Abstract
Potential redundancy among members of the CCAAT/enhancer-binding protein (C/EBP) family in myeloid cells is indicated by the ability of C/EBPβ to replace C/EBPα in vivo, by the expression of granulocyte colony-stimulating factor receptor (G-CSFR) on C/EBPα−/− cell lines, and by our finding that as with C/EBPα–estrogen receptor (C/EBPα-ER), either C/EBPβ-ER or C/EBPδ-ER can induce terminal granulopoiesis in 32D cl3 cells. To assess the consequences of globally inhibiting C/EBPs, we employed KαER, containing a Kruppel-associated box (KRAB) transrepression domain, the C/EBPα DNA-binding domain, and an ER ligand-binding domain. C/EBPs have a common DNA-binding consensus, and activation of KαER repressed transactivation by endogenous C/EBPs 50-fold and reduced endogenous G-CSFR expression. In 32D cl3 cells coexpressing exogenous G-CSFR, activation of KαER prevented and even reversed myeloperoxidase, lysozyme, lactoferrin, and C/EBPε RNA induction by G-CSF. In contrast, induction of PU.1 and CD11b, a gene regulated by PU.1 but not by C/EBPs, was unaffected. A KαER variant incapable of binding DNA owing to an altered leucine zipper did not affect 32D cl3 differentiation. Transduction of KαER into murine hematopoietic progenitor cells suppressed the formation of granulocyte colony-forming units, even in cytokines that enable C/EBPα−/−progenitors to differentiate into neutrophils. The formation of macrophage and of granulocyte-macrophage colony-forming units were also inhibited, but erythroid burst-forming units grew normally. Thus, in 32D cl3 cells and perhaps normal progenitors, C/EBPs are required for granulopoiesis beyond their ability to induce receptors for G-CSF and other cytokines. One requisite activity may be activation of the C/EBPε gene by C/EBPα, as either C/EBPα-ER or C/EBPβ-ER rapidly elevated C/EBPε RNA in 32D cl3 cells in the presence of cycloheximide but not actinomycin D.
Introduction
The CCAAT/enhancer-binding protein (C/EBP) transcription factors homodimerize and heterodimerize via their C-terminal leucine zipper domains and bind DNA as obligate dimers via the adjacent basic regions.1,2 C/EBPα, C/EBPβ, and C/EBPδ have N-terminal transactivation domains, and translation initiation from internal methionines produces truncated, dominant-inhibitory polypeptides that retain the bZIP domain.3-5 C/EBPε has both transactivation and transrepression domains,6 and C/EBPγ and CHOP are dominant inhibitory by virtue of their ability to dimerize with other C/EBPs and their lack of intact basic regions.7 8
Within hematopoiesis, full-length C/EBPα, C/EBPβ, and C/EBPδ are predominantly expressed in the granulocyte, monocyte, and eosinophil lineages.9-11 C/EBPα is the most prominent isoform detected in immature granulocytes,9,12 whereas C/EBPε is found in later-stage granulocytes as well as in T cells.13CHOP is detected only in granulocytic cells subjected to stress, such as DNA damage,14 and C/EBPγ expression in this lineage is not well characterized.
C/EBPα−/− mice lack neutrophils and eosinophils, but retain monocytes, lymphocytes, erythroid cells, and immature myeloblasts.15 Fetal liver cells from these mice lack granulocyte colony-stimulating factor receptor (G-CSFR) RNA, consistent with the ability of C/EBPα to transactivate the G-CSFR promoter.16 Transduction of G-CSFR or interleukin-6 (IL-6) receptor complementary DNA (cDNAs) into C/EBPα−/− fetal liver cells largely restores their ability to generate neutrophils in vitro in response to G-CSF or IL-6.17C/EBPβ−/− mice retain all of the hematopoietic lineages, and hematopoietic defects in mice lacking C/EBPδ or both C/EBPβ and C/EBPδ were not severe.18-20C/EBPε−/− mice also retain neutrophils, although they lack secondary granules.21 22
The presence of several C/EBPs in the myeloid lineages suggests that related family members might compensate in vivo for the lack of a single isoform, just as GATA family members partially compensate for the lack of GATA-1.23 Potential redundancy is evident from the ability of C/EBPβ to compensate for the loss of C/EBPα in hepatocytes24 and granulocytes (Y.-H. Lee, written personal communication, June 2001), from the ability of both C/EBPα and C/EBPε to direct the granulocytic differentiation of myeloblastic cell lines,11,25,26 from findings presented here indicating that C/EBPβ or C/EBPδ can direct 32D cl3 cell maturation, and from the expression of high-levels of G-CSFR messenger RNA (mRNA) in EML cell lines lacking C/EBPα.27
To determine the effect of global inhibition of C/EBP-regulated genes on granulopoiesis, we developed a potent dominant-inhibitory protein, Kruppel-associated box (KRAB)–C/EBPα–estrogen receptor (KαER), in which the C/EBPα DNA-binding domain is linked to both a KRAB transrepression domain and the ER ligand-binding domain. We pursued this strategy after failing to observe phenotypic changes in 32D cl3 lines expressing truncated C/EBPα or C/EBPβ proteins, those lacking their N-terminal transactivating domains. The C/EBP family members bind with similar affinity to a common DNA consensus site.28Therefore, when activated by 4-hydroxytamoxifen (4HT), KαER is expected to interfere with transactivation by each C/EBP. KαER reduced expression of endogenous G-CSFR in 32D cl3 cells and rendered these cells unresponsive to G-CSF. In 32D cl3 lines expressing exogenous G-CSFR, KαER arrested granulopoiesis at the myeloblast stage, whereas a variant incapable of binding DNA had no effect. When transduced into murine hematopoietic progenitors, KαER inhibited formation of granulocyte colony-forming units (CFU-Gs), macrophage CFUs (CFU-Ms), and granuloctye-macrophage CFUs (CFU-GMs), even in the presence of IL-3 or GM-CSF, cytokines that allowed development of these colonies from C/EBPα−/− cells.17 Thus, in 32D cl3 cells and perhaps in normal progenitors, C/EBPs are required for granulopoiesis beyond their ability to induce the expression of cytokine receptors. To identify relevant target genes, we evaluated the effect of C/EBP inhibition and activation on several myeloid differentiation markers and transcription factors. Uniquely, lysozyme and C/EBPε RNA levels were both suppressed by KαER and rapidly induced by C/EBPαwildtype (WT)–ER or C/EBPβWT-ER in the presence of cycloheximide.
