Key Points
LSC17 predicts the outcome independently of ELN 2022, age, and WBC count in adult patients with AML aged <60 years, treated intensively.
Combined with MRD, LSC17 helps identify a subset of patients with NPM1-mutated AML with excellent long-term outcome.
Abstract
Whether the LSC17 gene expression can improve risk stratification in the context of next generation sequencing–based risk stratification and measurable residual disease (MRD) in patients with intensively treated AML has not been explored. We analyzed LSC17 in 504 adult patients prospectively treated in the ALFA-0702 trial. RUNX1 or TP53 mutations were associated with higher LSC1 scores while CEBPA and NPM1 mutations were associated with lower scores. Patients with high LSC17 scores had a lower rate of complete response (CR) in a multivariable analysis (odds ratio, 0.41; P = .0007), accounting for European LeukemiaNet 2022 (ELN22), age, and white blood cell count (WBC). LSC17-high status was associated with shorter overall survival (OS) (3-year OS: 70.0% vs 52.7% in patients with LSC17-low status; P < .0001). In a multivariable analysis considering ELN22, age, and WBC, patients with LSC17-high status had shorter disease-free survival (DFS) (hazard ratio [HR], 1.36; P = .048) than those with LSC17-low status. In 123 patients with NPM1-mutated AML in CR, LSC17-high status predicted poorer DFS (HR, 2.34; P = .01), independent of age, WBC, ELN22 risk, and NPM1-MRD. LSC-low status and negative NPM1-MRD identified a subset comprising 48% of patients with mutated NPM1 with a 3-year OS from CR of 93.1% compared with 60.7% in those with LSC17-high status and/or positive NPM1-MRD (P = .0001). Overall, LSC17 assessment refines genetic risk stratification in adult patients with AML treated intensively. Combined with MRD, LSC17 identifies a subset of patients with NPM1-mutated AML with excellent clinical outcome.
Introduction
Despite the active reduction of disease burden with intensive chemotherapy, relapses often occur in acute myeloid leukemia (AML), and their prognosis remains dismal.1 Recurrent cytogenetic lesions and gene mutations play key roles in the prognostic stratification of patients with AML at diagnosis, but their predictive power remains limited.2 The European LeukemiaNet (ELN) 2022 genetic risk stratification, which accounts for 14 cytogenetic lesions and 13 gene mutations, is the standard risk stratification tool in patients with newly diagnosed AML.3 In addition, detection of measurable residual disease (MRD) after the first chemotherapy course may refine risk stratification.4 In NPM1-mutated AML, the most frequent genetic AML subset, MRD is assessed via the quantification of NPM1 mutant transcripts.2,4 However, MRD is, by definition, not able to guide upfront treatment decisions, such as modulation of dose intensity or addition of drugs during induction therapy, both of which affect long-term outcomes of patients with AML.5,6 Therefore, novel disease-related prognostic factors available at diagnosis are necessary.
The LSC17 prognostic score is a 17-gene expression signature based on the combination of extensive functional in vivo studies of patient-derived leukemic stem cells (LSCs) and biostatistical analyses.7 The LSC17 score has been shown to have prognostic value in patients treated with intensive chemotherapy in multiple AML cohorts.7-9 However, to our knowledge, to date, no study has addressed its prognostic impact in the context of the ELN 2022 risk stratification and or its integration with MRD assessment. Here, we report the first clinical and genetic evaluation of the LSC17 score in a multicentric cohort of 504 patients prospectively treated in the ALFA-0702 trial and define the respective prognostic contribution of LSC17 score, ELN22 risk classification, and NPM1 mutation–based MRD.
Methods
Patients and treatment
From March 2009 to September 2013, 713 patients aged between 18 and 59 years with previously untreated de novo AML, excluding acute promyelocytic and core binding factors leukemias, were included in the prospective multicenter ALFA-0702 study (NCT00932412).10 The study focused on 504 patients with centralized genetic and LSC17 data (supplemental Figure 1).11
Informed consent was obtained before inclusion in accordance with the Declaration of Helsinki. The study protocol was approved in December 2008 by the institutional review board of the French Regulatory Agency and the ethics committee Sud-Est IV, France (ID: 08/099).
In the ALFA-0702 trial, patients achieving complete response (CR), defined per international criteria,12 or CR with incomplete platelet recovery (CRp; all CR criteria except for platelet count < 100 × 109/L)13 after a daunorubicin and cytarabine timed-sequential induction chemotherapy (with or without a high-dose cytarabine and idarubicin salvage course) with protocol-defined nonfavorable risk (supplemental Table 1) were eligible for allogeneic hematopoietic cell transplantation (allo-HCT) in the first CR/CRp. In the absence of an HLA-matched donor (a sibling or 10/10 matched unrelated donor) or if otherwise ineligible for an allo-HCT, patients were randomly assigned to receive either 3 cycles of high-dose cytarabine (HDAC) or 3 cycles or clofarabine associated with intermediate-dose cytarabine (CLARA) as consolidation regimen. Patients with protocol-defined favorable risk were not randomly assigned to a group but received 3 HDAC cycles. Results of the CLARA randomization have been published previously.10
Genetic classification and LSC17 score at diagnosis
Pretreatment bone marrow samples were obtained for 656 patients in ALFA-0702 and centrally stored at the Lille University Hospital. Screening for FLT3 internal tandem duplications (FLT3-ITDs) via fragment analysis, targeted sequencing of 41 genes recurrently mutated in AML, and detecting recurrent gene fusion via ligation-dependent reverse transcriptase polymerase chain reaction (PCR) were performed as previously reported.11
Nanostring assay and LSC17 score
Expression of the LSC17 signature genes was determined via NanoString nCounter PlexSet Analysis (NanoString Technologies, Seattle, WA), using the standard CodeSet nCounter GEx gene expression assay, with 88 samples per run and a custom CodeSet.7 A volume of 5 μL containing 150 ng total RNA from mononuclear cells of AML samples was incubated with 20 μL of reporter probes and 5 μL of capture probe mix at 65°C for 16 hours. Each run included an internal control of 300 fM of 26 predefined synthetic control oligonucleotides (Integrated Technologies, Ashford, United Kingdom). Subsequent steps per the manufacturer’s protocol were followed (Plexset Reagent for Gene Expression User Manual). Raw digital images were processed, with final barcoded counts tabulated in reporter code count output files. For quality control and normalization purposes, default settings were used for GX Analysis of the nSolver Software (version 4.0). In brief, reporter code count files and reporter library files were imported into nSolver. Standard positive control normalization was performed. The output files from nSolver were read into R for further normalization and data processing, as detailed by Ng et al.7 Specifically, normalized counts using the predefined internal control oligonucleotides and housekeeping genes were then log2-transformed and scaled. The LSC17 score was computed for each AML based on the LSC17 genes weighted based on the regression coefficients, estimated as follows: LSC17 score = (DNMT3B × 0.0874) + (ZBTB46 × −0.0347) + (NYNRIN × 0.00865) + (ARHGAP22 × −0.0138) + (LAPTM4B × 0.00582) + (MMRN1 × 0.02 58) + (DPYSL3 × 0.0284) + (KIAA0125 × 0.0196) + (CDK6 × −0.0704) + (CPXM1 × −0.0258) + (SOCS2 × 0.0271) + (SMIM24 × −0.0226) + (EMP1 × 0.0146) + (NGFRAP1 × 0.0465) + (CD34 × 0.0338) + (AKR1C3 × −0.0402) + (GPR56 × 0.0501).7
NPM1 mutation–based MRD
Types A, B, or D NPM1 mutant transcript levels were quantified at diagnosis and after the first induction course, using a mutation-specific real-time quantitative PCR assay, as previously described.14 Rare NPM1 mutant alleles were quantified via droplet digital PCR, as previously described.15 A 4-log reduction in the peripheral blood MRD after the first induction course was used to define MRD negativity, a threshold previously shown to indicate allo-HCT benefit in patients with both typical (A/B/D) and nontypical NPM1 mutations.11,14
Statistical analysis
Comparisons of LSC17 score values were performed using the Wilcoxon test, with adjustments per the Benjamini-Hochberg method. Continuous LSC17 score was dichotomized using its median value in the total cohort. Characteristics of populations LSC17-high and LSC17-low statuses were compared using the Fisher exact test. Multivariable analyses of categorial variables were performed with logistic regressions for CR/CRp and MRD positivity. Overall survival (OS) and OS from CR/CRp were defined from inclusion in the ALFA-0702 trial or time of CR/CRp achievement, respectively, to death or last follow-up. Disease-free survival (DFS) was defined from the date of CR/CRp to the date of relapse or death (whichever came first) or until the last follow-up. Nonrelapse mortality (NRM) and cumulative incidence of relapse (CIR) were studied since the date of CR/CRp, considering death and relapse as competing events. The follow-up period was estimated using the reverse Kaplan-Meier method. Survival was analyzed using the Kaplan-Meier method. LSC17 statuses were compared using the log-rank test for OS and DFS. For categorical baseline clinical and genetic covariables, the LSC17 status impact on OS and DFS was evaluated with a univariable Cox model in each subgroup per the covariable, and the significance of the interaction between LSC17 status and the covariable was analyzed using a Cox model accounting for the interaction term. Multivariable analyses were performed using Cox proportional hazard models accounting for conventional prognostic factors (age, log10-transformed white blood cell (WBC) count, and ELN22 risk stratification). The proportional hazard assumption was validated via visual inspection and testing of Schöenfeld residuals.16 Multicollinearity was inspected by studying the variance inflation factor, considering a variance inflation factor >4 as unacceptable.17 CIR and NRM were analyzed as competing events, with comparisons based on LSC17 status, using Gray test and multivariable analyses, with Fine and Gray model accounting for the same variables. We studied the impact of allo-HCT on OS from CR/CRp, considering allo-HCT as a time-dependent covariable, and analyzed the interaction with LSC17 status in a multivariable Cox model accounting for ELN22. As a sensitivity analysis, multivariable analyses of OS, DFS, and CIR were repeated with LSC17 dichotomized at an optimal cut-off based on the Youden index (YI) for the time-dependent receiver operating characteristic (ROC) curve for OS at 3 years.18,19 Survival analyses were performed in for the NPM1-mutated subgroup with the additional covariable of NPM1-MRD in multivariable models. Discrimination performances of risk stratification strategies were assessed using time-dependent areas under the ROC curves (ROC-AUC) for OS from CR, DFS, and CIR computed at 3 years.18 All statistical analyses were performed with R software, version 4.1.2.
Results
Baseline clinical and biological characteristics
The study population included 504 patients with AML (237 female and 267 male), with a median age of 48 years (range, 18-60 years). Of the 504 study participants, 187 (37%) patients had a NPM1 mutation. Genetic risk per the ELN22 was favorable in 142 (28%) patients (with NPM1 mutation n = 114 [80%]; with in-frame bZIP CEBPA mutation, n = 28 [20%]), intermediate in 152 (30%), and adverse in 205 (41%) patients. Clinical and biological characteristics of the 504 patients with AML are described in Table 1.
Clinical and genetic features associated with the LSC17 score
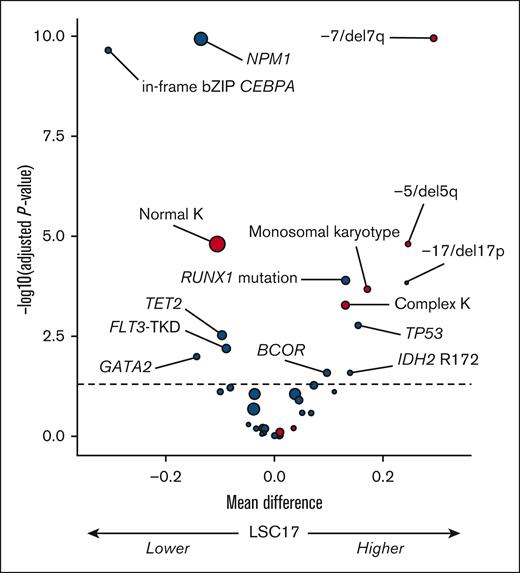
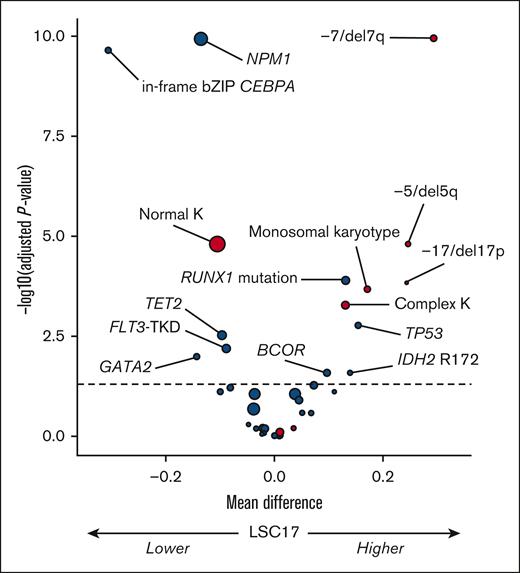
Firstly, we analyzed the influence of recurrent genetic factors (ie, those present in ≥3% of patients) on the LSC17 score as a continuous variable in the total study population (N = 504) (Figure 1). In univariable analysis, NPM1 mutation (median difference [md] = −0.14; adjusted P value [q] < .0001); in-frame bZIP CEBPA mutation, md = −0.31; q < .0001), and, to a lesser extent, FLT3-TKD (md = −0.09; q = .006), TET2 mutation (md = −0.10; q = .003), and GATA2 (md = −0.14; q = .01) were associated with lower LSC17 scores. In contrast, RUNX1 (md = 0.13; q < .0001), TP53 (md = 0.15; q = .002), BCOR (md = 0.10; q = .01), and IDH2 R172 (md = 0.14; q = .01) mutations were associated with higher LSC17 scores. FLT3-ITD, regardless of allelic ratio, had no influence on the LSC17 status (md = 0.04; q = .09). Regarding recurrent cytogenetic lesions, the presence of monosomy 7 or del(7q) (md = 0.29; q < .0001), monosomy 5 or del(5q) (md = 0.25; q < .0001), monosomy 17, del(17p) or abnormal 17p (md = 0.24; q = .0001), monosomal karyotype (md = 0.17; q = .0002), and complex karyotype (md = 0.13; q = .0005) were each associated with higher LSC17 scores in univariable analyses, whereas normal karyotype was associated with a lower LSC17 score (md = −0.11; q < .0001).
Volcano plot of the association between baseline genetic factors on the LSC17 score in the total study population (N = 504). The x-axis represents the difference between mean values of LSC17 scores in patients with or without the variable of interest. The y-axis represents the significance of the Wilcoxon test comparing the 2 groups, adjusting for multiple testing with the Benjamini-Hochberg method (dashed line corresponding to 0.05). Red and blue circles indicate cytogenetic and gene mutations variables, respectively; size is correlated with number of patients with the variable of interest. K, karyotype.
Volcano plot of the association between baseline genetic factors on the LSC17 score in the total study population (N = 504). The x-axis represents the difference between mean values of LSC17 scores in patients with or without the variable of interest. The y-axis represents the significance of the Wilcoxon test comparing the 2 groups, adjusting for multiple testing with the Benjamini-Hochberg method (dashed line corresponding to 0.05). Red and blue circles indicate cytogenetic and gene mutations variables, respectively; size is correlated with number of patients with the variable of interest. K, karyotype.
In all subsequent analyses, we dichotomized the continuous LSC17 score, using the median value from the total cohort (0.60). The baseline characteristics of the 252 patients assigned to the LSC17-low and LSC17-high groups are summarized in Table 2. Expectedly, compared with patients in the LSC17-low group, those in the LSC17-high group were older (P = .002), had lower WBC counts at diagnosis (P = .03), and more frequently had complex or monosomal karyotype (P = .003 and P = .001, respectively). Abnormalities of chromosome 17p (P = .0004) and inv(3)/t(3;3) (P = .04) as well as FLT3-ITD (P = .02), RUNX1 (P = .0003), and TP53 (P = .04) mutations were more frequent, whereas NPM1 (P < .0001) and in-frame bZIP CEBPA mutations (P < .0001) were less frequent in the LSC17-high group.
Impact of LSC17 status on hematologic remission achievement after induction
Overall, after the timed-sequential induction course, 400 patients (79%) achieved CR/CRp, 25 (5%) patients died before bone marrow assessment, and 79 (16%) were not in CR/CRp. The proportion of patients with high LSC17 scores who obtained CR/CRp after this first induction course was 69% (n = 175, including 163 CR and 12 CRp) compared with 89% (n = 225: CR, 209 and CRp, 16) in those with low LSC17 scores (P < .0001). In a multivariable logistic regression, the LSC17-high status predicted lower odds of achieving CR/CRp after a single induction course (odds ratio [OR], 0.41; 95% confidence interval [CI], 0.24-0.68; P = .0007) independently of ELN22 risk stratification (considering favorable risk as a reference, intermediate risk: OR, 0.23; 95% CI, 0.08-0.55; P = .002 and adverse risk: OR, 0.12; 95% CI, 0.05-0.29; P < .0001), higher WBC count (log10 scale, OR, 0.78; 95% CI, 0.54-1.13; P = .19) and older age (per 10-years of age; OR, 1.15; 95% CI, 0.94-1.41; P = .18) (Table 3). Of the 79 patients who failed to achieve CR/CRp after 1 course, 17 did not receive the salvage course, 3 died during the salvage course, 28 failed to achieve CR/CRp after salvage, and 31 patients achieved CR/CRp after the salvage course. Thus, overall, 431 (86%) patients achieved CR/CRp after 1 or 2 courses (supplemental Figure 1); 230 (91%) and 201 (80%) patients in the LSC17-low and LSC17-high groups, respectively, achieved CR/CRp (P = .0003). Finally, in a logistic regression on CR/CRp after 1 or 2 courses accounting for age, sex, WBC count, and ELN22 risk, there was a trend for lower odds of CR/CRp in patients with high LSC17 scores (OR, 0.60; 95% CI, 0.33-1.04; P = .07).
Impact of LSC17 status on survival
In the total study population with a median follow-up of 4.2 years (interquartile range, 3.4-5.0 years), 212 patients received an allo-HCT in first CR/CRp, and there were 150 relapses and 208 deaths. The 3-year OS was 61.3% (95% CI, 57.1-65.7). For the 431 patients achieving CR/CRp after 1 or 2 courses, the 3-year DFS was 57.2% (95% CI, 52.7-62.1).
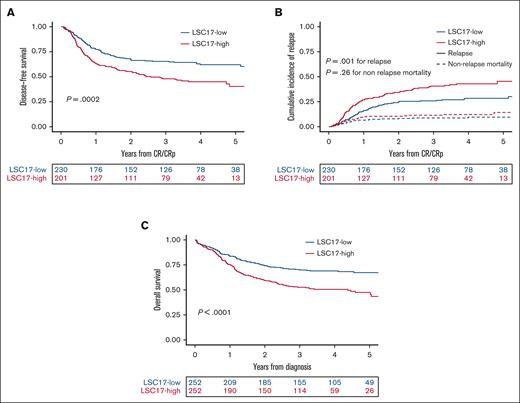
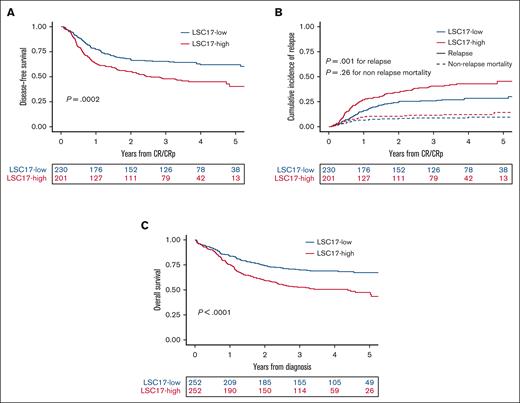
In univariable analysis, 3-year DFS was 65.4% (95% CI 59.5-71.9) in the 230 patients with low LSC17 scores and 48.0% (95% CI, 41.5-55.4) in the 201 patients with high LSC17 scores (P = .0002; Figure 2A). The LSC17-high cohort had a higher CIR (P = .001) but a similar NRM (P = .26) compared with patients in the LSC17-low cohort (Figure 2B). In multivariable analyses, LSC17-high status was associated with inferior DFS (hazard ratio [HR], 1.36,; 95% CI, 1.002-1.86; P = .048) independent of age (per 10 years of age, HR, 1.20; 95% CI, 1.05-1.37; P. =.008), WBC count (log10 scale, HR, 1.38; 95% CI, 1.10-1.74; P = .006), and ELN22 (HR, 1.78; 95% CI, 1.19-2.66; P = .005 for intermediate risk and HR, 2.31; 95% CI, 1.54-3.46; P < .0001 for adverse risk) (Table 4). When analyzing relapse mortality and NRM as competing events, there was a higher CIR (subdistribution HR [SHR], 1.47; 95% CI, 1.03-2.09; P = .034) among the LSC17-high cohort considering age (per 10 years of age, SHR, 1.19; 95% CI, 1.02-1.40; P = .029), WBC count (log10 scale, SHR, 1.44; 95% CI, 1.10-1.88; P = .009), and ELN22 (SHR, 1.54; 95% CI, 0.97-2.42; P = .066 for intermediate risk, and SHR, 1.89; 95% CI, 1.21-2.95; P = .005 for adverse risk) (supplemental Table 2).
Impact of LSC17 status on prognosis. (A) DFS and (B) CIR and NRM (as competing risks) in 431 patients achieving CR or CRp after 1 or 2 induction courses per the dichotomic LSC17 status. (C) OS from diagnosis per LSC17 status in the total study population (N = 504).
Impact of LSC17 status on prognosis. (A) DFS and (B) CIR and NRM (as competing risks) in 431 patients achieving CR or CRp after 1 or 2 induction courses per the dichotomic LSC17 status. (C) OS from diagnosis per LSC17 status in the total study population (N = 504).
Three-year OS was 69.9% (95% CI, 64.5-75.9) in patients with LSC17 scores and 52.7% (95% CI, 46.8-59.2) in those with high LSC17 scores (P < .0001; Figure 2C). In a multivariable analysis, there was a trend toward shorter OS (HR, 1.27; 95% CI, 0.94-1.72; P = .11) among patients in the LSC17-high cohort considering age (per 10 years of age HR, 1.28; 95% CI, 1.12-1.46; P = .0003), WBC count (log10 scale HR, 1.48; 95% CI, 1.19-1.83; P = .0004), and ELN22 risk (HR, 2.85; 95% CI, 1.80-4.51; P < .0001 for intermediate risk and HR, 3.93; 95% CI, 2.49-6.21; P < .0001 for adverse risk, considering favorable risk as the reference).
For a sensitivity analysis, we repeated the evaluation of LSC17 impact with a different cut-off: we determined the YI on a time-dependent ROC curve for 3-year OS (0.4916) and reclassified the patients accordingly (LSC17-highYI: 67%, n = 340 vs LSC17-lowYI: 33%, n = 164). LSC17 status using YI had predictably a stronger impact on OS (HR, 1.80; 95% CI, 1.21-2.67; P = .004), but there was no difference in terms of DFS and CIR (HR, 1.60; 95% CI, 1.11-2.31; P = .012 and SHR, 1.54; 95% CI, 1.02-2.33; P = .038, respectively).
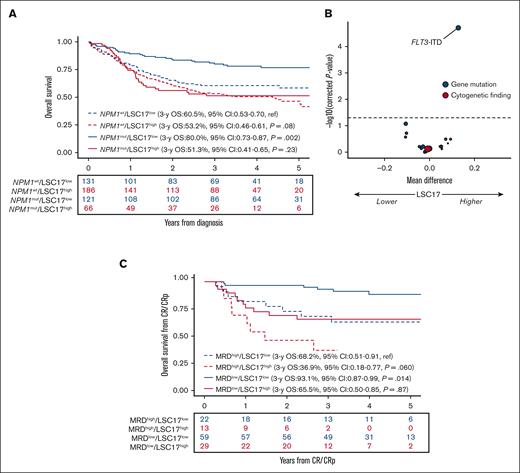
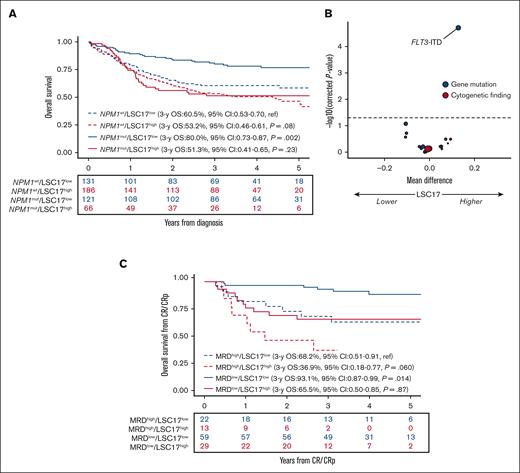
Of the 431 patients reaching CR/CRp after 1 or 2 courses, 212 (49%) patients received transplantation in the first CR/CRp. Considering allo-HCT as a time-dependent covariate in the Cox model, accounting for ELN22 risk, there was no significant interaction between allo-HCT in the first CR/CRp and LSC17 status with the OS from CR/CRp or DFS (P = .32 and P = .15, respectively). In patients with an ELN22 nonfavorable risk status who were in CR/CRp (n = 288), the DFS significantly improved after allo-HCT (HR, 0.57; 95% CI, 0.40-0.81; P = .0018). Similarly, this benefit was present in populations with LSC17-low and -high statuses(n = 117, HR, 0.47; 95% CI, 0.26-0.86; P = .013 and n = 171, HR, 0.63; 95% CI, 0.41-0.97; P = .035 respectively). In multivariable models accounting for ELN22 risk, age, WBC count, LSC17 status, and consolidation type (HDAC vs CLARA), there was no interaction between LSC17 status and consolidation type (HR, 1.68; 95% CI, 0.67-4.23; P = .27 for OS from CR/CRp; and HR, 1.51; 95% CI, 0.70-3.24; P = .29). Regarding the genetic subgroups, there was an interaction between NPM1 mutation and LSC17 status in multivariable Cox models accounting for LSC17 status and NPM1 mutation on OS and DFS (P = .026 and P = .017 respectively) (supplemental Figure 2). In a univariable Cox model, the impact of LSC17 status on OS was significant in patients with NPM1-mutated AML (HR, 2.71; 95% CI, 1.63-4.53; P = .0001) but did not reach statistical significance in patients with AML with wild-type NPM1 (HR, 1.35; 95% CI, 0.96-1.89; P = .08; Figure 3A). We, therefore, focused on patients with NPM1-mutated AML.
Impact of LSC17 in NPM1-mutated AML. (A) Impact of LSC17 status on OS from diagnosis per NPM1 status (N = 504). (B) Volcano plot of the association between baseline genetic factors on the LSC17 score in patients with NPM1-mutated AML (n = 187). The x-axis represents the difference between mean values of LSC17 scores in patients with or without the variable of interest. The y-axis represents the significance of the Wilcoxon test comparing the 2 groups, adjusting for multiple testing with the Benjamini-Hochberg method (dashed line corresponding to 0.05). Red and blue circles indicate cytogenetic and gene mutations variables, respectively; size is correlated with number of patients with the variable of interest. (C) Impact of LSC17 status (LSC17 high, red; LSC17 low, blue) and MRD status (negative, solid line; positive, dotted line) on OS from CR/CRp in NPM1-mutated AML (n = 123).
Impact of LSC17 in NPM1-mutated AML. (A) Impact of LSC17 status on OS from diagnosis per NPM1 status (N = 504). (B) Volcano plot of the association between baseline genetic factors on the LSC17 score in patients with NPM1-mutated AML (n = 187). The x-axis represents the difference between mean values of LSC17 scores in patients with or without the variable of interest. The y-axis represents the significance of the Wilcoxon test comparing the 2 groups, adjusting for multiple testing with the Benjamini-Hochberg method (dashed line corresponding to 0.05). Red and blue circles indicate cytogenetic and gene mutations variables, respectively; size is correlated with number of patients with the variable of interest. (C) Impact of LSC17 status (LSC17 high, red; LSC17 low, blue) and MRD status (negative, solid line; positive, dotted line) on OS from CR/CRp in NPM1-mutated AML (n = 123).
LSC17 status in NPM1-mutated AML
In the 187 patients with NPM1-mutated AML, a higher LSC17 score was only associated with FLT3-ITD mutations (md = 0.13; q < .0001) but not with DNMT3A mutations (md = −0.0007; q = .74; Figure 3B). Of 187 patients with mutated NPM1, 176 (94%) achieved CR/CRp after the first induction course (CR, n = 164; CRp, n = 12), including 116 of the 121 (96%) patients with LSC17-low and 60 of the 66 (90%) patients with LSC17-high scores (P = .20). Of those 176 patients with NPM1-mutated AML who achieved CR/CRp after the first induction course, 123 (66%) had an MRD assessment on NPM1 transcripts after this first course; MRD was negative in 88 (72%) of those patients. The lack of impact of LSC17 status on achievement of NPM1 MRD negativity in univariate analysis (P = .68) was confirmed in a multivariable logistic regression accounting for ELN22 risk, age, and WBC count (P = .95) (supplemental Table 3). In a multivariable Cox model, LSC17-high status at diagnosis predicted poorer DFS (HR, 2.34; 95% CI, 1.23-4.48; P = .010) independently of age (per 10-years of age, HR, 1.17; 95% CI, 0.80-1.71; P = .41), higher WBC (log10 scale, HR, 1.04; 95% CI, 0.60-1.81; P = .88), ELN22 risk (HR, 1.18; 95% CI, 0.59-2.36; P = .64 for intermediate risk, and HR, 1.77; 95% CI, 0.59-5.23; P = .31 for adverse risk, considering favorable risk as the reference), and MRD positivity (HR, 3.36; 95% CI, 1.81-6.24; P = .0001; supplemental Table 4). In a similar multivariate model for OS from CR/CRp, LSC17-high status predicted poorer OS from CR/CRp (HR, 2.31; 95% CI, 1.09-4.91; P = .029) independently of age (per 10 years of age, HR, 1.44; 95% CI, 0.92-2.25; P = .11), higher WBC counts (log10 scale, HR, 1.47; 95% CI, 0.77-2.81; P = .24), ELN22 risk (HR, 2.02; 95% CI, 0.92-4.46; P = .08 for intermediate risk; and HR, 2.58; 95% CI, 0.79-8.38; P = .11 for adverse risk, considering favorable risk as the reference), and MRD positivity (HR, 2.66; 95% CI, 1.29-5.48; P = .008). Of note, there was no significant interaction between LSC17 status and MRD positivity in either model (DFS, P = .88; OS from CR/CRp, P = .34).
The 3-year DFS of patients with LSC17-low status and negative MRD was 84.7% (95% CI, 75.9-94.4) compared with 49.9% (95% CI, 39.0-63.8) in patients with LSC17-high status and/or positive MRD (P = .0001). The 3-year OS from CR/CRp was 93.1% (95% CI, 86.8-99.9) in patients with LSC17-low status and negative MRD compared with 60.7% (95% CI, 49.8-74.0) in those with LSC17-high status and/or positive MRD (P = .0001). The outcome of each LSC17/MRD subset is reported in Figure 3C. The combination of MRD and LSC17 status outperformed the ELN22 risk stratification in terms of discrimination performance (3-year OS from CR: ROC-AUC, 0.800; 95% CI, 0.708-0.892 vs ROC-AUC, 0.671; 95% CI, 0.566-0.777; supplemental Table 5).
In patients with NPM1-mutated AML with LSC17-low status and negative MRD (n = 59), 16 patients received an allo-HCT in the first CR/CRp. In patients with LSC17-high status and/or positive NPM1-MRD (n = 64), 20 patients received transplantation in the first CR/CRp. In patients with negative NPM1-MRD, we confirmed the previously reported lack of benefit from allo-HCT both in terms of DFS and OS from CR/CRp, regardless of LSC17 status.14 In the small population of patients with positive MRD (n = 35, 28.5%), there was no significant interaction between LSC17 status and allo-HCT in first CR/CRp for both DFS and OS from CR/CRp.
Discussion
In this prospective cohort of 504 adult patients with AML treated with intensive chemotherapy in the ALFA-0702 trial,10,11,14 we confirmed and extended the association between higher LSC17 scores and recurrent adverse cytogenetic or molecular alterations, and the poorer outcome of patients with LSC17-high status, independent of age, WBC count, and ELN 2022 risk stratification. Analyzing, for the first time to our knowledge, the clinical relevance of LSC17 in the context of MRD assessment, we identified the additive prognostic contribution of NPM1-MRD and LSC17 in patients with NPM1-mutated AML. Notably, combining these 2 criteria helped identify a subset comprising 48% of patients with NPM1-mutated AML in complete remission with excellent long-term survival.
Extending previous approaches aimed at correlating expression of stemness programs with clinical outcome in AML,20-22 the LSC17 gene expression signature was shown in an initial report on adult patients with AML to predict survival independently of cytogenetics, NPM1 and FLT3 mutations.7 This finding was confirmed in pediatric AML.9 Another study in adult patients with AML treated with various induction and consolidation regimens demonstrated that the LSC17 score could predict survival independently of the 2017 edition of the reference ELN risk stratification.8
Alhough LSCs have been identified as a crucial determinant of relapse in AML, the gold-standard quantitative assessment by limiting-dilution xenotransplantation is not feasible in routine clinical practice. Flow cytometry LSC quantification has been shown to predict chemoresistance in AML.23-26 Besides issues in the prospective definition of a uniform phenotype for LSCs across AML samples,26-28 studies have shown that the relapse-initiating potential of LSCs may not be confined to a phenotypically defined subset of cells, rendering quantification of key genes in this program attractive for clinical applications.29 Given the recent efforts in standardizing assessment of the LSC17 score with a laboratory-developed clinical assay, it is plausible that its routine application in clinical laboratories will become feasible in the coming years.30
Our study in 504 uniformly and intensively treated adult patients with AML confirmed and extended the known association between higher LSC17 scores and adverse-risk cytogenetic or genetic lesions, including monosomy 7, del(17p) RUNX1, TP53, FLT3-ITD, and ASXL1 gene mutations. Conversely, favorable genetic events such as NPM1 mutations and in-frame bZIP mutated CEBPA were strongly associated with a lower LSC17 score. The impact of such genetic lesions on expression of the genes contributing to the LSC17 score may occur as cis or trans. For example, the LSC17 gene CDK6 is located in 7q21, whereas GPR56 expression has previously been shown to be regulated by MECOM and thus may be overexpressed in patients with inv(3)/t(3;3) AML.31
In recent years, MRD has become an emerging prognostic factor in AML,32,33 and its role in risk stratification is increasing.3,4 Postremission therapeutics can now be guided by MRD kinetics.34,35 For example in NPM1-mutated AML, the benefit of allo-HCT is restricted to patients with slow MRD kinetics or with persisting MRD positivity.14,36 However, predicting relapse risk before MRD assessment at remission is needed to tailor induction therapy, because significant improvements in long-term outcomes have been obtained through modification of the conventional 7 + 3 induction regimen by escalating anthracycline doses or6,37 adding gemtuzumab-ozogamicin38-40 or FLT3 inhibitors in selected patient populations.41,42 Here, we demonstrated that patients with LSC17-high status have a lower CR/CRp rate and higher risk of relapse than those with LSC17-low status. Future studies are needed to determine whether specific interventions during the induction phase could mitigate this chemoresistance, turning LSC17 into a predictive and not only prognostic biomarker.
Consistent with previous findings,7 we did not identify a specific interaction between LSC17 status and allo-HCT. Indeed, a higher proportion of LSCs at diagnosis, via flow cytometry, has been associated with worse prognosis in patients undergoing allo-HCT in the first remission,43 suggesting that allo-HCT does not specifically alleviate the poor prognosis of AMLs with high stemness traits.
In NPM1-mutated AML, relapse risk is well correlated with MRD reduction.14,32,44 In our study, we showed that assessment of LSC17 adds additional prognostic information to NPM1-MRD, possibly because these relapses are initiated by a small population of cells below the current limit of MRD detection. Improvement of sensitivity allowed for via droplet digital PCR or limited-cell fluorescence-activated cell sorting with sequencing might enable the detection of these smaller clones and especially those with stemness potential.45,46 By combining NPM1-MRD and LSC17 score, we identified a subgroup of patients representing ∼50% of NPM1-mutated AMLs, with LSC17-low status at diagnosis and negative MRD after induction chemotherapy, with long-term survival of >90%. Although the study was limited by small patient numbers, we did not identify a subset of patients with NPM1-mutated AML for whom LSC17 status at diagnosis would affect MRD-based allo-HCT indications.
Further work should be conducted to validate this simple stratification of patients with NPM1 mutation in the era of FLT3 inhibitors and oral azacitidine maintenance.42,47 Whether flow-based quantification of LSCs can substitute the LSC17 score also warrants investigation.24,48 Future studies are also required to investigate the interplay of LSC17 with other MRD tools, including flow cytometry and next generation sequencing.4,33 The applicability of LSC17 assessment in patients who are unfit treated with nonintensive strategies also deserves further investigation. Finally, the implementation of these gene expression biomarkers in clinical practice is expected to further fulfill the promise of precision medicine for leukemias.
Acknowledgments
The authors thank all investigators and clinical research associates from the ALFA group. R.I. is supported by grants from Association Laurette Fugain, Association Princesse Margot, Association pour la Recherche contre le Cancer, Fondation Leucémie Espoir, and Ligue Contre le Cancer. N.D. is supported by the North-West Canceropole (GIRCI AAP-AE 2018). C.P., M.C., and N.D. were supported by a grant from the French National Cancer Institute (INCa-PRTK2016). This study was funded by the French National Cancer Institute and French Health Ministry (PHRC 2007/1911 and PRTK TRANSLA10-060).
Authorship
Contribution: N.D., C.V., M.F., and M.C. performed NanoString assay and analyzed data; L.F., C.P., and N.D. performed the sequencing analyses and interpreted the genomic data; L.F. and N.D. performed and interpreted MRD analyses; L.V. and R.I. assembled and analyzed the data, and drafted the manuscript; H.D., M.C., N.D., and R.I. supervised the study; S.d.B., M.H., P.-Y.D., C.R., C.B., E.L., D.L., Juliette Lambert, and K.C.-L. provided clinical data; Jérôme Lambert supervised the management of the clinical data and the statistical analyses; C.T. centrally reviewed cytogenetics data; and all authors revised the manuscript and accepted its final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Raphael Itzykson, Service Hématologie Adultes, Hôpital Saint-Louis, 1 Avenue Claude Vellefaux, F-75010 Paris, France; e-mail: raphael.itzykson@aphp.fr; and Nicolas Duployez, Biology and Pathology Center, Professor Leclercq Blvd, F-59000 Lille, France; e-mail: nicolas.duployez@chu-lille.fr.
References
Author notes
∗M.C., R.I., and N.D. are joint senior authors and contributed equally to this study.
This study was presented in abstract form at the 64th annual meeting of the American Society of Hematology, New Orleans, LA, 11 December 2022.
Original data are available on request from the corresponding author, Nicolas Duployez (nicolas.duployez@chru-lille.fr).
The full-text version of this article contains a data supplement.