Abstract
Hematopoietic stem cells from different strains of mice vary widely with respect to their cell cycle activity. In the present study we used complementary genetic and genomic approaches to identify molecular pathways affecting this complex trait. We identified a major quantitative trait locus (QTL) associated with variation in cell proliferation in C57BL/6 and DBA/2 mice to a 10 centimorgan (cM) region on chromosome 11. A congenic mouse model confirmed that a genomic interval on chromosome 11 in isolation confers the proliferation phenotype. To detect candidate genes we performed subtractive hybridizations and gene arrays using cDNA from highly enriched stem cells from parental strains. Intriguingly, a disproportionate number of differentially expressed genes mapped to chromosome 11 and, more specifically, these transcripts occurred in 3 distinct clusters. The largest cluster colocalized exactly with the cell cycling QTL. Such clustering suggested the involvement of genetic variation that affects higher-order chromosomal organization. This hypothesis was reinforced by the fact that differentially expressed genes mapped to recombination “coldspots,” as a consequence of which clustered genes are collectively inherited. These findings suggest the functional interdependence of these closely linked genes. Our data are consistent with the hypothesis that this isolated cell cycle QTL does not result from a mutation in a single gene but rather is a consequence of variable expression of a collection of highly linked genes.
Introduction
Lifelong replenishment of short-lived circulating blood cells is ensured by a relatively small population of lymphohematopoietic stem and progenitor cells residing in the bone marrow.1 Mechanisms regulating proliferation of these primitive cells during steady-state blood cell production are poorly understood. It has long been assumed that most hematopoietic stem cells are quiescent for prolonged periods and do not actively contribute to mature blood cell production.2-5 However, this concept has been challenged by in vivo bromodeoxyuridine labeling studies that indicated that in fact most, if not all, stem cells in the mouse will divide at least once during a period of 3 to 4 weeks, suggesting that all stem cells participate in steady-state blood cell production.6-8 In addition to a plethora of cell-extrinsically acting cytokines, such as the ligands for c-kit, flt3/flk2, and c-mpl, that have been demonstrated to affect kinetics of stem and progenitor cells,9-11 superimposed on such a growth factor network intrinsic genetic factors play an important role in this process as well. We have shown by analyzing various inbred strains of mice that the rate of progenitor cell cycling varies dramatically from strain to strain. Intriguingly, we found that strains with rapidly cycling stem and progenitor cells (eg, AKR/J, C3H, DBA) invariably had a shorter mean life span than strains with more slowly proliferating cells (B6, 129/J).12,13 To assess whether these 2 traits were causally related, we have mapped quantitative trait loci (QTL) that were associated with variation in cell cycling in a panel of BXD recombinant inbred mice and compared these with loci associated with mean life span.14 Indeed, 2 loci, occurring on chromosome 7 and 11, respectively, showed suggestive linkage with both traits.15
The heritable component of quantitative traits, such as variation in cell cycling, is thought to result from the additive and epistatic interactions of multiple genetic loci. The ensuing nonmendelian segregation pattern has significantly hampered the identification of individual genes associated with quantitative traits.16,17Strategies such as the production of congenic or consomic strains, carrying only the QTL or chromosome of interest, have been proposed to circumvent this problem.18-20 In the present study we followed this approach, and by employing a variety of complementary genetic and genomic approaches we have searched for molecular pathways affecting hematopoietic progenitor cell proliferation.
Materials and methods
Mice
Eight- to 12-week-old female recombinant inbred BXD-33, -34, -35, -38, -39, -40 and -42/TyJ mice were purchased from the Jackson Laboratory (Bar Harbor, ME). C57BL/6 (B6), DBA/2 (DBA), and B6D2F1 (BDF1) mice were all from Harlan (Harlan Nederland, Horst, The Netherlands).
Generation of backcross animals and the use of a marker-assisted selection protocol to produce congenic mice
To produce N2 backcross animals, male BDF1 mice were crossed with female B6 mice at the Central Animal Facility of the University of Groningen. To generate congenic mice, we used a marker-assisted selection protocol18 and selected male carriers of DBA alleles at the region of interest on chromosome 11 that otherwise showed the lowest frequency of DBA alleles throughout the genome, using 57 MapPairs primers (Research Genetics, Huntsville, AL). For genetic linkage analyses, mice were genotyped with an additional 15 markers on chromosome 11 (D11Mit226, -135, -20, -154, -131, -349, -278, -177, -212, -285, -67, -132, -224, -181, and -338). Polymerase chain reaction (PCR) conditions were according to the manufacturer's suggestions, and DNA was resolved on 3.5% to 4% agarose gels (Gibco, Life Technologies, Breda, The Netherlands).
All mice were kept in sterile microisolators and were fed sterile food and water ad libitum.
Progenitor and stem cell assays
The percentage of hematopoietic progenitors in S phase was determined using standard semisolid granulocyte-macrophage colony-forming unit (CFU-GM) assays supplemented with recombinant rat stem cell factor (a gift from Amgen, Thousand Oaks, CA) and recombinant murine granulocyte-macrophage colony-stimulating factor (GM-CSF) (a gift from Behringwerke, Marburg, Germany). Prior to their use in the progenitor assay, bone marrow cells were incubated with or without the S phase–specific cytotoxic drug hydroxyurea (200 μg/mL; Sigma, St Louis, MO) for 1 hour. Cells were washed twice, and CFU-GM assays were subsequently performed using standard procedures. Cultures were evaluated after 7 days. The fraction of progenitors that was killed by hydroxyurea incubation was calculated and was considered to have been in the S phase of the cell cycle. Cobblestone area–forming cell (CAFC) assays to quantify progenitor and stem cell frequencies were performed exactly as described previously.13 Both assays were chosen for consistency and to allow comparison with our previously published data.12,13 15
Competitive repopulation assay and chimerism analysis
Recipient C57BL/6.SJL.Ptprca(Ly5.1+/CD45.1+) were conditioned with a sublethal dose of 4 Gy total body γ-irradiation administered in 1 single dose 24 hours prior to transplantation. Recipient mice received a dose of 3 × 106 unfractionated bone marrow cells derived from wild-type B6 (n = 6) or congenic B6.DBAScp2 (n = 6) donors (both strains are Ly5.2+/CD45.2+). To determine the percentage donor blood cell contribution, mice were bled from the retro-orbital sinus. Erythrocytes were eliminated by hypotonic lysis, and the remaining leukocytes were incubated with saturating amounts of phycoerythrin-conjugated anti-Ly5.1/CD45.1 antibodies and fluorescein isothiocyanate (FITC)–labeled anti-Ly5.2/CD45.2 antibodies (Pharmingen, San Diego, CA). Fluorescence was determined using a dual laser FACSCalibur System (Becton Dickinson Immunocytometry Systems, San Jose, CA).
Linkage analysis
In this study we used 3 independent data sets. First, we phenotyped an additional set of 7 newly derived BXD strains21 and added these data to our database, which includes information on 26 BXD strains that we have previously used to search for cell cycle QTL.15 In addition, we derived 2 novel datasets consisting of 60 N2 and 43 N6 backcross animals. For linkage analysis we used MapManager QT software22 (version b28ppc, available at ftp://mcbio.med.buffalo.edu/pub/MapMgr/QT). We determined the probability of obtaining the observed likelihood ratio statistic (LRS) values by random chance using the permutation test. This test, which is implemented in MapManager QT software, was performed with 2000 permutations at 1 centimorgan (cM) intervals (genome-wide for the BXD set and restricted to chromosome 11 for the N2 and N6 backcross).
Stem cell purification
Hematopoietic stem cells were purified using flow cytometry exactly as described before, from pooled marrow cells obtained from 10 B6 and 10 DBA mice.23 Sorted cells were identified by the absence of a panel of lineage-specific cell surface markers (CD3, CD5, CD8, CD11b/Mac-1, CD45R/B220, and Ly-6G/Gr-1, collectively referred to as Lin−) and the presence of the Sca-1 and c-kit antigen. Viable (propidium iodide–negative) Lin−Sca-1+c-kit+ cells were sorted using a dual-laser FACS Vantage instrument (Becton Dickinson Immunocytometry Systems).
Subtractive hybridization
Total RNA was isolated from 105Lin−Sca-1+c-kit+ B6 and DBA cells using Trizol (Gibco). DNAse-treated RNA was reverse transcribed (Superscript II, Gibco), and cDNA was amplified using the SMART PCR cDNA synthesis kit (Gibco). Bidirectional subtractive hybridizations using B6- and DBA-amplified stem cell cDNA as “tester” and “driver,” reciprocally, were performed with the PCR-Select cDNA subtraction kit (Gibco). All procedures were carried out according to the manufacturer's suggestions.
Cloning of differentially expressed transcripts
Subtracted cDNA samples were randomly cloned in pCR2-Topo or pCR4-Topo (Invitrogen, Hoogkerk, The Netherlands). A total number of 240 clones (about 50% originating from the B6 minus DBA subtraction and 50% derived from the reverse DBA minus B6 subtraction) were sequenced, and BLAST searches were performed to identify the transcripts.
Gene array analysis
Amplified but unsubtracted B6 and DBA cDNA samples were labeled with [33P]-dCTP (deoxycytidine triphosphate). Two mouse filter gene arrays (GF400a, Research Genetics), each containing 5184 cDNA fragments, were hybridized with B6- and DBA-labeled cDNA samples. Hybridization signals were identified using a Storm phospor imager system (Molecular Dynamics), and appropriately configured files were analyzed with Pathways Software (Research Genetics). After signal normalization, only spots for which the intensity of either B6 or DBA signal was 3-fold above background were considered. Both in B6 and DBA samples about 12% of the transcripts were found to be expressed in hematopoietic stem cells. We selected the 100 transcripts from both B6 and DBA that showed most pronounced differential hybridization intensities for further study.
Mapping of differentially expressed genes
We tried to determine the chromosomal localization of all (ie, 440) nonredundant differentially expressed transcripts in silico using a variety of publicly available databases (predominantlyhttp://www.informatics.jax.org andhttp://www.ncbi.nlm.nih.gov/UniGene/). We were successful in assigning the chromosomal location to 156 transcripts. Some transcripts were mapped by synteny to their map position in the human genome.
RT-PCR analysis
To independently confirm that transcripts were differentially expressed, we studied the expression pattern of a selected number of chromosome 11 genes by reverse transcriptase (RT)–PCR. To this end purified stem cells or unfractionated bone marrow cells were isolated from an additional cohort of B6 and DBA mice. Also, cells from B6.DBAScp2 congenic mice were used. The primers that were used to amplify differentially expressed transcripts were designed with sequence information as present in GenBank.
Results
Genetic mapping of cell proliferation loci
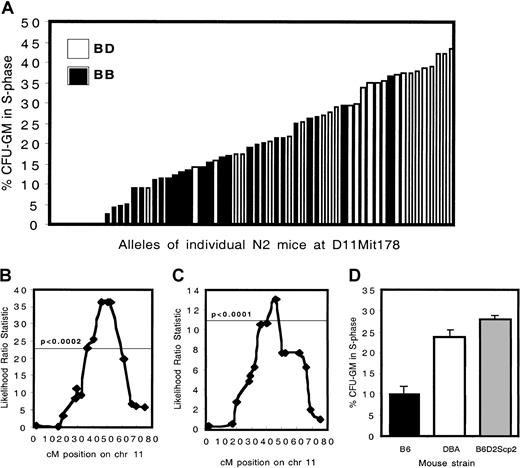
To map quantitative trait loci associated with variation in cell cycle, we performed a genome-wide linkage analysis using cell cycling phenotypes (ie, fraction of CFU-GMs in S phase) of 33 BXD recombinant inbred strains (analysis of 26 of these strains was previously reported15). We found 2 markers, D7Mit178 and D11Mit131, that showed suggestive linkage (Table1) and refer to these QTL asStem cellproliferation-1 and -2, respectively. For both loci the presence of DBA alleles was associated with higher cell proliferation rates.
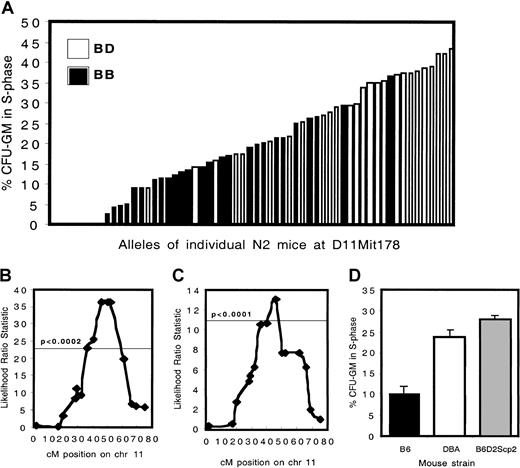
To confirm linkage, a set of 60 (B6 × BDF1) backcross animals was generated. A distinctive effect of the presence of D alleles at D11Mit178 (at 47 cM) was observed. The phenotype of mice carrying 2 B alleles (n = 34; mean percentage S phase cells, 12.4%) was significantly different (P < 2.5 × 10−9) compared with mice carrying 1 B and 1 D allele (n = 26; mean percentage S phase cells, 30.2%) (Figure1A-B).
Cell cycling phenotypes and genetic analysis of C57BL/6 × B6D2F1 backcross mice.
(A) The percent progenitors in S phase for 60 N2 animals is depicted. Animals carrying 2 B alleles at D11Mit178 are indicated by ▪, whereas the ■ show heterozygous mice carrying 1 B and 1 D allele. (B) Interval mapping analysis of the N2 generation. The horizontal line indicates the statistical threshold for highly significant linkage, obtained by performing a permutation test with 2000 permutations at 1 cM intervals, on chromosome 11 only. (C) Interval mapping analysis of the N6 generation. (D) The percentage of CFU-GMs in S phase in wild-type B6 (n = 26), DBA (n = 29), and in congenic B6.DBA2Scp2 animals (n = 17) is shown.
Cell cycling phenotypes and genetic analysis of C57BL/6 × B6D2F1 backcross mice.
(A) The percent progenitors in S phase for 60 N2 animals is depicted. Animals carrying 2 B alleles at D11Mit178 are indicated by ▪, whereas the ■ show heterozygous mice carrying 1 B and 1 D allele. (B) Interval mapping analysis of the N2 generation. The horizontal line indicates the statistical threshold for highly significant linkage, obtained by performing a permutation test with 2000 permutations at 1 cM intervals, on chromosome 11 only. (C) Interval mapping analysis of the N6 generation. (D) The percentage of CFU-GMs in S phase in wild-type B6 (n = 26), DBA (n = 29), and in congenic B6.DBA2Scp2 animals (n = 17) is shown.
In contrast to homozygous BXD mice, heterozygous backcross animals provided no evidence for the involvement of Scp1, located on chromosome 7 (data not shown). The backcrossing strategy was continued to derive a congenic mouse strain in which the effect ofScp2 could be investigated in isolation. In N6 mice highly significant linkage was observed with an interval extending from 39 cM to 49 cM on chromosome 11, documenting the strength of this isolated locus (more than 99% of the genome of these animals is of B6 origin) (Figure 1C). Homozygous B6.DBAScp2 congenic animals were produced, which, as expected, showed a high fraction of cells in S phase (Figure 1D). The difference between B6 and DBA (P < 1.4 × 10−6) and between B6 and B6.DBA2Scp2(P < 1.2 × 10−7) was highly significant. The difference between DBA and B6.DBA2Scp2 was not significant (P < .088).
Stem cell pool size is not affected in B6.DBAScp2congenic mice
Previous studies by several groups, including ours, have mapped loci regulating stem cell frequency to chromosomes 1, 12, and 18.13,24 25 These chromosomal locations suggested that stem cell frequency is maintained by a different set of genes than those affecting cell cycling, mapping to chromosome 11. Our congenic mice offered the possibility of directly assessing whether stem cell frequency and proliferation are regulated by distinct genetic pathways. We quantified cobblestone area–forming cell (CAFC) frequencies in bone marrow from B6, DBA, and B6.DBAScp2 mice after 7, 21, and 35 days in culture (Figure 2). As expected, CAFC frequency was substantially higher in DBA compared with B6 bone marrow. However, stem and progenitor cell frequency in the B6.DBAScp2 congenics was identical to B6 values. This demonstrates that Scp2 is not involved in maintaining stem cell pool size.
Stem cell frequency is not affected by the Scp2 locus on chromosome 11.
CAFC assays were performed to quantify the number of progenitors (CAFC day 7 [A]) and more primitive CAFC day 21 and CAFC day 35 cell subsets (B,C) in B6, DBA, and B6.DBA2Scp2 bone marrow cells.
Stem cell frequency is not affected by the Scp2 locus on chromosome 11.
CAFC assays were performed to quantify the number of progenitors (CAFC day 7 [A]) and more primitive CAFC day 21 and CAFC day 35 cell subsets (B,C) in B6, DBA, and B6.DBA2Scp2 bone marrow cells.
B6.DBAScp2stem cells are defective in long-term repopulating ability
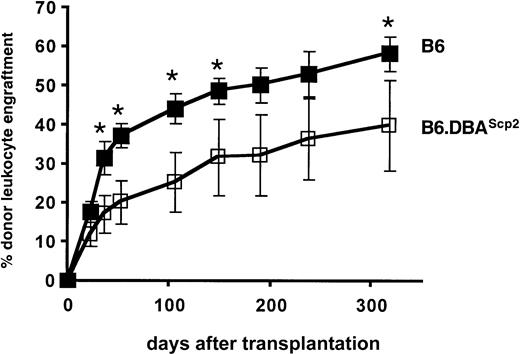
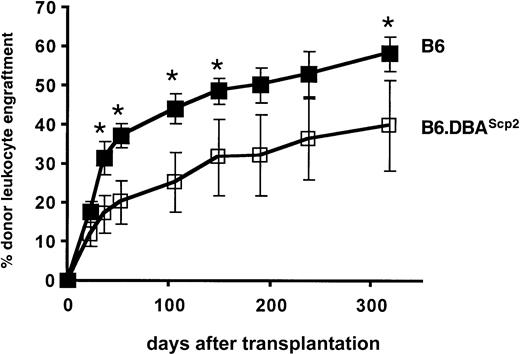
In embryo and radiation chimeras in which DBA and B6 stem cells coexist, highly cycling DBA stem cells, although initially functioning appropriately, cease to contribute to the process of blood cell formation during aging.26-28 It has been shown that DBA stem cells can be reactivated after secondary transplantation, demonstrating that DBA stem cells are present but remain fully dormant in the unmanipulated, aged chimeras.29 Conversely, more quiescent B6 stem cells have a major competitive advantage over the long term and will eventually outcompete DBA stem cells. Our previous data showing a correlation between cell cycling and mouse life span12 15 support a model in which enhanced cellular senescence is induced by high cell turnover rates and thus would be controlled by Scp2. The availability of B6.DBAScp2 congenic mice allowed us to address this question directly by testing the competitive repopulation potential of B6.DBAScp2 stem cells compared with wild-type B6 stem cells. C57BL/6.SJL.Ptprca(Ly5.1+/CD45.1+) mice were sublethally irradiated and underwent transplantation with 3 × 106unfractionated B6 or B6.DBAScp2(Ly5.2+/CD45.2+) bone marrow cells. B6.DBAScp2 stem cells showed a reduced repopulating potential (Figure 3).
Repopulation potential of transplanted stem cells is affected by the Scp2 locus on chromosome 11.
B6 or B6.DBA2Scp2 bone marrow cells were intravenously transplanted in sublethally (4 Gy) irradiated B6.SJL-CD45.1 congenic recipients. The percentage of donor-derived (CD45.2+) leukocytes was determined by flow cytometry. Data represent mean values ±1 SD from 2 independent experiments. Asterisks indicate significant differences between the 2 treatment groups (P < .05, t test).
Repopulation potential of transplanted stem cells is affected by the Scp2 locus on chromosome 11.
B6 or B6.DBA2Scp2 bone marrow cells were intravenously transplanted in sublethally (4 Gy) irradiated B6.SJL-CD45.1 congenic recipients. The percentage of donor-derived (CD45.2+) leukocytes was determined by flow cytometry. Data represent mean values ±1 SD from 2 independent experiments. Asterisks indicate significant differences between the 2 treatment groups (P < .05, t test).
Because B6 and B6.DBAScp2 bone marrow cells contained an equal number of stem cells (Figure 2), differences in engraftment kinetics must have resulted from qualitative stem cell differences. Although we cannot rule out the possibility that unknown minor histocompatibility antigens may (partially) have induced lower levels of chimerism, no such genes are known to map to the congenic chromosome 11 interval, and therefore we favor the concept that high rates of cell turnover are associated with impaired stem cell functioning.
Clustering of candidate genes
By cotransplanting DBA and B6 stem cells into BDF1 recipients, we have previously shown that Scp2 acts cell autonomously.28 Therefore, to detect candidate genes for this locus, we identified differentially expressed genes in highly enriched (and consequently highly identical) Lin−Sca-1+c-kit+ stem cells isolated by flow cytometry from 10 pooled B6 and DBA bone marrow samples using bidirectional subtractive hybridization and gene array analysis. After subtractive hybridization, 240 B6- and DBA-derived cDNA fragments were cloned and identified by sequence analysis.
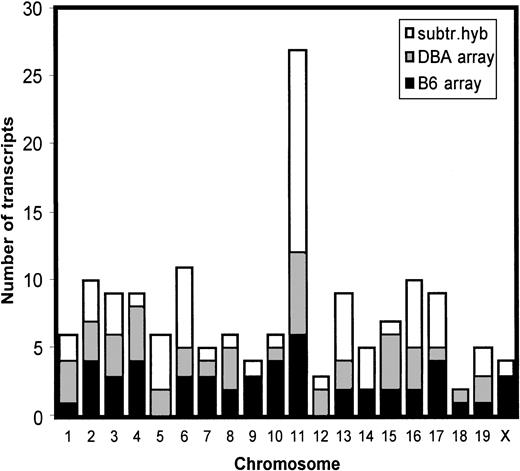
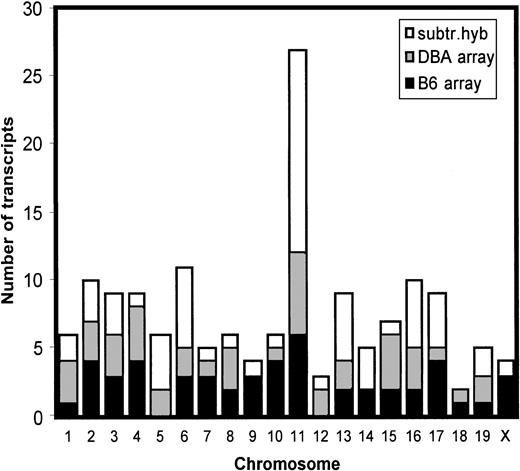
In parallel, unsubtracted B6 and DBA cDNAs were separately hybridized with DNA arrays. A total of 200 most strongly differentially expressed transcripts were analyzed. We were able to assign the chromosomal localization to 156 transcripts. Interestingly, a disproportionate number of these differentially expressed transcripts was located on chromosome 11 (Figure 4).
Genomic distribution of differentially expressed genes.
Transcripts that were shown to be differentially expressed in B6- and DBA-purified hematopoietic stem cells using bidirectional subtractive hybridization or gene array analysis were not randomly distributed across the genome but, rather, mapped preferentially to chromosome 11.
Genomic distribution of differentially expressed genes.
Transcripts that were shown to be differentially expressed in B6- and DBA-purified hematopoietic stem cells using bidirectional subtractive hybridization or gene array analysis were not randomly distributed across the genome but, rather, mapped preferentially to chromosome 11.
A total of 28 genes were shown to map to the average sized chromosome 11, which represents about 18% of all differentially expressed transcripts for which a chromosomal map position could be assigned (Table 2). The observed excess of chromosome 11 genes is significantly different from expected random distribution (P < 1 × 10−4). In addition, these transcripts were not evenly distributed across chromosome 11 but occurred in 3 distinct clusters (Table 2, Figure5).
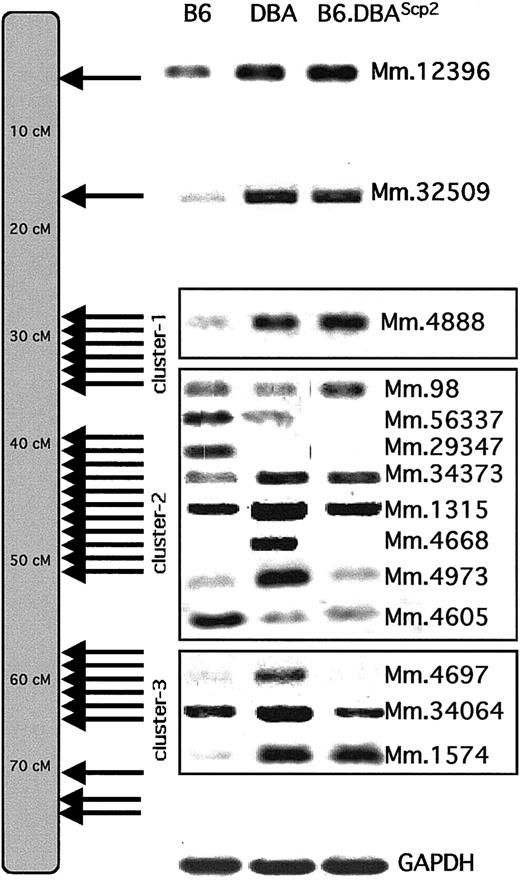
Clustering of differentially expressed transcripts on chromosome 11.
The chromosomal localization of all differentially expressed chromosome 11 transcripts shows that they occurred in 3 distinct clusters (for exact centimorgan position, see Table 2). To confirm the differential expression patterns, RT-PCR analyses were performed on a random selection of transcripts using B6, DBA, or B6.DBA2Scp2 cDNA. Transcripts are identified by their Unigene (Mm) cluster number.
Clustering of differentially expressed transcripts on chromosome 11.
The chromosomal localization of all differentially expressed chromosome 11 transcripts shows that they occurred in 3 distinct clusters (for exact centimorgan position, see Table 2). To confirm the differential expression patterns, RT-PCR analyses were performed on a random selection of transcripts using B6, DBA, or B6.DBA2Scp2 cDNA. Transcripts are identified by their Unigene (Mm) cluster number.
Assuming a total genome size of about 1500 cM, the distance between the 156 differentially expressed transcript on average is expected to be 156/1500 equals about 10 cM. We considered transcripts to be clustered if multiple genes mapped to an interval less than 10 cM.
The first cluster centered around 30 cM (6 transcripts were identified that map to this cluster). The largest cluster (containing 11 genes) extended from about 40 to about 50 cM and coincided exactly with theScp2 interval (compare Figure 1B and C). The most telomeric cluster covered a region from about 60 to about 78 cM and contained 9 transcripts.
To confirm the differential expression pattern of transcripts first identified by subtractive hybridization or array analysis, we performed RT-PCR experiments using cDNA obtained from B6, DBA, and B6.DBAScp2 bone marrow or stem cells (Figure 5).
When we randomly checked expression of other genes that map closely to those identified by subtractive hybridization or array analysis, we were able to show distinct expression levels in a significant number of them as well (such as Epx and Stat5a). This indicates that the collection of variably expressed genes within the clusters can be extended.
Differentially expressed genes map to recombination coldspots
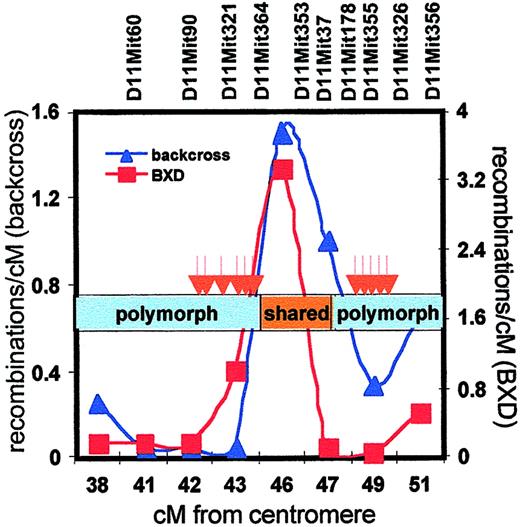
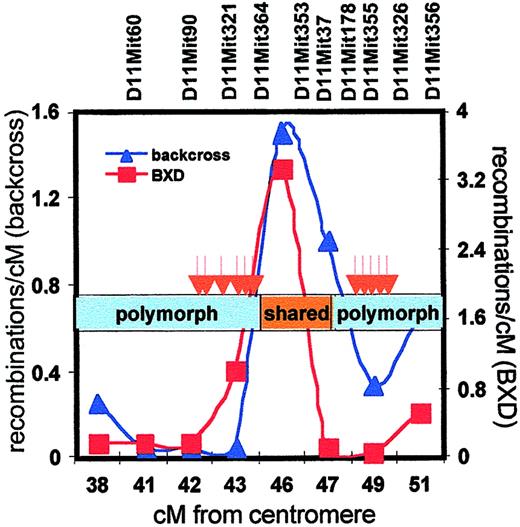
A more detailed analysis of the Scp2 cluster showed that differentially expressed transcripts mapped either to a region from 42 to 45 cM or around 49 cM, whereas no transcripts were found from 45 to 48 cM (Table 2, Figure 5) Also, RT-PCR analysis of randomly selected genes known to map to the silent interval failed to reveal any expression differences (data not shown). Such clustering of differentially expressed genes suggested the involvement of genetic variation affecting higher-order chromosomal organization. Because most laboratory mice have been derived from a limited number of selected founders, there is a number of shared haplotypes across the genome in B6 and DBA mice.30 When we assessed the extent of shared haplotypes in the Scp2 interval (by comparing polymorphism at a series of microsatellite repeats), we found evidence of extensive polymorphism from 38 to 45 cM and again from 47 to 52 cM, but the silent region from 45 to 47 cM in which we failed to detect differentially expressed genes turned out to be identical in both strains (Figure 6).
Meiotic recombinations, haplotype sharing, and differential gene expression in the Scp2 cluster.
The frequency of meiotic recombinations within the Scp2interval was determined in the backcross panel that was used to breed congenic B6.DBAScp2 mice (blue line, left y-axis) and in the BXD recombinant inbred panel (data obtained fromhttp://mickey.utmem.edu/MMfiles/MMlist.html; red line, right y-axis). The region of shared haplotypes of B6 and DBA mice (45 to 47 cM) is indicated by the orange box, whereas distinct haplotypes are shown in blue. Differentially expressed stem cell genes (red arrows) clustered in the polymorphic regions in which recombination was severely suppressed.
Meiotic recombinations, haplotype sharing, and differential gene expression in the Scp2 cluster.
The frequency of meiotic recombinations within the Scp2interval was determined in the backcross panel that was used to breed congenic B6.DBAScp2 mice (blue line, left y-axis) and in the BXD recombinant inbred panel (data obtained fromhttp://mickey.utmem.edu/MMfiles/MMlist.html; red line, right y-axis). The region of shared haplotypes of B6 and DBA mice (45 to 47 cM) is indicated by the orange box, whereas distinct haplotypes are shown in blue. Differentially expressed stem cell genes (red arrows) clustered in the polymorphic regions in which recombination was severely suppressed.
We made an interesting observation by assessing meiotic recombination frequencies in the Scp2 region using our own backcross panel and the existing BXD panel. In both datasets, recombinations in theScp2 cluster occurred preferentially in the silent region, in which the DBA and B6 genomes are highly homologous. In contrast, recombinations in the bordering regions were strongly suppressed (Figure 6).
Discussion
In this study we demonstrate convergence of a collection of differentially expressed stem cell genes with a locus that is strongly associated with variation in hematopoietic cell turnover between B6 and DBA mice. Differentially expressed transcripts mapped to a polymorphic genomic region that is characterized by suppressed recombination frequencies.
To exclude the possibility that clustering resulted from a preponderance of hematopoietic genes in these regions, we randomly picked 150 genes from the Stem Cell Data Base (http://stemcell.princeton.edu/), which contains a large collection of B6 stem cell–specific transcripts.31 We found no evidence of clustering in this dataset (data not shown), reinforcing the notion that it is the genetic difference between the 2 strains that causes the clustering. Also, the extent of overrepresentation of chromosome 11 transcripts cannot be attributed to a potentially increased gene density on this chromosome.32
Most fundamental to our studies is the issue of how variation in progenitor cell proliferation is induced. Two alternative hypotheses can be provided. First, allelic variation of one of the genes in theScp2 interval may confer the cell cycling trait. Possible candidates, based upon known functional activity and/or an appropriate (ie, DBA-like) expression pattern in B6.DBAScp2 congenic mice, includeplatelet-activating factor acetylhydrolase,ribosomal protein S6 kinase,33 andTbox2.34
If this “single-gene” hypothesis is correct, the additional differentially expressed transcripts that are closely linked to the underlying gene are not contributing to the phenotype, and their map position is entirely coincidental. Although we cannot exclude this, we favor an alternative but more provocative explanation: Variation in cell cycle does not result from a single gene but rather is the consequence of variable expression of a collection of highly linked genes in stem cells from both parental strains. Genotype-independent phenotypic variability (ie, reduced penetrance) in the N2, but particularly in the more homogenous N6 generation, would be explained by such a model. The fact that the Scp2 gene cluster occurs in a genomic interval in which meiotic recombination is suppressed implies that the cluster is transmitted to the next generation in its entirety. It is therefore tempting to speculate that the strong linkage disequilibrium points to a functional interdependence of genes within the cluster.35 Interestingly, several studies have recently documented findings in other model systems that are reminiscent of the data that we show here. First, Caron et al showed that in the human genome gene expression occurs in highly localized “ridges.”36 In addition, suppression of recombination over a large segment has been reported for the cytokine cluster on human chromosome 5q,37 a region that has shown strong linkage to various immunologic quantitative traits.38,39Intriguingly, the human cytokine cluster is homologous to a segment of mouse chromosome 11 at about 30 cM, colocalizing with one of the clusters we report here. Very similarly, an extreme degree of long-range linkage disequilibrium has been reported for a locus on human chromosome 17q21, containing BRCA1.40 Again, this segment is homologous to the most telomeric cluster (about 60 cM) we report here. Collectively these data indicate that the selective forces that result in effective recombination suppression have been conserved in human and mouse.
It has generally been assumed that the heritable component of quantitative traits results from additive and epistatic interactions of multiple loci, each locus containing a single mutated gene.16,20 In this respect it is interesting that in our study a number of DBA-derived genes showed a B6-like expression pattern in the B6.DBAScp2 congenic animals (eg,Zfp147, Mm.4973). This confirms the presence of specific modifying loci in the B6 genome that may prevent the penetrance of a bona fide QTL. Loss of the expected phenotype in congenic mice has repeatedly been observed.41 42
It has been notoriously difficult to identify genes underlying QTL.17 43 Our studies suggest a novel concept that may explain the difficulty of associating individual genes with quantitative traits. Variation at a specific locus may result in altered basal expression of many genes in this region, and it is only in the context of the expression pattern of neighboring genes that biologic variation is induced. We speculate that the model we put forward may also apply to certain tumorigenic chromosomal abberations (translocations, deletions, insertions, inversions) in which the altered chromosomal structure affects the expression of a large number of adjacent genes that collectively induce a (subtle) growth advantage.
The authors thank Anke van den Berg, Geert Harms, Joost Kluiver, and Peter Terpstra for technical assistance and advice.
Prepublished online as Blood First Edition Paper, May 24, 2002; DOI 10.1182/blood-2002-03-0808.
Supported by the Dutch Cancer Society (RUG 2000-2183), a fellowship from the Royal Netherlands Academy of Arts and Sciences (KNAW) to G.d.H., National Institutes of Health grant AG16653 to G.V.Z., and a fellowship of the Deutsche Akademie der Naturforscher Leopoldina funded by the Bundesministerium fuer Bildung und Forschung to H.G.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Gerald de Haan, Department of Stem Cell Biology, University of Groningen, A. Deusinglaan 1, 9713 AV Groningen, The Netherlands; e-mail: g.de.haan@med.rug.nl.

![Fig. 2. Stem cell frequency is not affected by the Scp2 locus on chromosome 11. / CAFC assays were performed to quantify the number of progenitors (CAFC day 7 [A]) and more primitive CAFC day 21 and CAFC day 35 cell subsets (B,C) in B6, DBA, and B6.DBA2Scp2 bone marrow cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/6/10.1182_blood-2002-03-0808/4/m_h81823123002.jpeg?Expires=1769293137&Signature=bGnEDOqqvireMvcUHfhnDvkuPVbF5s2arNJ7UcWEwmFzslElgF0-vsz~X-7WrZr0Gr16MYRwGfYFNxfVJ1YRYfSWlNSyc9KGSBRtE9oNargu6VrJ6qGrcqS7VFrjqdV422n8IWA6odnBKM7HDmpcK4mOf1~pNuqQGMZpuDsCQATXmW8R4D1PyZWfOLXpjEToMH1u26rzOZAqQIxQEUG6pY4RcRK4PR0Y8UyDEtup4S5DjfTiXmFiCaQ0RMfhxdS6PzHM6UmBGFWcGTVXxu6bOjBhbiLBcTuY6vwwmnw6YIRlRz2EITTyDAfQCPR~1riiESqoH8yg5Q~kb-Z7gEDchw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 2. Stem cell frequency is not affected by the Scp2 locus on chromosome 11. / CAFC assays were performed to quantify the number of progenitors (CAFC day 7 [A]) and more primitive CAFC day 21 and CAFC day 35 cell subsets (B,C) in B6, DBA, and B6.DBA2Scp2 bone marrow cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/6/10.1182_blood-2002-03-0808/4/m_h81823123002.jpeg?Expires=1769996956&Signature=r9a2V6BXf19z3wBAMDpfZ2valBoALJbzrlyUtIDxEVWk-tfL0jugOfHMTPmMF5TW7RdGgk6ZM~uXEARF2H0kYg5T~WDM6uedzHzr3Yqx0GlYsyYC0A3pPiFv8ntdr1UCbp01GBqRyEYd08cvD1ghCMaRpI~H7Ct7r~q3gesRrNPFku1UP3M9Jui4wJ~ALEoSzcPucxxRfXd2nXO8mldPQca7l8SiR5bMBCQsTBK1P~4x7gpqOgBfQ6bfj6j4u0cO8IpOwsnJ5CvBG~XpYf1Pq4w3aggOHhF1NbHH8xwi2tCEruXmRBiJN3SjaJL53LERVKDyzLxxmxnTIfjjZaDwGg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)