Materials and methods
Cell culture, transduction, and proliferation assays
The 32D cl3 cells29 were cultured in Iscoves modified Dulbecco medium (IMDM) with 10% heat-inactivated fetal calf serum (HI-FCS), 1 ng/mL IL-3 (R&D Systems, Minneapolis, MN), and penicillin (100 units/mL)/streptomycin (100 μg/mL). To induce differentiation, the cells were washed twice with phosphate-buffered saline and transferred to IMDM with 10% HI-FCS and 20 ng/mL human G-CSF (Amgen, Thousand Oaks, CA). We used 200 nM 4HT, 1 μM estradiol, and 0.1% ethanol as a vehicle control. We added 50 μg/mL cycloheximide or 10 μg/mL actinomycin D 30 minutes before adding the estradiol. NIH 3T3 and ψCRE cells30 were cultured in Dulbecco modified Eagle medium (DMEM) with 10% heat-inactivated calf serum. The ψCRE cells were transfected by calcium phosphate precipitation, and pools of transfectants were obtained by selection with 2 μg/mL puromycin or 1.0 mg/mL G418 (total). BOSC2331 and Phoenix-A cells32 were cultured in DMEM with 10% HI-FCS and were transfected for 5 hours with the use of Lipofectamine 2000 (Gibco-BRL, Gaithersburg, MD), at a ratio of 2 μg DNA per 6 μL lipid for each 60-mm dish. The 32D cl3 cells were transduced by coculture for 2 days with 4 μg/mL polybrene (Aldrich, Milwaukee, WI) and subconfluent ψCRE packaging cells irradiated to 3000 cGy. After transduction with pBabePuro-KαER or pBabePuro–Kα-1,2Val-ER (pBabePuro–KVER), subclones were isolated by limiting dilution in 2 μg/mL puromycin. Puromycin-resistant 32D–KαER-1 and 32D–KVER-2 cells were in turn transduced with pLNCX–G-CSFR, and 32D-KαER/G-CSFR (GR) and 32D-KVER/GR subclones were isolated by limiting dilution in 1.2 mg/mL G418. Morphology was assessed by Wright-Giemsa staining of cytospins. Viable cell counts were obtained by enumerating, by means of a hemocytometer, cells that exclude trypan blue dye. Bromodeoxyuridine/propidium iodide staining and fluorescent-activated cell sorter (FACS) analysis and fluorescein isothiocyanate (FITC)–annexin V binding assays were performed as described.33 34
Plasmids and transient transfection
The KαER cDNA was constructed by linking cDNA segments encoding amino acids 1 through 89 of KOX1,35 amino acids 191 through 356 (MluI to NcoI) of rat C/EBPα,36 and the tamoxifen-responsive murine ERα ligand-binding domain.37 An additional 6 base pairs (bp), 5′-GGATCC-3′, are present between the KOX1 and C/EBPα segments. This cDNA was ligated into the polylinker of pBabePuro38 as aBamH1/SalI fragment. The C/EBPα segment in this plasmid was replaced with an MluI-NcoI fragment derived from pMSV–C/EBP-1,2Val,39 to generate pBabePuro-KVER. The p(C/EBP)2TKLUC has been described,14 as has pMSV-C/EBPα-WT.39 The C/EBPβWT-ER and C/EBPδWT-ER fusion proteins, containing the estradiol-responsive human ERα ligand-binding domain, were kindly provided by Z. Cao and S. McKnight (Tularik, San Francisco, CA) and were ligated into the polylinker of pBabePuro. The pBabe-ER was constructed by deleting the 1062-bpNcoI-NcoI fragment, encoding C/EBPα residues 2 through 355, from pBabePuro-C/EBPαWT-ER. The pLNCX-G-CSFR has been described.40 The KαER cDNA was also ligated into the MIG retroviral vector, containing the murine stem cell virus (MSCV) long terminal repeat followed by a polylinker, an internal ribosome entry site, and the enhanced green fluorescent protein (EGFP) cDNA (kindly provided by L. Cheng, Johns Hopkins University, Baltimore, MD). Transient transfection of 32D cl3 or NIH 3T3 cells, luciferase assays, and β-galactosidase (βGal) assays were performed as described.41The p–cytomegalovirus (pCMV)–βGal was included in each transfection as an internal control.
Western, Northern, and FACS analyses
Preparation of total cellular protein and RNA, Western blotting, and filter stripping were performed as described.41 Each figure represents the same blot probed sequentially. Murine and human ERα antisera (MC-20 and HC-20; Santa Cruz Biotechnology, Santa Cruz, CA) were employed at 1:1000. The murine myeloperoxidase (MPO), lactoferrin (LF), lysozyme, PU.1, G-CSFR, C/EBPε, and β-actin cDNA probes have been described.40,42-44Assessment of G-CSFR expression was performed as described.44 In brief, human G-CSF was biotinylated by means of a kit (Pierce, Rockford, IL). Then, 2 × 106cells were washed and incubated in 100 μL on ice with 5 μg/mL biotin–G-CSF in the presence or absence of 1000-fold excess, unlabeled G-CSF. The cells were then washed 3 times, incubated with 10 μg/mL streptavidin-phycoerythrin (PE), washed, fixed with 1% paraformaldehyde in 10 mM Hepes, pH 7.4, and analyzed by FACS. CD11b expression was assessed with the use of FITC–anti-CD11b antibody or rat immunoglobulin G2b isotype control, with the use of 2 μL of each to stain 106 cells in 100 μL (BD Pharmingen, San Diego, CA).
Transduction and analysis of murine hematopoietic progenitors
MIG-KαER was transfected into Phoenix-A cells, and supernatant collected 48 hours later was used to transduce ψCRE cells in the presence of 4 μg/mL polybrene. At 7 days later, GFP+cells were isolated by flow cytometry and expanded. GFP+cells were then again selected, to yield a uniformly GFP+packaging line. Marrow cells for transduction were obtained from the long bones of C57BL/6 mice that had been treated by tail vein injection with 150 mg/kg 5-fluorouracil (5-FU) 3 days earlier. Fetal liver cells were obtained at day 14.5 of gestation from timed matings and were rendered into a single suspension by passage through a 25-gauge needle. Isolated marrow cells were prestimulated for 48 hours with 10 ng/mL IL-3, 50 ng/mL IL-6, and 100 ng/mL stem cell factor (SCF) in IMDM with 10% HI-FBS and penicillin (100 units/mL)/streptomycin (100 μg/mL), and then cocultured with irradiated CRE–MIG–KαER cells in the same media in the presence of 8 μg/mL polybrene for an additional 72 hours in 100-mm dishes. To stimulate erythroid progenitors from fetal liver, we employed DMEM with 15% HI-FBS, 1% bovine serum albumin (BSA), 1.9 mM NaHC03, 0.1 mM β-mercaptoethanol, 128 μg/mL transferrin, 1 μM dexamethasone, 1 μM β-estradiol, 3 U/mL erythropoietin (Epo), and 100 ng/mL SCF, as described.45 During this 72-hour period, 3 mL BOSC23 supernatant obtained from cells transiently transfected with MIG-KαER was added to the coculture at both 24 and 48 hours. Transduced cells were then rinsed with media and recovered in prestimulation media for 1 day. GFP+ marrow or fetal liver cells were then isolated by flow cytometry and cultured in Marrow-Gro methylcellulose (Quality Biologicals, Gaithersburg, MD), including IMDM, 32.5% HI-FBS, 0.11 mM β-mercaptoethanol, 1.1% BSA, penicillin (100 units/mL)/streptomycin (100 μg/mL), and either 20 ng/mL G-CSF, 10 ng/mL macrophage CSF (M-CSF), 10 ng/mL GM-CSF, 10 ng/mL IL-3, or the combination of 3 U/mL Epo and 5 μg/mL insulin. Each 35-mm dish was seeded with 2 to 4 × 103 transduced marrow cells or 2 × 104 transduced fetal liver cells, with 200 nM 4HT or the ethanol vehicle, and colony numbers were assessed on days 8 through 10 by light microscopy. Several colonies of each type were plucked, cytospun, and subjected to Wright-Giemsa staining to confirm the accuracy of morphologic assignments.
Results
C/EBPβ or C/EBPδ induce granulopoiesis in 32D cl3 cells
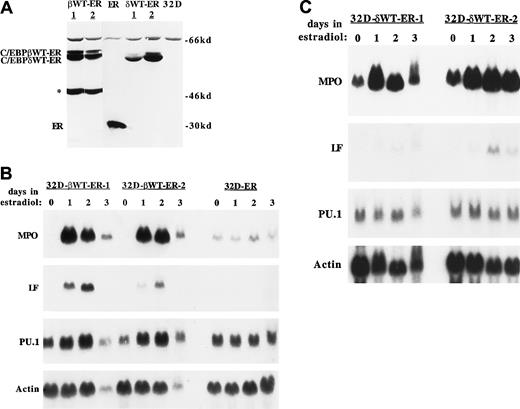
C/EBPβWT-ER, C/EBPδWT-ER, and the ER segment alone were introduced into 32D cl3 cells by retroviral transduction. Subclones obtained by limiting dilution were assessed for expression of the introduced proteins by Western blotting, with the use of extracts corresponding to equivalent numbers of cells (Figure1A). A slowly migrating, nonspecific band is evident in each extract, including the extract derived from parental 32D cl3 cells. The 2 subclones expressing full-length C/EBPβWT-ER also express a prominent shorter form (indicated by an asterisk in Figure 1A) reactive with the ER antisera. As the ER segment is at the C-terminal end of the fusion protein, this shorter form probably results from the use of an internal ATG.4 Only one subclone was obtained in which the expression of the ER segment alone was similar to that of C/EBPβWT-ER and C/EBPδWT-ER.
Effect of C/EBPβ or C/EBPδ on granulopoiesis.
C/EBPβ or C/EBPδ induces granulopoiesis. (A) Total cellular proteins corresponding to 1 × 106 cells, from subclones transduced with pBabePuro-C/EBPβWT-ER (βWT-ER), pBabePuro-ER (ER), or pBabePuro-C/EBPδWT-ER (δWT-ER) or from parental 32D cl3 cells (32D) were subjected to Western blotting with the use of a human ER antiserum. The position of a truncated C/EBPβWT-ER isoform is indicated by an asterisk. (B) 32D-C/EBPβWT-ER-1 cells, 32D-C/EBPβWT-ER-2 cells, and 32D-ER cells were exposed to 1 μM estradiol for 0, 1, 2, or 3 days. Total RNAs prepared each day were subjected to Northern blotting for MPO, LF, PU.1, and β-actin by sequential probing of the same blot. (C) 32D-C/EBPδWT-ER-1 and 32D-C/EBPδWT-ER-2 cells were analyzed similarly.
Effect of C/EBPβ or C/EBPδ on granulopoiesis.
C/EBPβ or C/EBPδ induces granulopoiesis. (A) Total cellular proteins corresponding to 1 × 106 cells, from subclones transduced with pBabePuro-C/EBPβWT-ER (βWT-ER), pBabePuro-ER (ER), or pBabePuro-C/EBPδWT-ER (δWT-ER) or from parental 32D cl3 cells (32D) were subjected to Western blotting with the use of a human ER antiserum. The position of a truncated C/EBPβWT-ER isoform is indicated by an asterisk. (B) 32D-C/EBPβWT-ER-1 cells, 32D-C/EBPβWT-ER-2 cells, and 32D-ER cells were exposed to 1 μM estradiol for 0, 1, 2, or 3 days. Total RNAs prepared each day were subjected to Northern blotting for MPO, LF, PU.1, and β-actin by sequential probing of the same blot. (C) 32D-C/EBPδWT-ER-1 and 32D-C/EBPδWT-ER-2 cells were analyzed similarly.
Morpologically, the cell lines expressing C/EBPβWT-ER or C/EBPδWT-ER differentiated to neutrophils when exposed to estradiol, whereas the 32D-ER cells remained myeloblastic (not shown). Expression of granulocytic markers in response to estradiol was evaluated in these cell lines by Northern blotting (Figure 1B-C). MPO reached maximal levels by 24 hours and LF peaked by 48 hours in the subclones expressing C/EBPβWT-ER or C/EBPδWT-ER, whereas the expression of these RNAs was not affected by estradiol in the 32D-ER line. This pattern of MPO and LF expression is consistent with that seen when parental 32D cl3 cells differentiate to neutrophils in response to G-CSF.29,46 The 32D-C/EBPβWT-ER lines were fully mature by day 2, accounting for the reduced level of all 4 RNAs on day 3, when the cells were undergoing apoptosis. These findings indicate that, as described for C/EBPα,25 expression of exogenous C/EBPβ or C/EBPδ is sufficient to induce terminal granulopoiesis in a diploid, factor-dependent myeloblastic cell line.
Of note, C/EBPβWT-ER activation led to increased endogenous PU.1 RNA expression, whereas PU.1 levels did not change when either 32D-C/EBPδWT-ER line was exposed to estradiol. C/EBPαWT-ER induced endogenous PU.1 expression even in the presence of cycloheximide.25 The PU.1 levels present in the 2 32D-C/EBPδWT-ER lines studied are apparently sufficient to allow terminal neutrophilic differentiation.
Development of a regulated, dominant-negative C/EBP
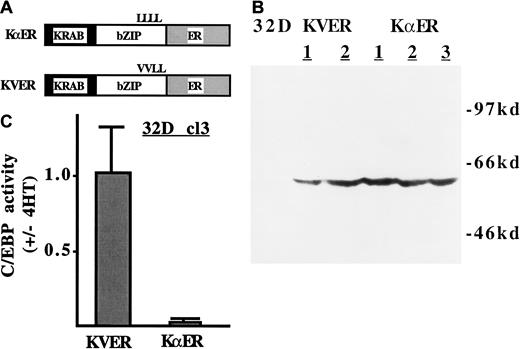
To determine the consequences of repressing genes regulated by C/EBP family members, we generated a fusion between an 89–amino acid KRAB transrepression domain (K), the C/EBPα DNA-binding domain (α), and a variant of the murine ER ligand-binding domain that is activated by 4HT, but not by estradiol. KαER is diagrammed in Figure2A. Also shown is the diagram of a variant, KVER, in which the first 2 leucines of the C/EBPα segment's leucine zipper have been mutated to valine. KVER is expected to be incapable of both dimerization and DNA-binding.1 The 32D cl3 cells were stably transduced with KαER and KVER. Exogenous protein was detected by Western blotting in several subclones (Figure2B). C/EBP activities in 32D–KαER-1 and 32D–KVER-2 cells were assessed by transient transfection of p(C/EBP)2TKLUC, a reporter plasmid containing 2 C/EBP-binding sites, a minimal thymidine kinase promoter, and the luciferase cDNA. In the presence of 4HT, KαER reduced the activity of p(C/EBP)2TKLUC in 32D cl3 cells approximately 50-fold, whereas KVER had no effect (Figure 2C). KαER also specifically inhibited activation by exogenous C/EBPα in NIH 3T3 cells (not shown). KVER did not affect 32D cl3 proliferation in IL-3 or in G-CSF, and KαER only mildly affected their proliferation in IL-3 (see below and data not shown). Therefore, KαER is a potent inhibitor of endogenous C/EBPs in the presence of 4HT and does not introduce nonspecific toxic effects.
An inducible dominant inhibitor of C/EBP-regulated genes.
(A) KRAB-C/EBPα-ER (KαER) and KRAB-C/EBPα-1,2Val-ER (KVER) are diagrammed. The KRAB segment is a transrepression domain derived from Kox-1. The C/EBPα segment contains the C/EBPα DNA-binding domain, but not its trans-activating domains. All C/EBP family members have a common DNA-binding consensus, and it is therefore expected that each will be inhibited by KαER. The ER segment is responsive to 4HT. In KVER, mutation of 2 leucines (L) to valine (V) prevents DNA binding. (B) Total cellular proteins corresponding to 1 × 106 cells, from 32D cl3 cells or from subclones transduced with pBabe-KVER or pBabe-KαER, were subjected to Western blotting with the use of a murine ER antiserum. (C) First, 5 × 106 32D–KαER-1 and 32D–KVER-2 cells proliferating in IL-3 were transfected with 20 μg p(C/EBP)2TKLUC and 1.0 μg pCMV-βGal with the use of diethylaminoethyl-dextran. One-half of each culture was then treated with 200 nM 4HT and the other half with 0.1% ethanol. Luciferase and βGal activities were assessed 2 days later. The ratio of reporter activity (activity with 4HT/activity without 4HT), normalized to the internal control, is shown for each condition (mean and SE from 2 experiments).
An inducible dominant inhibitor of C/EBP-regulated genes.
(A) KRAB-C/EBPα-ER (KαER) and KRAB-C/EBPα-1,2Val-ER (KVER) are diagrammed. The KRAB segment is a transrepression domain derived from Kox-1. The C/EBPα segment contains the C/EBPα DNA-binding domain, but not its trans-activating domains. All C/EBP family members have a common DNA-binding consensus, and it is therefore expected that each will be inhibited by KαER. The ER segment is responsive to 4HT. In KVER, mutation of 2 leucines (L) to valine (V) prevents DNA binding. (B) Total cellular proteins corresponding to 1 × 106 cells, from 32D cl3 cells or from subclones transduced with pBabe-KVER or pBabe-KαER, were subjected to Western blotting with the use of a murine ER antiserum. (C) First, 5 × 106 32D–KαER-1 and 32D–KVER-2 cells proliferating in IL-3 were transfected with 20 μg p(C/EBP)2TKLUC and 1.0 μg pCMV-βGal with the use of diethylaminoethyl-dextran. One-half of each culture was then treated with 200 nM 4HT and the other half with 0.1% ethanol. Luciferase and βGal activities were assessed 2 days later. The ratio of reporter activity (activity with 4HT/activity without 4HT), normalized to the internal control, is shown for each condition (mean and SE from 2 experiments).
A dominant-negative C/EBP reduces endogenous G-CSFR expression in 32D cl3 cells
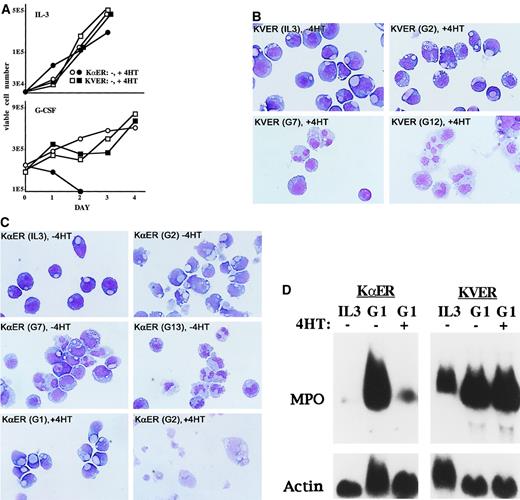
The proliferation of 32D-KVER and 32D–KαER cells was assessed in IL-3 or G-CSF, in the presence or absence of 4HT (Figure3A). The 4HT did not alter the growth of 32D-KVER cells, whereas 32D–KαER cells exposed to 4HT mildly slowed their proliferation in IL-3 and rapidly died in G-CSF. KVER cells differentiated to neutrophils in the presence of 4HT and G-CSF (Figure3B). The finding that KVER did not alter the differentiation of 32D cl3 cells indicates that the KRAB domain does not produce nonspecific toxicities. In the absence of 4HT, the KαER cell lines differentiated to neutrophils in response to G-CSF, whereas in the presence of 4HT they developed an apoptotic nuclear morphology by day 2 (Figure 3C). Induction of apoptosis in 32D-KαER cells transferred to G-CSF in the presence of 4HT was confirmed with the use of FITC–annexin V (not shown). Apoptosis was not induced when KαER was activated in 32D cl3 cells proliferating in IL-3; the slowing observed resulted from modest inhibition of G1 to S progression (not shown). Induction of MPO RNA by G-CSF was prevented by activation of KαER but not KVER, confirming that KVER with its KRAB domain did not prevent differentiation in response to G-CSF (Figure 3D).
Effect of C/EBP when 32D cl3 cells are cultured in G-CSF.
C/EBP inhibition leads to apoptosis without differentiation when 32D cl3 cells are cultured in G-CSF. (A) First, 2 × 10432D–KαER-1 (circles) or 32D–KVER-2 (squares) cells were cultured in IL-3 either without 4HT (open) or with 4HT (filled), and viable cell counts were obtained daily. Results of a typical experiment are shown (top). Then, 2 × 105 32D–KαER-1 or 32D–KVER-2 cells were transferred to G-CSF without or with 4HT, and viable cell counts were again obtained daily (bottom). (B) Morphology of 32D–KVER-2 cells in IL-3 and after transfer to G-CSF for 2, 7, or 12 days (G2, G7, G12) with 4HT. Wright-Giemsa stain, original magnification ×100. (C) Morphology of 32D–KαER-1 cells in IL-3, after transfer to G-CSF for 2, 7, or 13 days (G2, G7, G13) without 4HT, or in G-CSF for 1 or 2 days (G1, G2) with 4HT. Wright-Giemsa stain, original magnification ×100. (D) Total cellular RNAs were prepared from 32D–KαER-1 or 32D–KVER-2 cells proliferating in IL-3 or 1 day after transfer to G-CSF, without or with 4HT. These RNAs, 20 μg per lane, were subjected to Northern blot analysis for MPO and β-actin.
Effect of C/EBP when 32D cl3 cells are cultured in G-CSF.
C/EBP inhibition leads to apoptosis without differentiation when 32D cl3 cells are cultured in G-CSF. (A) First, 2 × 10432D–KαER-1 (circles) or 32D–KVER-2 (squares) cells were cultured in IL-3 either without 4HT (open) or with 4HT (filled), and viable cell counts were obtained daily. Results of a typical experiment are shown (top). Then, 2 × 105 32D–KαER-1 or 32D–KVER-2 cells were transferred to G-CSF without or with 4HT, and viable cell counts were again obtained daily (bottom). (B) Morphology of 32D–KVER-2 cells in IL-3 and after transfer to G-CSF for 2, 7, or 12 days (G2, G7, G12) with 4HT. Wright-Giemsa stain, original magnification ×100. (C) Morphology of 32D–KαER-1 cells in IL-3, after transfer to G-CSF for 2, 7, or 13 days (G2, G7, G13) without 4HT, or in G-CSF for 1 or 2 days (G1, G2) with 4HT. Wright-Giemsa stain, original magnification ×100. (D) Total cellular RNAs were prepared from 32D–KαER-1 or 32D–KVER-2 cells proliferating in IL-3 or 1 day after transfer to G-CSF, without or with 4HT. These RNAs, 20 μg per lane, were subjected to Northern blot analysis for MPO and β-actin.
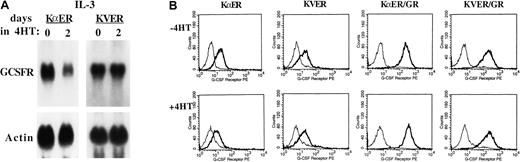
As C/EBPα−/− mice have greatly reduced G-CSFR expression and responsiveness, the effect of KαER and KVER on the expression of endogenous G-CSFR RNA was assessed (Figure4A). Activation of KαER, but not KVER, for 2 days reduced G-CSFR RNA levels several-fold in 32D cl3 cells proliferating in IL-3. To determine the effect of C/EBP inhibition on cell surface expression of the G-CSFR, 32D–KαER-1 or 32D–KVER-2 cells, cultured without or with 4HT for 2 days, were incubated with biotin–G-CSF in the presence or absence of an excess of unlabeled G-CSF. Bound biotin–G-CSF was then detected by means of streptavidin-PE and FACS analysis (Figure 4B, left panels). Activation of KαER reduced surface G-CSFR expression approximately 3-fold, whereas activation of KVER had no effect. Reduced G-CSFR expression probably accounts for the apoptosis observed when 32D-KαER cells are cultured with 4HT and G-CSF.
Effect of C/EBP inhibition on endogenous and exogenous G-CSFR expression.
Inhibition of C/EBPs reduces endogenous, but not exogenous, G-CSFR expression. (A) Total cellular RNAs were prepared from 32D–KαER-1 and 32D–KVER-2 cells proliferating in IL-3, after exposure to 4HT or the ethanol vehicle for 2 days. These RNAs were subjected to Northern blotting for G-CSFR and β-actin. (B) The 32D–KαER-1, 32D–KVER-2, 32D-KαER/GR, and 32D-KVER/GR cells were cultured with 4HT or the ethanol vehicle for 2 days. Then, 2 × 106 cells from each culture were incubated with biotin–G-CSF in the absence (thick lines) or presence (thin lines) of 1000-fold excess unlabeled G-CSF. The cells were then incubated with streptavidin-PE, fixed, and subjected to FACS analysis.
Effect of C/EBP inhibition on endogenous and exogenous G-CSFR expression.
Inhibition of C/EBPs reduces endogenous, but not exogenous, G-CSFR expression. (A) Total cellular RNAs were prepared from 32D–KαER-1 and 32D–KVER-2 cells proliferating in IL-3, after exposure to 4HT or the ethanol vehicle for 2 days. These RNAs were subjected to Northern blotting for G-CSFR and β-actin. (B) The 32D–KαER-1, 32D–KVER-2, 32D-KαER/GR, and 32D-KVER/GR cells were cultured with 4HT or the ethanol vehicle for 2 days. Then, 2 × 106 cells from each culture were incubated with biotin–G-CSF in the absence (thick lines) or presence (thin lines) of 1000-fold excess unlabeled G-CSF. The cells were then incubated with streptavidin-PE, fixed, and subjected to FACS analysis.
A dominant-negative C/EBP blocks differentiation in response to exogenous G-CSFR
The 32D–KαER-1 and 32D–KVER-2 cells were transduced with the human G-CSFR. Then, two 32D-KαER/GR sublcones (designated KαER/GR and KαER/GR2) and one 32D–KVER/GR subclone were isolated and subjected to further analysis. Expression of exogenous G-CSFR was confirmed by means of biotin–G-CSF and FACS analysis (Figure 4B, right panels and data not shown for the 32D-KαER/GR2 clone). Exogenous G-CSFR is expressed more than 10-fold higher than endogenous G-CSFR in these lines, and its expression was not affected by addition of 4HT (Figure 4B).
To assess their proliferation, 32D-KαER/GR cells were seeded at 2 × 105 cells/mL in G-CSF, with and without 4HT, and viable cell counts were made daily (Figure5A). This subclone did not accumulate after day 1 in 4HT, but continued to proliferate for 2 additional days in its absence. The 32D-KαER/GR2 cells behaved similarly, and 32D-KVER/GR cell accumulation in G-CSF was unaffected by 4HT (not shown). Thus, exogenous G-CSFR signals protected 4HT-treated 32D-KαER cells from rapid apoptosis upon transfer from IL-3 to G-CSF.
Effect of C/EBP inhibition on differentiation in response to exogenous G-CSFR.
Inhibition of C/EBPs blocks and even reverses differentiation in response to exogenous G-CSFR. (A) First, 2 × 10532D-KαER/GR cells were transferred to G-CSF without 4HT (○) or with 4HT (●). Viable cell counts performed daily are shown. (B) Cells from these same cultures were cytospun and subjected to Wright-Giemsa staining. Cells are shown in IL-3, after transfer to G-CSF in the absence of 4HT for 2, 4, or 6 days (G2, G4, G6), or after transfer to G-CSF in the presence of 4HT for 1, 2, 4, or 8 days (G1, G2, G4, G8). Original magnification ×100. (C) The 32D-KαER/GR cells were transferred to G-CSF without or with 4HT. A subset of cells cultured in the absence of 4HT for 3 days were then exposed to 4HT (−/+ d3). Total cellular RNAs prepared on days 0, 1, 2, 3, 4, 5, and 6 were subjected to Northern blot analysis, 20 μg per lane, for MPO, lysozyme (Lys), LF, C/EBPα, C/EBPβ, C/EBPε, and β-actin.
Effect of C/EBP inhibition on differentiation in response to exogenous G-CSFR.
Inhibition of C/EBPs blocks and even reverses differentiation in response to exogenous G-CSFR. (A) First, 2 × 10532D-KαER/GR cells were transferred to G-CSF without 4HT (○) or with 4HT (●). Viable cell counts performed daily are shown. (B) Cells from these same cultures were cytospun and subjected to Wright-Giemsa staining. Cells are shown in IL-3, after transfer to G-CSF in the absence of 4HT for 2, 4, or 6 days (G2, G4, G6), or after transfer to G-CSF in the presence of 4HT for 1, 2, 4, or 8 days (G1, G2, G4, G8). Original magnification ×100. (C) The 32D-KαER/GR cells were transferred to G-CSF without or with 4HT. A subset of cells cultured in the absence of 4HT for 3 days were then exposed to 4HT (−/+ d3). Total cellular RNAs prepared on days 0, 1, 2, 3, 4, 5, and 6 were subjected to Northern blot analysis, 20 μg per lane, for MPO, lysozyme (Lys), LF, C/EBPα, C/EBPβ, C/EBPε, and β-actin.
To further assess differentiation, 32D-KαER/GR cells from the cultures depicted in Figure 5B were also subjected to daily morphologic analysis (Figure 5B). By day 6 in G-CSF, without 4HT, cells with the mature, doughnut-shaped nuclei commonly seen in murine neutrophils were evident, as were other cells with maturing, kidney-shaped nuclei. Primary granules, staining red, were also present. In sharp contrast, in the presence of G-CSF and 4HT, the 32D-KαER/GR cells underwent an initial morphologic change by day 2, but then did not mature further, ultimately dying. No primary granules were evident. The 32D-KαER/GR2 cells behaved similarly. Although the morphology of KαER/GR cells in 4HT resembles monocytes, they do not express increased levels of M-CSF receptor RNA or of F4/80, a macrophage marker (not shown).
The 32D-KαER/GR cells were cultured in G-CSF with and without 4HT, and total cellular RNAs were prepared. These RNAs were subjected to Northern analysis for the expression of several myeloid differentiation markers and transcription factors (Figure 5C). Induction of the MPO, lysosome, LF, and C/EBPε RNAs by G-CSF was prevented by activation of KαER; basal expression of C/EBPα was reduced; and C/EBPβ was induced but to much lower levels. In a separate experiment, induction of C/EBPδ was reduced, but only about 2-fold (not shown). The 4HT was also added to a group of cells that had been exposed to G-CSF alone for 3 days. Total RNAs were prepared from this culture on days 4, 5, and 6, and these were also subjected to Northern blotting (Figure 5C, −/+ d3). Delayed addition of 4HT reversed induction of each of the RNAs that had occurred in response to G-CSF, although the effect on C/EBPβ was significantly delayed. Morphologically, cells exposed to delayed addition of 4HT on day 3 or 4 did not mature to neutrophils (not shown).
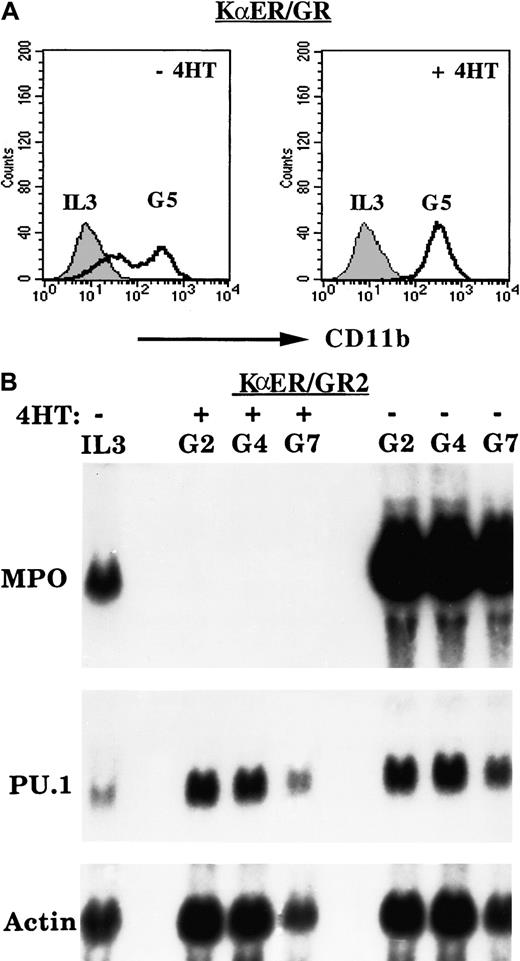
Dominant inhibition of C/EBPs does not prevent induction of CD11b or PU.1 by G-CSF
The 32D-KαER/GR cells were cultured in G-CSF with and without 4HT, and CD11b surface expression was assessed daily by FACS analysis. Activation of KαER did not inhibit, and in fact increased, the average CD11b expression in 2 separate experiments. Results from a representative experiment are shown in Figure6A. CD11b expression was not affected by addition of 4HT to 32D-KVER/GR cells in G-CSF (not shown). The 32D-KαER/GR2 cells were cultured in G-CSF with and without 4HT, and total cellular RNAs were prepared on days 0, 2, 4, or 7. These RNAs were subjected to Northern analysis for MPO, PU.1, and β-actin (Figure 6B). In the absence of 4HT, MPO levels were strongly induced by G-CSF, whereas even basal expression of MPO was eliminated by addition of 4HT, as was seen with 32D-KαER/GR cells. Despite potent inhibition of endogenous C/EBP activities, G-CSF induced PU.1 RNA in 32D-KαER/GR-2 cells (Figure 5D) and in 2 experiments with 32D-KαER/GR cells (not shown). Induction of PU.1 may account for the observed induction of CD11b by G-CSF even in the presence of activated KαER.
Effect of C/EBP inhibition on induction of CD11b or PU.1 by G-CSF.
Inhibition of C/EBPs does not prevent induction of CD11b or PU.1 by G-CSF. (A) KαER/GR cells were cultured in G-CSF with and without 4HT, and the expression of CD11b was assessed by FACS analysis daily. Results from a representative experiment on days 0 and 5 are shown. (B) The 32D-KαER/GR2 cells were cultured in G-CSF without or with 4HT, and total cellular RNAs prepared on days 0, 2, 4, and 7 were subjected to Northern blotting for MPO, PU.1, and β-actin.
Effect of C/EBP inhibition on induction of CD11b or PU.1 by G-CSF.
Inhibition of C/EBPs does not prevent induction of CD11b or PU.1 by G-CSF. (A) KαER/GR cells were cultured in G-CSF with and without 4HT, and the expression of CD11b was assessed by FACS analysis daily. Results from a representative experiment on days 0 and 5 are shown. (B) The 32D-KαER/GR2 cells were cultured in G-CSF without or with 4HT, and total cellular RNAs prepared on days 0, 2, 4, and 7 were subjected to Northern blotting for MPO, PU.1, and β-actin.
Dominant C/EBP inhibition reduces the number of myeloid colonies from marrow
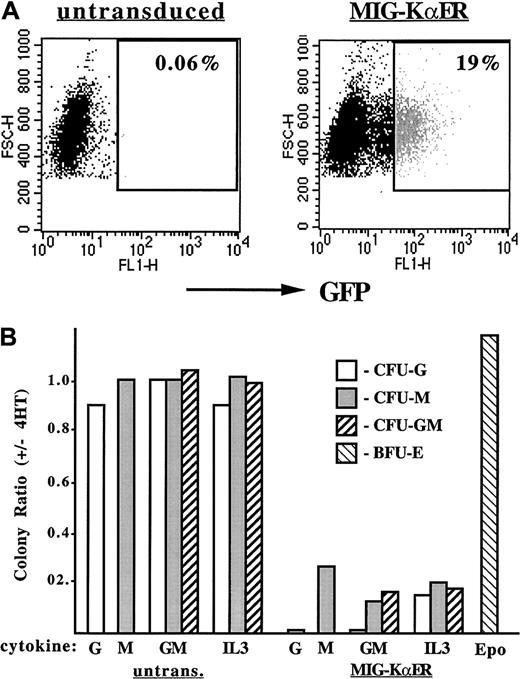
To assess the effect of KαER on normal hematopoietic myeloid progenitors, murine marrow was transduced with MIG-KαER, which expresses both KαER and EGFP from a single mRNA. The CRE–MIG-KαER packaging line produced 5 × 105 infectious particles per milliliter, on the basis of transduction of NIH 3T3 cells. Typically, approximately 20% of marrow cells expressed high levels of GFP (Figure7A). This fraction was isolated by flow cytometry and then cultured in methylcellulose with G-CSF, M-CSF, GM-CSF, or IL-3, with and without 4HT. 4HT did not affect the growth of myeloid colonies, CFU-Gs, CFU-Ms, and CFU-GMs, from untransduced marrow, but markedly reduced the yield of each of these myeloid CFUs from marrow cells transduced with KαER, in 2 experiments. Results from a representative experiment are shown in Figure 7B. As our yield of erythroid burst-forming units (BFU-Es) from transduced marrow cells was very low, we assessed the affect of KαER on the growth of BFU-Es from transduced day-14.5 fetal liver cells (Figure 7B). BFU-E yields were similar with and without 4HT in 2 determinations.
Effect of C/EBP inhibition on the growth of myeloid colonies from murine marrow.
Inhibition of C/EBPs prevents the growth of myeloid colonies from murine marrow. (A) FACS analysis of murine marrow isolated from 5-FU–treated mice either untransduced (left) or transduced (right) with MIG-KαER. Forward scatter is shown on the y-axis, and GFP expression is shown on the x-axis. Particles with fewer than 280 units of forward scatter were excluded from this analysis, as they are likely to represent cell debris. The gate set for isolation of GFP+ cells from the transduced marrow is also shown. Note that 19% of the transduced cells were GFPbright and that cells with modest or low levels of GFP expression were not collected. (B) The ratio, with and without 4HT, of myeloid CFUs obtained in response to the indicated cytokines, for untransduced or transduced marrow cells, and of erythroid burst-forming units (BFU-Es) from fetal liver cells is shown for a typical experiment. G indicates G-CSF; M, M-CSF; GM, GM-CSF; IL3, IL-3; and Epo, erythropoietin and insulin. An average of 13 CFU-Gs, 130 CFU-Ms, and 23 CFU-GMs per 8 × 103 GFP+-transduced cells were obtained in response to GM-CSF, in the absence of 4HT. BFU-E yields from transduced fetal liver cells averaged 96 per 4 × 104transduced cells.
Effect of C/EBP inhibition on the growth of myeloid colonies from murine marrow.
Inhibition of C/EBPs prevents the growth of myeloid colonies from murine marrow. (A) FACS analysis of murine marrow isolated from 5-FU–treated mice either untransduced (left) or transduced (right) with MIG-KαER. Forward scatter is shown on the y-axis, and GFP expression is shown on the x-axis. Particles with fewer than 280 units of forward scatter were excluded from this analysis, as they are likely to represent cell debris. The gate set for isolation of GFP+ cells from the transduced marrow is also shown. Note that 19% of the transduced cells were GFPbright and that cells with modest or low levels of GFP expression were not collected. (B) The ratio, with and without 4HT, of myeloid CFUs obtained in response to the indicated cytokines, for untransduced or transduced marrow cells, and of erythroid burst-forming units (BFU-Es) from fetal liver cells is shown for a typical experiment. G indicates G-CSF; M, M-CSF; GM, GM-CSF; IL3, IL-3; and Epo, erythropoietin and insulin. An average of 13 CFU-Gs, 130 CFU-Ms, and 23 CFU-GMs per 8 × 103 GFP+-transduced cells were obtained in response to GM-CSF, in the absence of 4HT. BFU-E yields from transduced fetal liver cells averaged 96 per 4 × 104transduced cells.
Lysozyme and C/EBPε RNAs are induced by C/EBPs in cycloheximide
KαER inhibited MPO induction by G-CSF, but C/EBPαWT-ER did not induce MPO RNA in the presence of cycloheximide, an inhibitor of RNA translation,25 suggesting that C/EBPs are necessary but not sufficient, for activating the MPO gene during granulopoiesis. As KαER also prevented or reduced lysozyme, C/EBPα, C/EBPβ, and C/EBPε induction by G-CSF, we assessed the effect of activating C/EBPαWT-ER or C/EBPβWT-ER for 8 hours in 32D cl3 cells on the expression of these RNAs, with and without cycloheximide. Extension of cycloheximide exposure beyond 8 hours results in nonspecific toxicity. The endogenous C/EBPα RNA was not induced and the C/EBPβ RNA was only minimally increased by C/EBPα-ER within 8 hours (not shown), as previously shown for the G-CSFR and LF RNAs.25 In contrast, both the lysozyme and C/EBPε RNAs were rapidly and strongly induced by C/EBPαWT-ER. C/EBPε was also strongly induced by C/EBPβWT-ER, whereas lysozyme was only mildy induced by this C/EBP isoform (Figure 8). Even in cycloheximide, estradiol induced the C/EBPε RNA in both cell lines and in a second experiment with C/EBPαWT-ER cells, but not to the same extent as it induced in the absence of cycloheximide. As cycloheximide alone consistently reduced actin RNA expression, 28S and 18S RNAs are shown (Figure 8) as a control for equality of loading. Cycloheximide increased the basal expression of lysozyme RNA in both cell lines. C/EBPαWT-ER induced lysozyme strongly and C/EBPβWT-ER induced lysozyme weakly even in cycloheximide, similar to the inductions seen with estradiol alone. Cycloheximide prevented induction of MPO RNA in each cell line, confirming the activity of this reagent in these experiments. Actinomycin D, an inhibitor of RNA polymerase, prevented induction of both C/EBPε and lysozyme by C/EBPαWT-ER (Figure 8), suggesting that the increases observed with estradiol are due to direct gene activation, and not to increased RNA stability. The difference in basal expression of MPO and C/EBPε between the 32D–αWT-ER and 32D–βWT–ER-1 cells employed in this experiment is typical of clonal variation seen among 32D cl3 subclones. Such variation is also evident when one subclone is cultured at different times, as is evident if one compares MPO expression in 32D–βWT-ER cells in Figures 1 and 8.
Effect of C/EBPs in cycloheximide versus actinomycin D on induction of lysozyme and C/EBPε.
Lysozyme and C/EBPε are induced by C/EBPs in cycloheximide but not actinomycin D. The 32D-C/EBPαWT-ER (32D–αWT-ER) or 32D–C/EBPβWT–ER-1 (32D–βWT-ER) cells proliferating in IL-3 were exposed to 1 μM estradiol (Est), 50 μg/mL cycloheximide (CHX), both, or neither for 8 hours. The 32D–C/EBPαWT-ER cells were also cultured with actinomycin D (Act) with and without estradiol. Total cellular RNAs were then isolated and subjected to Northern blotting for lysozyme, C/EBPε, and MPO. An ethidium stain of ribosomal RNAs is also shown as a control for RNA integrity and loading (bottom panels).
Effect of C/EBPs in cycloheximide versus actinomycin D on induction of lysozyme and C/EBPε.
Lysozyme and C/EBPε are induced by C/EBPs in cycloheximide but not actinomycin D. The 32D-C/EBPαWT-ER (32D–αWT-ER) or 32D–C/EBPβWT–ER-1 (32D–βWT-ER) cells proliferating in IL-3 were exposed to 1 μM estradiol (Est), 50 μg/mL cycloheximide (CHX), both, or neither for 8 hours. The 32D–C/EBPαWT-ER cells were also cultured with actinomycin D (Act) with and without estradiol. Total cellular RNAs were then isolated and subjected to Northern blotting for lysozyme, C/EBPε, and MPO. An ethidium stain of ribosomal RNAs is also shown as a control for RNA integrity and loading (bottom panels).
Discussion
The major finding of this study is that C/EBPs are required for granulopoiesis beyond their ability to induce the expression of cytokine receptors. In addition, we have taken advantage of our ability to regulate C/EBP activities in 32D cl3 cells, both positively and negatively, to determine the role that C/EBPs play in the expression of several endogenous myeloid differentiation markers and transcriptional regulators.
C/EBPs activities increase when 32D cl3 myeloblasts are transferred from IL-3 to G-CSF,14 and overexpression of either C/EBPα, C/EBPβ, or C/EBPδ in 32D cl3 cells induces terminal neutrophilic differentiation. While these findings implicate C/EBPs as master regulators of granulopoiesis, introduction of an exogenous transcription factor may not reflect in vivo circumstances. The phenotypes of mice lacking one or more C/EBP isoforms provide additional insight into the roles that endogenous C/EBPs play in granulopoiesis. Most strikingly, neutrophils from C/EBPε−/− mice do not contain secondary granules,22 and C/EBPα−/− neonates lack G-CSF–responsive progenitors and mature granulocytic cells.15 In contrast, G-CSF−/− mice, G-CSFR−/− mice, and G-CSFR−/−/IL-6 receptor−/− mice retain significant numbers of marrow neutrophils,47-49 indicating that C/EBPα contributes more to granulopoiesis than the activation of the genes encoding one or both of these cytokine receptors. Yet IL-3, GM-CSF, or exogenous G-CSFRs or IL-6 receptors allowed C/EBPα−/− progenitors to generate large numbers of neutrophils, and an EML cell line lacking C/EBPα expresses the G-CSFR and can mature into neutrophils, indicating that C/EBPα is not required for granulocytic maturation.17 27 Perhaps in the latter paradigms, other C/EBPs compensate for the lack of C/EBPα more effectively than they do in vivo.
To further characterize the role of endogenous C/EBPs in granulopoiesis, we developed 32D cl3 lines expressing a dominant-negative C/EBP, KαER. The ability of KαER to be regulated and to interfere with the multiple C/EBPs avoids compensatory effects that can arise in gene knockouts or other paradigms. Specificity of KαER is indicated by the ability of 32D cl3 cells to proliferate near control rates in IL-3 in the presence of KαER, by the ability of 32D cl3 cells to proliferate normally in IL-3 or G-CSF in the presence of KVER, and by the observation that CD11b and PU.1 expression and CMV promoter activity were not suppressed by KαER. On the other hand, expression of KαER might be capable of repressing the transcription of C/EBP-regulated genes that would otherwise function at or near control levels in the absence of endogenous C/EBPs. Therefore, the effects of KαER on myeloid differentiation are potentially exaggerated. Nevertheless, activation of KαER may well mimic the effect of deleting multiple C/EBP genes and is certainly a valuable tool for identifying C/EBP genetic targets.
Activation of KαER strongly inhibited the expression of endogenous G-CSFR RNA, consistent with the inability of 32D cl3 cells expressing CHOP to survive in G-CSF and with the finding that C/EBPα directly activates the G-CSFR promoter.14,16 Activation of C/EBPαWT-ER in 32D cl3 cells did not increase G-CSFR RNA levels until day 3, suggesting that while C/EBPs may be required for basal expression of this RNA in progenitors and in myeloblasts, additional factors present in maturing granulocytes enable its further induction.25
In the presence of exogenous G-CSFR signals, KαER prevented the induction of C/EBPε and both early and late granulocytic markers, reduced basal expression of C/EBPα, attenuated the induction of C/EBPβ and C/EBPδ, prevented the development of a neutrophilic morphology, and rapidly reversed the expression of the MPO and lysozyme RNAs. These effects were not due to nonspecific, cytotoxic interactions as KVER did not affect 32D cl3 cell proliferation or differentiation. Thus, C/EBPs are necessary for granulopoiesis beyond their ability to regulate expression of cytokine receptors.
We surveyed the effect of C/EBP inhibition and activation on a group of myeloid differentiation markers and transcriptional regulators. The lysozyme and C/EBPε RNAs were strongly induced by C/EBPαWT-ER in the presence of cycloheximide, but not actinomycin D, and their induction by G-CSF was inhibited by KαER. Such complementary results were obtained only with these 2 RNAs, not with the MPO, LF, G-CSFR, C/EBPα, C/EBPβ, or PU.1 RNAs. These findings suggest that the endogenous murine lysozyme and C/EBPε genes are directly regulated by C/EBPs. The avian lysozyme gene enhancers at −6.1 and −2.7 kilobase are each regulated by C/EBPs,50,51 but regulation of the murine lysozyme gene by C/EBPs has not been previously demonstrated. The rat C/EBPε promoter contains potential C/EBP-binding sites at −157 and −454, and the human gene contains a near-perfect C/EBP consensus 324 bp upstream of the initiation site of the more highly used Pβ promoter.43 52 Future experiments will determine whether C/EBPα or C/EBPβ activates the C/EBPε gene via these or other binding sites.
As with the G-CSFR RNA, C/EBPαWT-ER did not rapidly induce the LF or C/EBPα RNAs, although C/EBPs have been implicated in the regulation of the promoters of these genes.53-55 Optimal transcription of the G-CSFR, LF, and C/EBPα genes apparently requires factors not already present in uninduced 32D cl3 cells. Similarly, MPO RNA is rapidly induced by C/EBPαWT-ER but not in the presence of cycloheximide, a finding that may reflect regulation of the MPO distal enhancer by both C/EBPs and PU.1 and the ability of C/EBPα-ER to induce PU.1 expression.25 56
Unexpectedly, KαER did not prevent induction of the PU.1 RNA by G-CSF. We previously found that C/EBPαWT-ER rapidly induces PU.1 RNA in both 32D cl3 and Ba/F3 cells, within 4 hours, in the presence of cycloheximide but not actinomycin D, suggesting that C/EBPα directly regulates the PU.1 gene. Apparently, the PU.1 gene can be induced via both C/EBP-dependent and C/EBP-independent pathways. Unlike C/EBPαWT-ER, PU.1-ER did not induce 32D cl3 granulopoiesis in IL-3.25 We have extended this finding by demonstrating that increased PU.1 levels combined with G-CSFR signals are insufficient for inducing neutrophilic differentiation in the absence of C/EBP activities. Also, just as PU.1-ER induced MPO RNA in IL-3, induction of PU.1 by G-CSF may account for the increased expression of CD11b observed in the context of global C/EBP inhibition.
As with C/EBPαWT-ER,25 KαER inhibited progression from the G1 to S cell-cycle phases in 32D cl3 cells (Figure 5A and data not shown). C/EBPα may inhibit the G1/S transition via interaction with E2F or with cdk2.57-60Interaction with cdk2 occurs via the C/EBPα basic region, and this segment is retained in KαER.60 Expression of p21WAF1/CIP1 or p27Kip1 in U937 cells induces monocytic markers.61 Similarly, induction of endogenous p27Kip1 by mimosine induced lysozyme, LF, and C/EBPε expression in 32D cl3 cells (Q.W. and A.D.F, unpublished data, May 2001). On the other hand, alterations that stimulate 32D cl3 proliferation during G1, such as expression of exogenous c-Myb, cdk4, or cyclin D2, prevent their differentiation in response to G-CSF.62 63 Thus, inhibition of G1 progression by KαER does not account for its ability to block 32D cl3 granulopoiesis, as slowed proliferation is expected to accelerate their differentiation.
KαER inhibited the growth of normal granulocyte and monocyte marrow progenitors, but not of erythroid progenitors, suggesting that the reduced numbers of CFU-Gs, CFU-Ms, and CFU-GMs did not result from general cell-cycle inhibition. On the other hand, undifferentiated, blastic colonies were not obtained after transduction with KαER, indicating that proliferation of myeloid progenitors was inhibited, perhaps via effects on myeloid-specific regulatory proteins. As inhibition was seen in G-CSF, M-CSF, GM-CSF, or IL-3, it is unlikely that loss of cytokine receptors accounts for the lack of myeloid colony growth. C/EBPα−/− mice retain monocytes,15and C/EBPβ compensates for the loss of C/EBPα in granulocytes (Y. H. Lee, written personal communication, June 2001). Perhaps C/EBPβ similarly compensates for loss of C/EBPα in the monocyte lineage, in C/EBPα−/− mice, whereas global C/EBP inhibition via expression of KαER uncovers an essential role of C/EBPs in monocyte development. Using transient transfection in 32D cl3 cells, we found that 10- to 30-fold lower levels of 4HT (6 to 20 nM) allowed partial C/EBP inhibition. Whether graded C/EBP activities distinguish monocyte- from granulocyte-lineage commitment in myeloid progenitors, just as PU.1 levels distinguish monocyte- and B-lineage development,64 remains to be determined. Exposure of 32D-KαER/GR cells to G-CSF and these doses of 4HT resulted in a portion of the cells differentiating to neutrophils without evident macrophage differentiation (not shown).
In summary, we have perturbed granulopoieis by globally inhibiting C/EBP-regulated genes. The multiplicity of C/EBPs made it necessary to employ a dominant-inhibitory protein for this purpose, and this complicates interpretation of the observations, owing to the possibility of protein-protein interactions. Despite this, we conclude that C/EBPs contribute to granulopoiesis via induction of lineage-specific markers, including MPO, lysozyme, and LF, via induction of transcription factors, including PU.1 and C/EBPε, and via induction of the G-CSFR. Future experiments will focus on identifying the role that each C/EBP isoform plays in these regulatory processes.
We thank W. Wang for technical assistance, J. Flook for assistance with flow cytometry, and L. Cheng for the MIG vector.
A.D.F. was supported by National Institutes of Health grant R01 HL62274. A.D.F. is a Scholar of the Leukemia and Lymphoma Society and also receives support from the Children's Cancer Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Alan D. Friedman, Johns Hopkins University, Cancer Research Bldg, Rm 253, 1650 Orleans St, Baltimore, MD 21231, MD; e-mail: adfrdman@jhmi.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal