantation after cytoreduction in vivo with multiple cycles of combination therapy, including the 2 light chain MM patients. Clonotypic transcripts were detected in the mobilized blood of all 17 patients, using both single-stage and nested RT-PCR (Table 2, Figures 3,4). In 7 of 17 patients, more than one isotype was detected in mobilized blood; this finding includes 5 of 15 patients expressing a clinical IgH monoclonal protein and 2 of 2 patients with a light chain phenotype. However, clonotypic IgM was detectable in mobilized blood of only 2 of 17 patients and only using the nested technique (Figures3,4).
The patterns of clonotypic IgM expression in the blood over time were examined in the 17 patients undergoing high-dose therapy with autologous transplantation. In 12 of 17 patients, clonotypic IgM was not detectable in the blood prior to high-dose therapy, but in 3 of these 12 patients clonotypic IgM appeared at some point following stem cell reinfusion. In 4 of 17 patients, IgM was detected prior to high-dose therapy and persisted following stem cell reinfusion (Figure4). In one patient (Figure 3), IgM transcripts were present prior to high-dose chemotherapy but were absent from PBMCs following stem cell reinfusion until the time of clinical relapse. In total, clonotypic IgM transcripts were found in 8 of 17 patients at some point following high-dose chemotherapy.
Peripheral blood consistently contains clonotypic cells expressing the clinical isotype
A total of 334 PBMC samples from 29 patients were analyzed (Table2, Figures 2-4). Of the 334 samples, 312 (93%) contained clonotypic message (strategy D). Among the 27 patients with an M-protein phenotype, the clinical isotype was detected in a large majority of samples (286 of 318) and was detectable in most cases using strategy A (281 of 318) (Figure 2). In the 2 light chain MM patients, multiple postswitch clonotypic isotypes were detectable using both strategy A and strategy B in the majority of samples. In 1 of 2 light chain patients, clonotypic IgM was not detected in PBMCs (0 of 10 samples); in the other light chain patient, clonotypic IgM was detected in 2 of 6 samples using strategy A and in 6 of 6 samples using strategy B.
Peripheral blood also contains clonotypic cells expressing nonclinical isotypes, including IgM
Multiple isotypes, including nonclinical isotypes, were detectable using strategy A in 70 (21%) of 334 samples (Figure 2), and in 127 (38%) of 334 samples using strategy B (Table 2, Figures 2-4). Clonotypic IgM transcripts were found in only 26 (8%) of 334 samples on single-stage RT-PCR (Figure 2), but after nested RT-PCR 83 (25%) of 334 samples had detectable clonotypic IgM (Figures 2-4).
Qualitative assessment of the nested RT-PCR results revealed varying patterns of isotype expression over time. In 7 of 29 patients, there was no expression of nonclinical isotypes in the blood at any time. In 5 of 29 patients, multiple clonotypic isotypes, including IgM, were found in every blood sample. In the remaining 17 of 29 patients, including the 2 light chain MM patients, the spectrum of expression of clonotypic isotypes was variable. In some patients, the presence and/or abundance of clonotypic IgM transcripts seemed to wax and wane in association with disease progression and remission (Figure 3). However, in the majority of patients this pattern was not evident. When Studentt test was used to assess the mean serum M-protein level in samples with or without clonotypic IgM transcripts, no statistically significant differences were found for individual patients or for the data set (334 blood samples) as a whole.
Circulating MM cells can express at relatively high frequency the clinical and nonclinical isotypes of the clonotypic immunoglobulin
The relative frequency of pre and postswitch clonotypic isotypes in blood from patient 1, who had clonotypic IgM detectable in strategy A (Figure 2), was estimated by sequencing the cloned RT-PCR product. RNA isolated from peripheral blood of patient 1 was amplified in a single-stage RT-PCR using strategy C, to amplify all transcripts of the Vh3 family, regardless of clonotype (Table3). For this patient, transcripts of Vh3 family were detected in conjunction with μ, δ, γ, and α constant chains. RT-PCR products were subcloned and sequenced to determine the frequency of clonotypic transcripts. The sequenced products included portions of the relevant constant region gene, validating the isotype designation. Sequencing analysis showed that clonotypic transcripts were detected in all isotype classes in this single-stage RT-PCR, with the highest representation in the clinical isotype (IgG) (Table 3). The detection of clonotypic transcripts among the nonclinical isotypes confirmed a relatively high frequency (18%-29%) among the aggregate transcripts in each isotype class.
Clinical isotype encoding the M protein is exclusively clonotypic; nonclinical isotypes with detectable clonotypic product in single-stage RT-PCR include frequent clonotypic sequences
| Primers used . | No. subclones sequenced . | No. clonotypic (%) . |
|---|---|---|
| Vh3 leader/μ | 28 | 8 (29) |
| Vh3 leader/δ | 27 | 7 (26) |
| Vh3 leader/γ | 24 | 24 (100) |
| Vh3 leader/α | 28 | 5 (18) |
| Primers used . | No. subclones sequenced . | No. clonotypic (%) . |
|---|---|---|
| Vh3 leader/μ | 28 | 8 (29) |
| Vh3 leader/δ | 27 | 7 (26) |
| Vh3 leader/γ | 24 | 24 (100) |
| Vh3 leader/α | 28 | 5 (18) |
RNA from multiple myeloma peripheral blood mononuclear cells was amplified in single-stage reverse transcriptase polymerase chain reaction (RT-PCR), using primers to the Vh family that characterizes the clonotypic immunoglobulin heavy chain (IgH) variable diversity joining region (VDJ) rearrangement in each patient, and primers to the indicated constant region (strategy C). The products of each PCR reaction were subcloned and sequenced. Sequencing of subclones included the 5′ end of exon 1 for each isotype, confirming the specificity of the reaction. The pattern of sequences for each isotype analysis gives a relative frequency of the clonotypic IgH VDJ representation for nonclinical (IgM, IgD, and IgA) as compared with the clinical isotype (IgG).
Measurement of the patient's burden of circulating clonotypic cells expressing IgM transcripts: defining the “IgM detection rate”
It was hypothesized that clonotypic cells expressing IgM or other nonclinical isotypes were playing an important role in myeloma progression and that quantification of these cells at diagnosis might therefore predict clinical outcomes. Direct quantitative assays were not feasible for this study. Instead, using RT-PCR strategy B, the presence or absence of nonclinical clonotypic transcripts was assessed in all PBMC samples obtained. For each patient the percentage of blood samples containing detectable clonotypic IgM transcripts was used as an overall indicator of the patient's load of circulating clonotypic IgM cells during the study period. This percentage is hereafter referred to as the IgM detection rate or IgM-DR.
The limitations of using IgM-DR rather than a single, quantitative measurement of clonotypic IgM cells are (1) the need for serial blood samples, (2) the lack of direct quantitation of cells or transcripts, (3) the potential influence of the timing of sample collection on the subsequent results, and (4) the difference in sample numbers. Patients with greater follow-up times are likely to contribute more samples than will patients with shorter follow-up. In the present study, the first 3 limitations have been addressed by obtaining multiple samples at regular intervals for long periods of follow-up, ensuring that the samples collected were truly representative of each patient's full disease course (see “Patients, materials, and methods”). The potential confounding effects of the number of samples collected with longer follow-up times have been treated statistically (see below). IgM-DR varied significantly in this group of 29 patients, with a mean of 35%, a range from 0% to 100%, and a large SD of 39%. IgM-DR is, therefore, a reasonable and discriminating marker of the overall burden of clonotypic cells expressing IgM transcripts in an individual patient. IgM-DR would not be useful for prognostication because it requires prolonged serial sampling, but it is valuable for exploring correlations between clonotypic IgM expression and clinical parameters.
Persistence of clonotypic IgM transcripts in the blood is associated with decreased survival
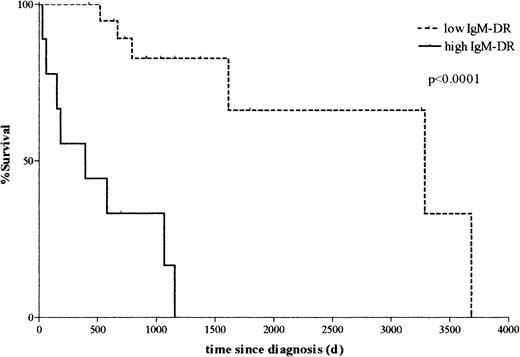
To assess the relationship between clinical MM disease and clonotypic IgM expression, a survival analysis was performed. Patients were divided into 2 groups: those with an IgM-DR of less than 50% (low IgM-DR) and those with an IgM-DR of 50% or more (high IgM-DR). These 2 groups represent the patients who had infrequently versus frequently detectable clonotypic IgM transcripts, respectively, in the peripheral blood. Kaplan-Meier curves plotting overall survival (OS) from the time of diagnosis were generated for the 2 groups and compared using the log-rank test. The group with low IgM-DR had significantly better OS than the group with high IgM-DR (median OS not reached in low IgM-DR group; 395 days in high IgM-DR group; P < .0001; Figure5). To test the robustness of this result, the analysis was repeated excluding patients at extremes of survival (< 1 year, n = 3; or > 5 years, n = 3). In this smaller data set, all patients provided at least 5 PBMC samples, with the majority providing 10 or more samples. The survival difference was confirmed with a high level of significance (median OS not reached in low IgM-DR group; 579 days in high IgM-DR group;P = .001), showing that the result was not an artifact of outlying data.
Persistent expression of clonotypic IgM is significantly correlated with decreased survival.
Kaplan-Meier survival curves comparing patients with detectable clonotypic IgM transcripts in less than 50% of blood samples (low IgM detection rate, or IgM-DR) versus patients with clonotypic IgM transcripts in 50% or more of blood samples (high IgM-DR). The group with low IgM-DR exhibits significantly better survival than the group with high IgM-DR (median survival not yet reached in low IgM-DR group; 395 days in high IgM-DR group; P < .0001 by the log-rank test).
Persistent expression of clonotypic IgM is significantly correlated with decreased survival.
Kaplan-Meier survival curves comparing patients with detectable clonotypic IgM transcripts in less than 50% of blood samples (low IgM detection rate, or IgM-DR) versus patients with clonotypic IgM transcripts in 50% or more of blood samples (high IgM-DR). The group with low IgM-DR exhibits significantly better survival than the group with high IgM-DR (median survival not yet reached in low IgM-DR group; 395 days in high IgM-DR group; P < .0001 by the log-rank test).
Using Cox univariate regression analysis, a strong negative correlation was found between IgM-DR and survival (P = .0005). In general, patients surviving for longer periods contributed more samples for analysis; therefore, a potentially confounding univariate correlation existed between survival and the number of samples (P = .02). However, when the number of samples obtained was included with IgM-DR in a 2-variable Cox multivariate survival analysis, IgM-DR remained a significant correlate of survival (P = .003), whereas the number of samples did not (P = .1). This result shows that the correlation between IgM-DR and survival was not due to a bias in the number of samples obtained.
Other known prognostic variables were univariate predictors of survival
Clinical parameters from the time of diagnosis that correlated with survival by Cox univariate regression analysis included hemoglobin (P = .02), albumin (P = .01), β2-microglobulin (P = .02), and the percentage of plasma cells in the marrow (P = .0007). Clinical parameters from the time of diagnosis that did not correlate with survival by univariate analysis included age, stage, serum creatinine, calcium, mode of therapy, and M-protein level.
IgM-DR correlated with other known prognostic variables
By using Pearson or Spearman correlation analysis as appropriate (see “Patients, materials, and methods”), there was a significant association between IgM-DR and several clinical parameters assessed at the time of diagnosis, including negative correlation with hemoglobin (P = .01), and positive correlations with β2-microglobulin (P = .02) and the percentage of plasma cells in diagnostic marrows (P = .0004). There was no significant correlation between IgM-DR and age, Durie-Salmon stage, serum albumin, creatinine, calcium, or M-protein levels, or with the type of therapy received.
IgM-DR was an independent correlate of survival
A stepwise multivariate Cox regression analysis of the factors influencing survival was performed to assess the independent influence of IgM-DR. The technique of substituting means for missing values was used. All variables that correlated with survival on univariate analysis (hemoglobin, albumin, β2-microglobulin, percentage of plasma cells in the marrow at diagnosis, and number of samples obtained) were included in the model.
In the resulting model, the factors found to independently correlate with survival were IgM-DR (P = .0005) and serum albumin level (P = .006). When the number of blood samples obtained was forcibly included in the model to account for potential confounding effects, it was not a significant correlate of survival (P = .1), but IgM-DR (P = .01) and albumin (P = .007) remained significant.
Ordinarily, β2-microglobulin is expected to be a powerful predictor of survival in MM. In this study, the baseline β2-microglobulin level was only available for 16 of 29 patients. This lack of statistical power likely explains why β2-microglobulin was not an independent predictor of survival in the multivariate analysis. For each of the other variables studied, missing data were limited to fewer than 3 cases.
Cells expressing nonclinical clonotypic isotypes engraft immunodeficient mice
In patients, clonotypic B cells expressing nonclinical isotypes appear to contribute to myeloma pathogenesis (Table 2, Figure 5). Although malignant cells might be expected to engraft in mice, persisting components of the nonmalignant parent clone for MM are likely to require antigen and T-cell interactions to survive for prolonged periods. Because human T cells are not detectable in these mice,26 and the unknown MM antigen is unlikely to be present in mice, xenografting was chosen to begin distinguishing these possibilities. To determine whether B cells expressing the nonclinical clonotypic isotypes engraft, freshly harvested cells from 5 myeloma patients with late-stage aggressive myeloma were xenografted into NOD SCID mice.26 The isotype profile of clonotypic cells was examined by using strategies A and B (Table4). All mice were engrafted as defined by the presence of clonotypic transcripts (strategy D). All mice had detectable clinical isotype (strategy A). Nonclinical isotypes, including one mouse with clonotypic IgM, were found in xenografts from only 2 patients using strategy A. In nested RT-PCR (strategy B), nonclinical isotypes were detectable in mice xenografted with cells from 4 of 5 patients. Clonotypic IgM was detectable in mice xenografted with cells from 2 of 5 patients. Analogous to patients who had transplantations, expression of clonotypic IgM could be transferred by a femoral BM graft from a primary to secondary recipients, indicating probable clonal expansion in the murine BM (Table 4, line 6). These results indicate that clonotypic cells expressing both clinical and nonclinical isotypes engraft immunodeficient mice.
Preswitch and postswitch clonotypic multiple myeloma cells engraft NOD SCID mice after primary and secondary xenotransplantation
| . | . | M protein . | CDR2/CDR3 . | PCR results from xenotransplanted mice . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Single-stage PCR . | Nested PCR . | ||||||||||
| IgM . | IgD . | IgG . | IgA . | IgM . | IgD . | IgG . | IgA . | ||||
| Patient 4 | 1° xenograft | IgG | 4/4 | 0/4 | 0/4 | 4/4 | 0/4 | 0/4 | 0/4 | 4/4 | 0/4 |
| Patient 5 | 1° xenograft | IgA | 5/5 | 0/5 | 0/5 | 0/5 | 5/5 | 0/5 | 0/5 | 1/5 | 5/5 |
| Patient 6 | 1° xenograft | IgA | 3/3 | 0/3 | 0/3 | 0/3 | 3/3 | 0/3 | 1/3 | 3/3 | 3/3 |
| Patient 7 | 1° xenograft | Lt chain | 3/3 | 0/3 | 0/3 | 2/3 | 2/3 | 1/3 | 0/3 | 1/3 | 1/3 |
| Patient 8 | 1° xenograft | IgA | 4/4 | 1/4 | 0/4 | 4/4 | 4/4 | 3/4 | 0/4 | 4/4 | 4/4 |
| Patient 8 | 2° xenograft | IgA | 2/2 | 0/2 | 0/2 | 0/2 | 2/2 | 1/2 | 0/2 | 0/2 | 2/2 |
| . | . | M protein . | CDR2/CDR3 . | PCR results from xenotransplanted mice . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Single-stage PCR . | Nested PCR . | ||||||||||
| IgM . | IgD . | IgG . | IgA . | IgM . | IgD . | IgG . | IgA . | ||||
| Patient 4 | 1° xenograft | IgG | 4/4 | 0/4 | 0/4 | 4/4 | 0/4 | 0/4 | 0/4 | 4/4 | 0/4 |
| Patient 5 | 1° xenograft | IgA | 5/5 | 0/5 | 0/5 | 0/5 | 5/5 | 0/5 | 0/5 | 1/5 | 5/5 |
| Patient 6 | 1° xenograft | IgA | 3/3 | 0/3 | 0/3 | 0/3 | 3/3 | 0/3 | 1/3 | 3/3 | 3/3 |
| Patient 7 | 1° xenograft | Lt chain | 3/3 | 0/3 | 0/3 | 2/3 | 2/3 | 1/3 | 0/3 | 1/3 | 1/3 |
| Patient 8 | 1° xenograft | IgA | 4/4 | 1/4 | 0/4 | 4/4 | 4/4 | 3/4 | 0/4 | 4/4 | 4/4 |
| Patient 8 | 2° xenograft | IgA | 2/2 | 0/2 | 0/2 | 0/2 | 2/2 | 1/2 | 0/2 | 0/2 | 2/2 |
NOD SCID indicates nonobese diabetic severe combined immunodeficiency; PCR, polymerase chain reaction; Ig, immunoglobulin; lt chain, light chain.
Single-stage polymerase chain reaction (PCR) used strategy A and nested PCR used strategy B. The single-stage CDR2/CDR3 primed reaction provides a control to confirm the presence of clonotypic messenger RNA in the patient peripheral blood mononuclear cells (PBMCs) used for xenografting to NOD SCID mice, using intracardiac injection, as previously described. Nonclinical clonotypic isotypes were detected only by nested reverse transcriptase (RT)-PCR (strategy B) in the patient PBMCs, suggesting they were quite infrequent cells. Results indicating the presence of nonclinical clonotypic transcripts are underlined. The majority of positive PCR reactions were for murine bone marrow (BM) and vertebral tumors, but spleen cells were also positive. A variety of evidence has suggested that strategies A and B, which are highly specific for the isotype being amplified, do not efficiently amplify clinical or nonclinical clonotypic isotypes in patients and in xenografted mice. This evidence suggests that the results in Table 4are likely to underestimate the true frequency of clonotypic isotypes. This suggestion is exemplified by the results for patient 4, where a single-stage CDR2/CDR3 (strategy D) strongly detected clonotypic transcripts (column 3), but none of the CDR3/nonclinical isotype combinations were able to detect clonotypic transcripts in these 4 mice, nor in all tissues from each mouse that were positive after PCR with CDR2/CDR3 primers (line 4). In contrast, use of a different strategy (PCR stage 1, Vh leader/constant region; stage 2, CDR2/CDR3) detected clonotypic IgM but not any other isotype in BM and spleen of these same mice (not shown). All 4 clonotypic isotypes were detectable in the peripheral cells harvested from each of the 5 patients and used for xenografting. For patient 8, pleural effusion cells that had exuded from a thoracic BM lesion were xenografted. The high clonogenicity of these cells, including detection of clonotypic IgM and nonclinical postswitch IgG using strategy A, suggests that the clonogenic multiple myeloma cells might be of BM origin. No Epstein-Barr virus (EBV) transcripts were detectable in tissues from any of these xenografted mice, eliminating the possibility that clonotypic IgM or any other clonotypic isotypes were derived from EBV transformants.
Discussion
This study describes the presence of MM cells expressing clinical and nonclinical clonotypic isotypes in the blood, BM, and G-CSF–mobilized blood from nearly all MM patients. Twenty-nine patients were followed for 2 to 107 months, pre and postautologous transplantation, to monitor clinical and nonclinical clonotypic isotypes. Over time, some patients had persistent expression of preswitch clonotypic transcripts in the blood despite therapy, including high-dose chemotherapy with stem cell rescue, suggestive of drug resistance. Other patients had only sporadic expression of nonclinical clonotypic isotypes during conventional chemotherapy and postautologous transplantation. The persistence of apparently drug resistant, preswitch clonotypic isotypes was associated with reduced survival and with more advanced disease at the time of diagnosis. For 8 of 17 patients, preswitch clonotypic transcripts were detected after transplantation, indicating that this component of the MM clone escapes therapy and/or is reinfused with the autograft. PBMCs expressing both pre and postswitch clonotypic isotypes were able to engraft immunodeficient mice, further emphasizing their potential clinical relevance. Because the pathology of MM is mediated by postswitch plasma cells, these results raise the possibility that IgH isotype class switching within the myeloma clone may accompany worsening disease.
In BM, the amount of clonotypic IgM message was relatively low, despite usually being obtained during relatively active periods of disease. IgM was detected only by nested RT-PCR in the majority of BM samples where it was found. Nonetheless, BM from the majority of patients harbored clonotypic cells expressing nonclinical IgH transcripts, including IgM. In contrast, IgM transcripts were rarely detectable in mobilized blood. This finding may have been because mobilized blood samples were always obtained at times of maximal disease regression. In support of this finding, for 10 patients who provided serial BM samples over time, we found that nonclinical clonotypic transcripts tend to be present in BM during periods of active disease and absent during periods of disease regression. Therefore, preswitch cells may not have been present in detectable numbers in the BM at the time of mobilization. In addition, it is possible that IgM-expressing cells did not mobilize well into the blood with chemotherapy and G-CSF. If these cells are truly absent from most mobilized blood collections, then their reappearance after transplantation must be due mainly to drug resistance rather than to autograft contamination.
At some points in time and with variable persistence, the full spectrum of preswitch and postswitch clonotypic isotypes was detectable in BM and PBMCs of nearly all patients tested. This study confirms, with a larger number of patients, published reports for PBMC and/or BM,7,8,24 indicating that the MM clone includes cells expressing the clonotypic IgH VDJ linked to μ, δ, γ, or α constant chains. Billadeau et al7 established that nonclinical clonotypic isotypes are expressed by CD38−CD45+ lymphocytes but not by plasma cells. Consistent with this finding, we found preswitch clonotypic transcripts in PBMCs having no detectable plasma cells and during minimal disease. Persistent clonotypic transcripts have been reported after transplantation.28,29 In our study, preswitch cells and nonclinical postswitch clonotypic isotypes were also detected after transplantation. In apparent contrast, Guikema et al30showed that post-transplantation samples lack nonclinical clonotypic isotypes, but their use of cryopreserved samples at only one time point may have compromised detection.
IgM-expressing clonotypic cells clearly reflect clinical disease activity. It remains to be determined whether these cells are malignant, or whether they compose a premalignant population whose behavior mirrors that of the malignancy, as discussed below. Xenotransplantations of primary human MM confirmed that cells expressing both clinical and nonclinical isotypes were able to engraft murine BM, including cells expressing clonotypic IgM. Clonotypic IgM-expressing cells localized in murine BM could be transplanted into BM of a secondary recipient. The ability of preswitch clonotypic cells to engraft immunodeficient mice provides support for the possibility that they may underlie myeloma progression in situ and after transplantation in patients.
Broadly speaking, there are at least 3 potential explanations for the persistence of nonclinical clonotypic isotypes in patients and in xenotransplanted mice. (1) The same B or plasma cell may express both preswitch and postswitch isotypes, for example, through some form of aberrant switch recombination events or trans-switching. (2) Discrete populations of preswitch and postswitch memory MM B cells may circulate in the blood and lodge in BM. (3) The MM progenitor cell may be a post-germinal center IgM-expressing memory B cell that undergoes persistent, directed isotype switching and differentiation to plasma cells. All models must accommodate the observations that MM exhibits hypermutated clonotypic variable regions having stringent intraclonal homogeneity11-14 and that disease progression may arise through switch recombination accompanied by chromosomal translocations.15-17,19 20
Model 1 assumes aberrant class-switch recombination with alternative RNA splicing or trans-switching–producing multiple isotypes with the same variable region within the same cell. Reports of neoplastic cells, or cell lines making multiple isotypes of the same specificity, support this possibility.31,32 However, Billadeau et al7 showed that nonclinical isotypes were found in the lymphoid fraction but not in the plasma cell fraction of MM BM. In addition, it is difficult to reconcile model 1 with our clinical observations unless more aggressive cellular behavior accompanies maximally aberrant alternative splicing of IgH transcripts.
Model 2 assumes distinct populations of post-germinal center memory B cells already committed to each isotype, with the malignant phenotype manifested only by the clinical isotype-expressing B and/or plasma cells. In this model, nonclinical isotype-expressing B cells may be nonmalignant or premalignant remnants of the parent clone giving rise to overt myeloma. Their rise and fall may be functionally tied to events within the MM clone, for example, in response to high cytokine levels. The negative clinical effect of nonclinical clonotypic isotypes may reflect aggressive stages of disease within clinical isotype-expressing cells. Alternatively, clonotypic B cells of the nonclinical isotypes may be immortalized but under negative regulation during periods when nonclinical clonotypic isotypes disappear. This model is inconsistent with the observed lack of correlation between preswitch transcripts and M-protein levels and with the xenograft capabilities of nonclinical isotype-expressing clonotypic cells. Both models 1 and 2 assume that a single isotype switch recombination event characterizes all members of the malignant clone.
Model 3 postulates that MM progenitors are preswitch memory B cells that generate postswitch plasma cells by continuous isotype switching, predominantly to the clinical isotype. This model is supported by the xenograft capability of cells expressing nonclinical isotypes and by their strong clinical correlation with reduced survival. If correct, preswitch MM B cells may selectively respond to cytokines regulating switching to the M-protein isotype, usually IgG isotypes or IgA, or they may be activated in the absence of cytokine receptor engagement. Division-related increases in class switching32 may promote a broader spectrum of clonotypic isotypes as well as increased switching to the clinical isotype. Model 3 predicts that multiple switch recombination junctions characterize B or plasma cells from a given patient. Provocatively, using Southern blot analysis, Palumbo et al25 identified multiple independent class switch recombinations occurring within the myeloma clone in 35% of patients, leading them to suggest that myeloma may originate from preswitch progenitors. Model 3 suggests that switch region translocations may contribute to disease progression rather than to originating events, a speculation supported by the lack of correlation between switch region translocations and prognosis shown by Ho et al.20
In summary, this work shows that preswitch and postswitch clonotypic isotypes are present at diagnosis, during therapy, and after transplantation. Members of the MM clone expressing nonclinical clonotypic preswitch and postswitch isotypes engraft mice. These observations suggest that myeloma may be a preswitch disease with pathologic complications arising from postswitch progeny. The persistence, and apparent drug resistance, of clonotypic IgM during/after therapy was significantly correlated with worsened survival. Persistent clonotypic IgM expression throughout disease also correlated significantly with more advanced disease at diagnosis, suggesting that drug resistant preswitch MM cells play a role in pretreatment phases of disease. These observations confirm the biological importance of cells expressing nonclinical clonotypic isotypes to the disease process in MM and suggest that the MM cells responsible for production of nonclinical clonotypic isotypes have a direct relationship to MM outcomes. Our results raise the possibility that some measure of clonotypic cells expressing preswitch IgH isotypes might be of independent prognostic value. With further study, the measurement of nonclinical clonotypic isotype expression may also prove to be a valuable marker of response to therapy or relapse and may, ultimately, identify new targets for therapy.
We thank the myeloma patients who so graciously contributed to this study. Yvon DeMossiac, Eva Pruski, Angie Battochio, Jennifer Carpenter, Tim Sousa, and Jennifer Szdylowski provided expert assistance.
Supported by grant RO1 CA80963 from the National Cancer Institute. A.J.S. was supported by a studentship from the Alberta Heritage Foundation for Medical Research.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Linda M. Pilarski, Cross Cancer Institute, 11560 University Ave, Edmonton, Alberta, T6G 1Z2, Canada; e-mail:lpilarsk@gpu.srv.ualberta.ca.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal