Abstract
SALL4, a human homolog to Drosophila spalt, is a novel zinc finger transcriptional factor essential for development. We cloned SALL4 and its isoforms (SALL4A and SALL4B). Through immunohistochemistry and real-time reverse-transcription-polymerase chain reaction (RT-PCR), we demonstrated that SALL4 was constitutively expressed in human primary acute myeloid leukemia (AML, n = 81), and directly tested the leukemogenic potential of constitutive expression of SALL4 in a murine model. SALL4B transgenic mice developed myelodysplastic syndrome (MDS)-like features and subsequently AML that was transplantable. Increased apoptosis associated with dysmyelopoiesis was evident in transgenic mouse marrow and colony-formation (CFU) assays. Both isoforms could bind to β-catenin and synergistically enhanced the Wnt/β-catenin signaling pathway. Our data suggest that the constitutive expression of SALL4 causes MDS/AML, most likely through the Wnt/β-catenin pathway. Our murine model provides a useful platform to study human MDS/AML transformation, as well as the Wnt/β-catenin pathway's role in the pathogenesis of leukemia stem cells.
Introduction
Myelodysplastic syndrome (MDS) is a hematologic disease marked by the accumulation of genomic abnormalities at the hematopoietic stem cell (HSC) level leading to pancytopenia, multilineage differentiation impairment, and bone marrow apoptosis.1 Mortality in this disease results from pancytopenia or transformation to acute myeloid leukemia (AML). AML is a hematologic cancer characterized by the accumulation of immature myeloid precursors in the bone marrow and peripheral blood. From the analysis of chromosomal translocation in bone marrow samples from AML patients, it is clear that transcription factors critical for hematopoiesis play an important role in leukemogenesis.2-5
The pathogenesis of AML is considered to involve multistep genetic alternations.6 Because only HSCs are considered to have the ability to self-renew, they are the best candidates for the accumulation of multistep, preleukemic genetic changes and transforming them into so-called leukemia stem cells (LSCs).7-9 Alternatively, downstream progenitors can acquire self-renewal capacity and give rise to leukemia. A good example is the Wnt/β-catenin signaling pathway, which has been associated with the self-renewal of normal HSCs and the granulocyte-macrophage progenitors (GMPs) of chronic myeloid leukemia (CML).10-15 LSCs are not targeted under current chemotherapy regimens and have been found to account for drug resistance and leukemia relapse.8,9 Hunting for genes or signaling pathways involved in leukemia self-renewal will promote the development of more effective leukemia treatments.
The SALL gene family, SALL1, SALL2, SALL3, and SALL4, was originally cloned on the basis of its DNA sequence homology to Drosophila spalt (sal).16-19 In Drosophila, spalt is a homeotic gene essential for development of posterior head and anterior tail segments. It plays an important role in tracheal development,20 terminal differentiation of photoreceptors, and wing vein placement.21 In humans, the SALL gene family is involved in normal development, as well as tumorigenesis.19,22-27 SALL proteins belong to a group of C2H2 zinc finger transcription factors characterized by multiple finger domains distributed over the entire protein.28 During the tracheal development of Drosophila, spalt is an activated downstream target of Wingless, a Wnt ortholog.29 Of interest, Sato et al30 demonstrated that SALL1 interacted with β-catenin by functioning as a coactivator, suggesting that the interaction between SALL and the Wnt/β-catenin pathway was bidirectional.
We report here on the identification of SALL4 isoforms and their constitutive expression in all human AML we examined. The direct impact of SALL4 expression in AML was tested in vivo. We show that constitutive expression of SALL4 in mice is sufficient to induce MDS-like symptoms and transformation to AML that is transplantable. We also demonstrate that SALL4 is able to bind β-catenin and activate the Wnt/β-catenin signaling pathway. SALL4 and β-catenin share similar expression patterns at different phases of CML. A potential mechanism accounting for the oncogenic role of SALL4 in LSCs is proposed.
Materials and methods
Molecular cloning
Plasmid construction and DNA sequencing were performed in accordance with standard procedures. For cloning of SALL4 isoforms, polymerase chain reaction (PCR) primers were designed, based on the genomic clone RP5-1112F19 (GenBank accession no. AL034420). SALL4 isoforms were cloned with the use of the Marathon-Ready cDNA library derived from human fetal kidney (BD Biosciences Clontech, Palo Alto, CA), according to the supplier's protocol. The amplified PCR products were cloned into a TA Cloning vector (Invitrogen, Carlsbad, CA), and the nucleotide sequences were determined by DNA sequencing.
Determination of SALL4 alternative splicing patterns in different tissues and Wnt/β-catenin downstream target gene expression (c-Myc and Cyclin D1)
Reverse transcription (RT)-PCR was used to evaluate mRNA expression patterns of SALL4 in adult tissues. A panel of 8 normalized first-strand cDNA preparations, derived from different adult tissues, was purchased from BD Biosciences Clontech. PCR amplification was performed in a 50-μL reaction volume containing 5 μL cDNA, 10 mM Tris HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 0.2 mM dNTPs, and 1.25 U Taq DNA polymerase (PerkinElmer Life Sciences, Boston, MA). After an initial denaturation at 94°C for 10 minutes, amplification was performed for 30 cycles under the following conditions: 30-second denaturation at 94°C, 30-second annealing at 55°C, and 30-second extension at 72°C. The last cycle was followed by a final 7-minute extension at 72°C. Amplification of glyceraldehyde phosphate dehydrogenase (GAPDH) mRNA was used to control for template concentration loading. The primer pairs for SALL4 isoforms were the following: primer A1, 5′-TCCGAGAACAGCCGCACTGAGATGGAAG-3′; primer B1, 5′-GTTCACTACATGACACACGGGGCG-3′; primer C1, 5′-ATGTCGAGGCGCAAGCAGGCGAAAC-3′; and primer D1, 5′-TTAGCTGACCGCAATCTTGTTTTCTTCC-3′. PCR products were electrophoretically separated on 1% agarose gel. DNA sequencing was also used to confirm amplification products. To determine the expressions of Wnt/β-catenin downstream target genes, RT-PCR was also used on bone marrow samples from wild-type control, and preleukemic and leukemic SALL4B transgenic mice. The primers for c-Myc were as follows: forward, 5′-TTT GTC TAT TTG GGG ACA GTG TT-3′; reverse, 5′-CAG CTT CTC CGA GAC CAG CTT GGC AGC-3′. The primers for Cyclin D1 were as follows: forward, 5′-CCT CTC CTG CTA CCG CAC AAC GCA C-3′; reverse, 5′-CTC TCA GGG TGA TGC AGA TTC TAT CTC-3′. Beta actin was used as a control; its primers were as follows: forward, 5′-GAC GAG GCC CAG AGC AAG AGA GG-3′; reverse, 5′-GTG ATG ACC TGG CCG TCA GGC AG-3′.
Antibody generation
The peptide MSRRKQAKPQHIN of human SALL4 was chosen for its potential antigenicity (amino acids 1-13) and used to prepare an antipeptide antibody. This region is also identical to that of mouse SALL4 so that the generated antibody could be expected to cross-react with mouse SALL4. SALL4 antipeptide antibody was produced in rabbits in collaboration with Lampire Biological Laboratories (Pipersville, PA).
Gel electrophoresis and Western blot analysis
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out in SDS 10% wt/vol polyacrylamide slab gels according to Laemmli, and the proteins were then transferred to nitrocellulose membranes. Immunoblotting of rabbit immune serum with the SALL4 antipeptide antibody (1:100) was performed with an electrochemiluminescence detection system as described by the manufacturer (Amersham Biosciences, Piscataway, NJ).
Leukemia and normal tissues
Leukemia and normal samples, either in paraffins or frozen in dimethylsulfoxide (DMSO), were collected from the files of the University of Texas M. D. Anderson Cancer Center (Houston) and the Dana-Farber Cancer Institute (Boston, MA), between 1998 and 2004 under approved institutional review board protocols. The diagnosis of all tumors was based on morphologic and immunophenotypic criteria according to the French-American-British (FAB) Classification for Hematopoietic Neoplasms. CD34+ fresh cells were purchased from Cambrex (Walkersville, MD).
Real-time quantitative RT-PCR
We used the TaqMan 5′ nuclease assay (Applied Biosystems, Foster City, CA) in these studies. Total RNA from purified CD34+ HSCs/hematopoietic progenitor cells (HPCs) from normal bone marrow and peripheral blood, 15 AML samples, and 3 leukemia cell lines were isolated with the RNeasy Mini Kit and digested with DNase I (Qiagen, Valencia, CA). RNA (1 μg) was reverse-transcribed in 20 μL with the use of Superscript II reverse transcriptase and a poly(dT)12-18 primer (Invitrogen). After the addition of 80 μL water and mixing, 5-μL aliquots were used for each TaqMan reaction. TaqMan primers and probes were designed with the use of Primer Express software version 1.5 (Applied Biosystems). Real-time PCR for SALL4 and GAPDH was performed with the TaqMan PCR core reagent kit (Applied Biosystems) and an ABI Prism 7700 Sequence Detection System (Applied Biosystems). The PCR reaction mixture contained 3.5 mM MgCl2; 0.2 mM each of deoxyadenosine triphosphate (dATP), deoxycytidine triphosphate (dCTP), and deoxyguanosine triphosphate (dGTP); 0.4 mM deoxyuridine triphosphate (dUTP); 0.5 μM forward primer; 0.5 μM reverse primer; 0.1 μM TaqMan probe; 0.25 U uracil DNA glycosylase; and 0.625 U AmpliTaq Gold polymerase in 1 × TaqMan PCR buffer. cDNA (5 μL) was added to the PCR mix, and the final volume of the PCR reaction was 25 μL. All samples were run in duplicate. GAPDH was used as an endogenous control. Thermal cycler conditions were 50°C for 2 minutes, 95°C for 10 minutes, and 45 cycles of 95°C for 0.30 minutes and 60°C for 1 minute. Data were analyzed with the use of Sequence Detection System software version 1.6.3 (Applied Biosystems). Results were obtained as threshold cycle (Ct) values. The software determines a threshold line on the basis of the baseline fluorescent signal, and the data point that meets the threshold is given as the Ct value. The Ct value is inversely proportional to the starting number of template copies. All measurements were performed in duplicate. TaqMan sequences include the following: GAPDH forward primer (5′-GAAGGTGAAGGTCGGAGTC-3′) and reverse primer (5′-GAAGATGGTGATGGGATTTC-3′); TaqMan probe (5′-CAAGCTTCCCGTTCTCAGCC-3′); SALL4A forward primer (5′-TGCAGCAGTTGGTGGAGAAC-3′) and reverse primer (5′-TCGGTGGCAAATGAGACATTC-3′); and SALL4B forward primer (5′-ACATCTCCGCGGTGGATGT-3′) and reverse primer (5′-TGCTCCGACACTTGTGCTTG-3′).
Design and construction of tissue arrays
Tissue arrays that included triplicate tumor cores from leukemia specimens were sectioned (5-μm thick). A manual tissue arrayer (Beecher Instruments, Silver Spring, MD) was used to construct the tissue arrays.
Immunohistochemistry
Immunohistochemical staining was performed according to standard techniques. Briefly, formalin-fixed, paraffin-embedded, 4-μm-thick tissue sections were deparafinized and hydrated. Heat-induced epitopes were retrieved with a Tris buffer (pH 9.9; Dako, Carpinteria, CA) and a rapid microwave histoprocessor. After incubation at 100°C for 10 minutes, slides were washed in running tap water for 5 minutes and then with phosphate-buffered saline (PBS; pH 7.2) for 5 minutes. Tissue sections were then incubated with anit-SALL4 antibody (1:200) for 5 hours in a humidified chamber at room temperature. After 3 washes with PBS, tissue sections were incubated with anti-mouse immunoglobulin G and peroxidase for 30 minutes at room temperature. After 3 washes with PBS, tissue sections were incubated with 3,3′-diaminobenzidine/H2O2 (Dako) for color development; hematoxylin was used to counterstain the sections. Cells were considered to be positive for SALL4 when they showed definitive nuclear staining.
Image acquisition
Images were visualized using an Olympus BH-2 microscope (Olympus, Tokyo, Japan), equipped with one of the following: a Dplan4 4×/0.10 numeric aperture (NA) air objective (Figures 2Bv, 4Bviii); an Splan10 10×/0.3 NA air objective (Figures 4Bii,v); an Splan 20 20×/0.46 NA air objective (Figure 4Diii-vi and 5Ai,ii); an Splan40 40×/0.70 NA air objective (Figures 2Bi-iv,vi, 4Biii,vi, and 4Bviii inset); or an Splan Apo60 60×/1.4 NA oil objective (Figures 4Ai-xvi, 4Bi,iv, 5Bi-iv; microscope immersion oil from Richard-Allan Scientific, Kalamazoo, MI). All micrographs were visualized using hematoxylin and eosin stain. All images including the gross-view pictures (Figures 4Bvii, 4Di-ii) were taken at room temperature using an Olympus Q-color 5 camera (model 32-0055B-128) and were processed using Adobe Photoshop (Adobe Systems, San Jose, CA).
Generation of transgenic mice
SALL4B cDNA, corresponding to the entire coding region, was subcloned into a pCEP4 vector (IntroGene; now Crucell, Leiden, The Netherlands) to create the cytomegalovirus (CMV)/SALL4B construct for the transgenic experiments. Subsequent digestion with SalI, which does not cut within the SALL4B cDNA, released a linear fragment containing only the CMV promoter, the SALL4 cDNA coding region, the simian virus 40 (SV40) intron, and polyadenylation signal without additional vector sequences.
Transgenic mice were generated via pronuclear injection performed in the transgenic mouse facility at Yale University. Identification of SALL4B founder mice and transmission of the transgene was determined by PCR analyses. Tissue expressions of transgene were confirmed by RT-PCR. The PCR primers used for the RT-PCR span the junction of the 5′ SALL4B cDNA to the CMV promoter (sense primer, 5′-CAG AGA TGC TGA AGA ACT CCG CAC-3′; antisense primer, 5′-AGC AGA GCT CGT TTA GTG AAC CG-3′).
Hematologic analysis
Complete blood cell counts with automated differentials were determined with a Mascot Hemavet cell counter (CDC Technologies, Oxford, CT). For progenitor assays, 1.5 × 104 bone marrow cells were plated in duplicate 1.25-mL methylcellulose cultures supplemented with recombinant mouse interleukin-3 (IL-3, 10 ng/mL), IL-6 (10 ng/mL), stem cell factor (SCF; 50 ng/mL), and erythropoietin (3 U/mL) (M3434; StemCell Technologies, Vancouver, BC). Colonies were recorded between days 7 and 14 (colony-forming unit-granulocyte [CFU-G], CFU-granulocyte-macrophage [CFU-GM], CFU-macrophage [CFU-M], CFU-granulocyte-erythrocyte-megakaryocyte-macrophage [CFU-GEMM], and burst-forming unit-erythroid [BFU-E]). Peripheral blood, bone marrow smears, and cytospin from pooled CFU cells were stained with Wright-Giemsa stain.
Flow cytometric analysis
Cells were stained with directly conjugated antibodies to Gr-1, Mac-1, B220, Ter119, c-kit, CD34, CD45, CD41, CD19, CD5, CD3, CD4, CD8, propidium iodide (PI), or annexin V (BD Biosciences Pharmingen, San Diego, CA). Ten thousand scatter-gated live cells were acquired on a FACScan and analyzed with CellQuest software (BD Biosciences Clontech).
Statistical analysis
Student t test was used for all the statistical analyses, assuming normal 2-tailed distribution and unequal variance.
Cell culture
HEK-293 cells (derived from human embryonic kidney) and cell lines KG.1 (derived from human acute myeloid leukemia), Kasumi-1 (derived from human myeloid leukemia AML-M2), and THP-1 (derived from human promonocytic leukemia AML-M4/M5) were purchased from the American Type Culture Collection (Manassas, VA). Cells were maintained at 37°C in a humidified environment with 5% carbon dioxide and 10% fetal serum.
Transfection
Transfection was performed with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. Cells were plated in 24-well plates at a density of approximately 1 × 105 cells/well. Cells were harvested 24 hours after transfection. Plasmid DNA for transient transfection was prepared with the Qiagen Plasmid Midi Kit.
β-Galactosidase and luciferase assays
The cells were extracted with 100 μL luciferase cell culture lysis reagent (Promega, Madison, WI) 24 hours after transfection. The β-galactosidase assay, performed with 15 μL cell extract, used the β-Galactosidase Enzyme Assay System (Promega) and the standard assay protocol provided by the manufacturer (except that 1 M Tris base was used as stopping buffer, instead of sodium carbonate). For the luciferase assay (Promega), 20 μL extract were used in accordance with the manufacturer's instructions. After subtraction of the background, luciferase activity (arbitrary units) was normalized to β-galactosidase activity (arbitrary units) for each sample.
Results
Molecular cloning of 2 alternatively splicing isoforms of human SALL4
Two full-length transcripts of SALL4 were isolated by 5′ and 3′ RACE-PCR (rapid amplification of the 5′ and 3′ cDNA ends-polymerase chain reaction) with the use of fetal human kidney Marathon-Ready cDNAs (BD Biosciences Clontech) as templates. Sequence analysis of the larger cDNA fragment isolated revealed a single, large open-reading frame, designated as SALL4A, that started from a strong consensus initiation sequence and was expected to encode 1053 amino acids. The other splicing variant of SALL4, designated SALL4B, lacked the region corresponding to amino acids 385 to 820 of the full-length SALL4A (Figure 1A). The putative protein encoded by SALL4B cDNA was expected to consist of 617 amino acids.
To rule out the possibility that these 2 apparent splicing variants might result from artifacts, we compared both variant mRNA sequences with corresponding sequences of the human genome. SALL4A contained all exons (1-4) (Figure 1A), whereas SALL4B lacked a large portion of the 3′ end of exon 2. Both exon-intron splice sites satisfied the G-T-A-G rule. Both splicing variants had the same translational reading frame, but SALL4B mRNA encoded a protein with internal deletion. SALL4A contained 8 zinc finger domains, while SALL4B had 3 zinc finger domains.
Expression pattern of the SALL4 isoforms in human tissues
The alternative splicing patterns of SALL4 were delineated by RT-PCR in a variety of human tissues. A fragment of the ubiquitous GAPDH gene cDNA was amplified as a control (Figure 1B). A 315-bp fragment representing the longer splice variant, SALL4A, was amplified in some tissues, achieving various expression levels. The SALL4B variant was present in every tissue at varying levels of expression.
Generation of SALL4 antibody and identification of SALL4 protein products
To identify SALL4 gene products and confirm the presence of SALL4 variants, we developed a polyclonal antibody against a synthetic peptide (amino acids 1-13) of SALL4. This region was chosen because it is common to both SALL4 variants. The affinity-purified SALL4 peptide antibody recognized specifically 2 endogenous proteins in a human kidney total lysate. The 2 proteins were approximately 165 kDa and 95 kDa, which were identical to the apparent molecular weights of overexpressed SALL4A and SALL4B in Cos-7 cells, respectively (Figure 1C). Western blotting with this antibody confirmed that the SALL4 isoforms had different tissue distributions (Figure 1Dii).
SALL4 has 2 isoforms. Alternative splicing generates 2 variant forms of SALL4 mRNA. (A) SALL4A and SALL4B vary in protein length and in the presence of different numbers of characteristic sal-like zinc finger domains. SALL4A (encoding 1067 amino acids) contains 8 zinc finger domains, while SALL4B (encoding 623 amino acids) has 3 zinc finger domains. Both variants have exons 1, 3, and 4, and SALL4A contains all exons from 1 to 4. However, SALL4B uses an alternative splice donor that results in deletion of the large 3′ portion of exon 2. (B) RT-PCR analysis of SALL4 variants in different human tissues. Four exons of SALL4 and their potential coding structures are illustrated, with arrows indicating the primers used for PCR amplification of the SALL4 transcripts (i). Tissue-dependent expression of SALL4 transcripts by RT-PCR (ii). A 315-bp expected product that was specific for SALL4A with primers A1 (exon 2) and B1 (exon 4) was amplified with cDNAs of various tissues. Primers D1 (exon 4) and C1 (exon 1) were used to amplify the 1851-bp expected product of SALL4B. Comparable amounts of cDNA were determined by GAPDH. (C) SALL4 protein products, SALL4A, and SALL4B identified by a SALL4 peptide antibody. Lysates from Cos-7 cells transiently expressing His-SALL4B (lane 1), His-SALL4A (lane 2), or control vector (lane 8), or lysates from different human tissues were resolved by 10% SDS-PAGE gel, transferred onto a nitrocellulose membrane, and probed with the N-terminal SALL4 peptide antibody.
SALL4 has 2 isoforms. Alternative splicing generates 2 variant forms of SALL4 mRNA. (A) SALL4A and SALL4B vary in protein length and in the presence of different numbers of characteristic sal-like zinc finger domains. SALL4A (encoding 1067 amino acids) contains 8 zinc finger domains, while SALL4B (encoding 623 amino acids) has 3 zinc finger domains. Both variants have exons 1, 3, and 4, and SALL4A contains all exons from 1 to 4. However, SALL4B uses an alternative splice donor that results in deletion of the large 3′ portion of exon 2. (B) RT-PCR analysis of SALL4 variants in different human tissues. Four exons of SALL4 and their potential coding structures are illustrated, with arrows indicating the primers used for PCR amplification of the SALL4 transcripts (i). Tissue-dependent expression of SALL4 transcripts by RT-PCR (ii). A 315-bp expected product that was specific for SALL4A with primers A1 (exon 2) and B1 (exon 4) was amplified with cDNAs of various tissues. Primers D1 (exon 4) and C1 (exon 1) were used to amplify the 1851-bp expected product of SALL4B. Comparable amounts of cDNA were determined by GAPDH. (C) SALL4 protein products, SALL4A, and SALL4B identified by a SALL4 peptide antibody. Lysates from Cos-7 cells transiently expressing His-SALL4B (lane 1), His-SALL4A (lane 2), or control vector (lane 8), or lysates from different human tissues were resolved by 10% SDS-PAGE gel, transferred onto a nitrocellulose membrane, and probed with the N-terminal SALL4 peptide antibody.
Constitutive expression of SALL4 mRNA in human primary AML and myeloid leukemia cell lines
Because another SALL gene family member SALL2 is involved in tumorigenesis, we examined SALL4 mRNA expression in AML. Expression of SALL4 was quantitatively investigated by real-time RT-PCR in bone marrow cells derived from AML samples (n = 15) and myeloid leukemia cell lines (n = 3) and compared with that of nonneoplastic hematopoietic cells from a purified CD34+ stem/progenitor pool (HSCs/HPCs purchased from Cambrex), normal bone marrow (n = 3), and normal peripheral blood (n = 3). With the use of isoform-specific primers (Figure 2A), we observed that during normal hematopoiesis, both SALL4 isoforms were down-regulated. In contrast, both SALL4A and SALL4B mRNA levels were constitutively expressed in 60% of AML samples and in all 3 cell lines. In the remaining 40% of AML samples, either SALL4A or SALL4B was constitutively expressed. Compared with normal hematopoiesis cells, leukemia samples had a wide range of SALL4A or SALL4B expression levels. This is probably due to the fact that the leukemia samples had a variable range of leukemic blasts population (40%-90%).
Constitutive expression of SALL4 protein in human primary AML
To investigate whether the observed aberrant SALL4 expression was also present at the protein level, we examined 81 AML samples, ranging from AML subtypes M1 to M5 (FAB classification): M1 (n = 20), M2 (n = 27), M3 (n = 8), M4 (n = 16), M5 (n = 3), and AML nonspecified (n = 7); and several samples of normal bone marrow, thymus, and spleen, as well as normal CD34+ HSCs/HPCs.
Normal bone marrow, spleen, and thymus showed no detectable SALL4 protein expression, and CD34+ HSCs/HPCs exhibited positive but weaker SALL4 protein staining; however, much stronger SALL4 expression was detected in the nuclei of leukemic cells (Figure 2Bvi). All 81 AML samples showed aberrant SALL4 expression, which was consistent with SALL4 mRNA expression levels demonstrated by real-time RT-PCR (Figure 2A). The strongest staining was seen in AML-M1 and -M2. Our data suggested that SALL4 was present in CD34+ HSCs/HPCs and down-regulated in mature granulocytes and lymphocytes. As a result, the constitutive expression of SALL4 in leukemia may have prevented the leukemic blasts from differentiating and/or gaining properties that were normally seen in HSCs, probably by interacting with additional mutations since leukemogenesis is a multistep pathologic process.
Generation of transgenic mice constitutively expressing full-length human SALL4B
To directly test whether constitutive expression of SALL4 is sufficient to induce AML, we generated a SALL4 transgenic mouse model. The CMV promoter was fused to cDNA that encoded the 617 amino acids of human SALL4B (Figure 3A), which was chosen because it was expressed in every tissue previously examined (Figure 1Dii). The CMV promoter was previously used to ectopically express human genes in most murine organs. RT-PCR amplification was performed to examine the expression of wild-type (WT), full-length SALL4B in the transgenic mice. A SALL4B transcript was detected in a variety of tissues from the transgenic mice, including brain, kidney, liver, spleen, lymph nodes, peripheral blood, and c-kit-positive population in the bone marrow (Figure 3B). Abnormal gaits and associated hydrocephalus 3 weeks after birth were observed in 20% of the transgenic mice from multiple lines; 60% had polycystic kidneys.
SALL4 expression in human primary AML and myeloid leukemia cell lines. (A) SALL4 mRNA expression in AML. Real-time PCR quantification of SALL4A and SALL4B normalized to GAPDH showed that both SALL4A and SALL4B were expressed in purified CD34+ cells, but SALL4A was rapidly down-regulated and SALL4B turned off in normal bone marrow (n = 3) and normal peripheral blood (n = 3) cells. In contrast, in 15 primary AML samples and 3 myeloid leukemia cell lines (Kasumi-1, THP-1, and KG.1), the expression of SALL4A or SALL4B, or both, failed to be down-regulated. The results were calibrated against the expression of SALL4A or SALL4B in purified CD34+ cells. Y-axis: Log scale on the relative quantification. (B) Constitutive expression of SALL4 protein in human AML (FAB M1-M5, n = 81) is demonstrated by immunohistochemical staining. No SALL4 expression was detected in normal bone marrow (i), normal thymus (ii), or normal spleen (iii). All cell nuclei remained blue. Nuclei of CD34+ HSCs/HPCs showed brown staining indicating SALL4 expression (iv); acute myeloid leukemia blasts showed similar staining (v, low power [× 4]) in microarray leukemia tissue samples. Each circle represents one leukemia sample. (vi) High-power (× 400) view of one leukemia sample shown in panel vii. The red arrows indicate positive nuclear staining.
SALL4 expression in human primary AML and myeloid leukemia cell lines. (A) SALL4 mRNA expression in AML. Real-time PCR quantification of SALL4A and SALL4B normalized to GAPDH showed that both SALL4A and SALL4B were expressed in purified CD34+ cells, but SALL4A was rapidly down-regulated and SALL4B turned off in normal bone marrow (n = 3) and normal peripheral blood (n = 3) cells. In contrast, in 15 primary AML samples and 3 myeloid leukemia cell lines (Kasumi-1, THP-1, and KG.1), the expression of SALL4A or SALL4B, or both, failed to be down-regulated. The results were calibrated against the expression of SALL4A or SALL4B in purified CD34+ cells. Y-axis: Log scale on the relative quantification. (B) Constitutive expression of SALL4 protein in human AML (FAB M1-M5, n = 81) is demonstrated by immunohistochemical staining. No SALL4 expression was detected in normal bone marrow (i), normal thymus (ii), or normal spleen (iii). All cell nuclei remained blue. Nuclei of CD34+ HSCs/HPCs showed brown staining indicating SALL4 expression (iv); acute myeloid leukemia blasts showed similar staining (v, low power [× 4]) in microarray leukemia tissue samples. Each circle represents one leukemia sample. (vi) High-power (× 400) view of one leukemia sample shown in panel vii. The red arrows indicate positive nuclear staining.
MDS-like features and AML in SALL4B transgenic mice
Monitoring of hematologic abnormalities in a cohort of 16 transgenic mice from all 6 lines revealed that all mice had apparent MDS-like features at ages 6 to 8 months. Increased number of immature blasts and many atypical and dysplastic white cells, including hypersegmented neutrophils and pseudo-Pelger-Huet-like cells, were seen on peripheral blood smears (Figure 4A). Nucleated red blood cells and giant platelets were also present, as well as erythroid and megakaryocyte dysplastic features, such as binucleate erythroid precursors and hypolobulated megakaryocytes.
Eight (50%) of these 16 mice eventually progressed to acute leukemia (Table 1). Leukemic infiltration of many organs, including lungs, kidneys, liver, spleen, and lymph nodes, emphasized the aggressiveness of the disease (Figure 4B). Leukemia blast cells were CD34+, and considered to be myeloid in origin because they were positive for c-kit, Gr-1, Mac-1, and myeloperoxidase; they were negative for B-cell (B220 and CD19), T-cell (CD4, CD8, CD3, and CD5), megakaryocytic (CD41), and erythroid (Ter119) markers (Figure 4C).
Generation of SALL4B transgenic mice. CMV/SALL4B transgenic construct and PCR analysis of transgenic line 507. (A) Schematic diagram of transgenic construct. The approximately 1.8-kb cDNA of SALL4B was subcloned into a pCEP4 vector, and the CMV/SALL4 construct was excited by digestion with SalI. (B) Tissue distribution of SALL4B in transgenic mice. The location of primers used for RT-PCR amplification is indicated by arrows in panel A. A primer specific for human SALL4B at the C-terminus was used as a 5′ primer, in combination with SV40-noncoding sequence-specific primers for RT-PCR of various tissues.
Generation of SALL4B transgenic mice. CMV/SALL4B transgenic construct and PCR analysis of transgenic line 507. (A) Schematic diagram of transgenic construct. The approximately 1.8-kb cDNA of SALL4B was subcloned into a pCEP4 vector, and the CMV/SALL4 construct was excited by digestion with SalI. (B) Tissue distribution of SALL4B in transgenic mice. The location of primers used for RT-PCR amplification is indicated by arrows in panel A. A primer specific for human SALL4B at the C-terminus was used as a 5′ primer, in combination with SV40-noncoding sequence-specific primers for RT-PCR of various tissues.
SALL4B-induced AML was transplantable
Aggressive fatal AML with onset at approximately 6 weeks developed in immunodeficient NOD/SCID mice after serial transplantation of SALL4B-induced AML cells by subcutaneous injection. The transplanted disease was positive for c-Kit and characterized by dissemination to multiple organs, with marked splenomegaly and hepatomegaly (Figure 4D). SALL4B expression was detectable in transplanted leukemic cells (data not shown).
SALL4B transgenic mice have an MDS-like/AML phenotype. (A) MDS-like changes in SALL4B transgenic mice. Giemsa staining of peripheral blood from normal, age-matched WT littermates showed normal neutrophils (i), and normal red blood cells and platelets (ii, black arrow). In transgenic mice, neutrophils were hypersegmented (v), and pseudo-Pelger-Huet-like atypical white cells were present (vi-viii), together with increased numbers of immature cells (ix-xi). Nucleate red cells (xii, red arrow), giant platelets (xiii, red arrow), and polychromasia (xiv) were also observed in the transgenic mice. A binucleate dysplastic erythrocyte (xv, red arrow) and a dysplastic megakaryocyte with a hypolobulated nucleus (xvi, red arrow) were found in the cytospin from transgenic mouse bone marrow. An erythroid precursor (iii) and a megakaryocyte (iv) from WT control animals are shown for comparison. (B) AML (AML is defined as blast count more than 20% in peripheral blood and/or bone marrow with multiple organ involvements) observed in SALL4B transgenic mice (mouse 25). Blasts were present in the peripheral blood (i, × 600), bone marrow biopsy specimen (ii, × 100; iii, × 400), bone marrow smear (iv, × 600), liver (v, × 100), lymph node (vi, × 400), and spleen (vii-viii, gross view; ix, × 100; and the inset, × 400). (C) Flow cytometric analysis of leukemia in SALL4B transgenic mice. Leukemia cells from bone marrow, spleen, and lymph nodes were positive for CD45 and myeloid markers such as c-kit, Gr-1, and Mac-1, and negative for B cells (B220), T cells (CD3), and erythrocytes (Ter119). Numbers in quadrants indicate the percentage of total cells. (D) Serial transplantation of SALL4B-induced AML to NOD/SCID mice. Gross picture (i-ii) and histology (iii-vi, × 200) on splenomegaly (i [black arrow], iii), hepatomegaly (i [double black arrows], iv), lymph node enlargement (ii [black arrow], v), and pale kidney (ii [double black arrows], vi) caused by leukemia infiltration in a NOD/SCID mouse 6 weeks after leukemia transplantation.
SALL4B transgenic mice have an MDS-like/AML phenotype. (A) MDS-like changes in SALL4B transgenic mice. Giemsa staining of peripheral blood from normal, age-matched WT littermates showed normal neutrophils (i), and normal red blood cells and platelets (ii, black arrow). In transgenic mice, neutrophils were hypersegmented (v), and pseudo-Pelger-Huet-like atypical white cells were present (vi-viii), together with increased numbers of immature cells (ix-xi). Nucleate red cells (xii, red arrow), giant platelets (xiii, red arrow), and polychromasia (xiv) were also observed in the transgenic mice. A binucleate dysplastic erythrocyte (xv, red arrow) and a dysplastic megakaryocyte with a hypolobulated nucleus (xvi, red arrow) were found in the cytospin from transgenic mouse bone marrow. An erythroid precursor (iii) and a megakaryocyte (iv) from WT control animals are shown for comparison. (B) AML (AML is defined as blast count more than 20% in peripheral blood and/or bone marrow with multiple organ involvements) observed in SALL4B transgenic mice (mouse 25). Blasts were present in the peripheral blood (i, × 600), bone marrow biopsy specimen (ii, × 100; iii, × 400), bone marrow smear (iv, × 600), liver (v, × 100), lymph node (vi, × 400), and spleen (vii-viii, gross view; ix, × 100; and the inset, × 400). (C) Flow cytometric analysis of leukemia in SALL4B transgenic mice. Leukemia cells from bone marrow, spleen, and lymph nodes were positive for CD45 and myeloid markers such as c-kit, Gr-1, and Mac-1, and negative for B cells (B220), T cells (CD3), and erythrocytes (Ter119). Numbers in quadrants indicate the percentage of total cells. (D) Serial transplantation of SALL4B-induced AML to NOD/SCID mice. Gross picture (i-ii) and histology (iii-vi, × 200) on splenomegaly (i [black arrow], iii), hepatomegaly (i [double black arrows], iv), lymph node enlargement (ii [black arrow], v), and pale kidney (ii [double black arrows], vi) caused by leukemia infiltration in a NOD/SCID mouse 6 weeks after leukemia transplantation.
Ineffective hematopoiesis and excessive apoptosis in SALL4B transgenic mice
Investigation of hematologic abnormalities in younger SALL4B transgenic mice (2-6 months old) revealed that their peripheral blood showed minimal myelodysplastic features but statistically significant leukopenia and neutropenia, as well as mild anemia (Table 2). To determine whether the cause of cytopenia in these transgenic mice was related to ineffective hematopoiesis, we studied their bone marrow. Bone marrow samples showed increased cellularity and an increased myeloid population (Figure 5A), compared with those of WT controls (Gr-1/Mac-1 double-positive population in SALL4B transgenic mice: 67% ± 16% [n = 10] vs WT: 55.3% ± 4% [n = 11]; P = .048).
As excessive apoptosis plays a central role in ineffective hematopoiesis in human MDS, we next examined apoptosis in SALL4 transgenic mice in vivo and in vitro. Increased apoptosis was observed in SALL4B transgenic mice on both primary bone marrow (annexin V-positive, PI-negative population in transgenic mice: 11% ± 4.48% [n = 10] vs WT: 6.15% ± 4.98% [n = 7]; P = .03) and day-7 CFUs (annexin V-positive, PI-negative population in transgenic mice: 20.1% ± 6% [n = 10] vs WT: 10.9% ± 4% [n = 7]; P = .002) (Figure 5A-B). These findings may account for the fact that despite an increased myeloid population in bone marrow, these transgenic mice had statistically significant low neutrophil counts in the peripheral blood, secondary to an ongoing ineffective myelopoiesis in their bone marrow. An increased population of immature cells was also noted in SALL4B transgenic mice on both primary bone marrow (c-kit-positive population in SALL4B transgenic mice: 10.2% ± 1.3%, [n = 14] vs WT: 6.5% ± 2.5% [n = 10]; P = .008) (Figure 5A) and day-7 CFUs (CD34+ population in SALL4B transgenic mice: 11% ± 2.2% [n = 8] vs WT: 6.3% ± 2.4% [n = 7]; P = .002) (Figure 5B). Similar numbers of total colonies were observed in SALL4B transgenic mice and WT controls (total colonies in SALL4B mice: 42 ± 29.5 [n = 10] vs WT: 39 ± 13.5 [n = 6]; P = .23). Statistically significant increased myeloid (CFU-GM in SALL4B transgenic mice: 53.6 ± 10.3 [n = 13] vs WT: 38.1 ± 3.1 [n = 8]; P = .002) and decreased erythroid (BFU-E in SALL4B transgenic mice: 7.8 ± 3.8 [n = 13] vs WT: 14.1 ± 2.7 [n = 8]; P = .001) colony populations (Figure 5Cii), however, were found in SALL4B transgenic mouse CFUs compared with those of WT controls, as has been reported in human MDS patients and other MDS mouse models.31-35 These observations suggest that the defect in SALL4B transgenic mice lies at the stem cell/progenitor level affecting hematopoietic differentiation.
Binding of SALL4A and SALL4B to β-catenin in vitro
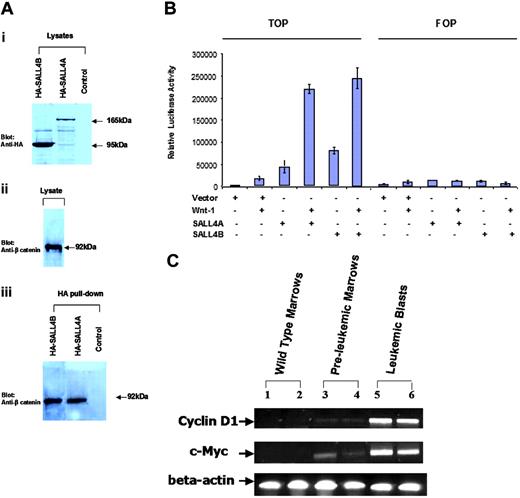
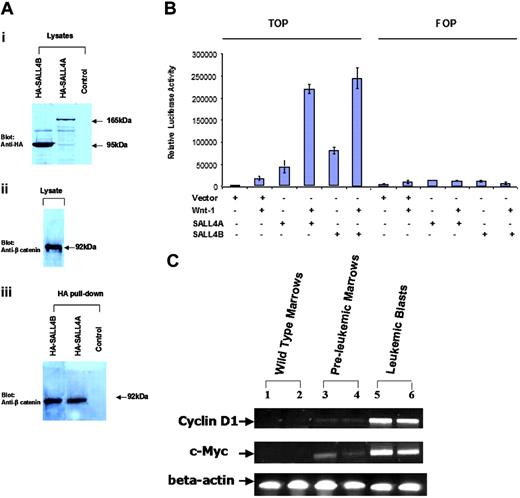
We next explored the potential signaling pathway that SALL4 may affect in leukemogenesis. In Drosophila, Wnt signaling controls spalt (sal) expression during tracheal morphogenesis. SALL1, another member of the SALL gene family, can interact with β-catenin. The high-affinity site for this interaction is located at the C-terminal double zinc finger domain. This region of SALL1 was found to be almost exactly identical to that of SALL4 (data not shown). This finding prompted us to investigate whether SALL4 was also able to bind β-catenin. We generated expression constructs of SALL4A and SALL4B tagged with hemagglutinin (HA). As shown in Figure 6A, endogenous β-catenin was pulled down by HA-SALL4A and HA-SALL4B, but not by HA alone.
Activation of the Wnt/β-catenin signaling pathway by both SALL4A and SALL4B
To investigate the functional effect of the interaction of the SALL4 isoforms with β-catenin, we used a luciferase reporter (TOPflash; Upstate USA) containing multiple copies of Wnt-responsive elements to determine the potential of SALL4A and SALL4B to activate the canonical Wnt signaling pathway. The FOPflash reporter plasmid with mutated Wnt-response elements was used as a negative control. This reporter construct has been shown to be efficiently stimulated by Wnt1 in a variety of cell lines.36-42 TOPflash or FOPflash reporter plasmid was transiently transfected in the HEK-293 cell line, in which both Wnt and its Wnt/β-catenin signal pathways were present. TOPflash reporter plasmid was also cotransfected with SALL4A or SALL4B. Significant activation of the Wnt/β-catenin signaling pathway by both SALL4A and SALL4B was indicated by increased luciferase activity in the TOPflash but not in the FOPflash reporter (Figure 6B).
Up-regulation of Wnt/β-catenin downstream target genes in SALL4B transgenic mice
We then studied the effect of overexpression of SALL4B on β-catenin/Tcf-dependent gene expression in SALL4B transgenic mice. Many genes are regulated by Wnt/β-catenin signaling pathway. We chose to study c-Myc and Cyclin D1 since both genes are transactivated by β-catenin/Tcf complex and involved in various human cancers including leukemia and lymphoma.43 We found that in contrast to the wild-type controls, the mRNA expression of both genes was significantly up-regulated in preleukemia bone marrows and leukemic blasts from SALL4B transgenic mice (Figure 6C). These findings strengthen our hypothesis that SALL4B contributes to leukemogenesis, probably through activation of the Wnt/β-catenin signaling pathway.
Ineffective hematopoiesis in SALL4B transgenic mice. (A) Comparison of bone marrow of SALL4B transgenic (i) and control mice (ii). SALL4B transgenic mouse bone marrow showed increased cellularity, myeloid population (Gr-1/Mac-1 double positive), immature population (c-kit positive), and apoptosis (annexin V positive, PI negative), compared with control WT mice. (B) Increased number of immature cells and apoptosis in CFUs from SALL4B transgenic mice. On day 7 of culture, a greater number of immature cells (ii-iv, red arrows) and apoptotic cells (ii-iv, double red arrows) were observed in transgenic mouse CFUs than in control CFUs (i). Consistent with this morphologic observation, there was increased apoptosis (annexin V positive, PI negative; v) and more CD34+ immature cells (vi). (C) Comparison of bone marrow CFUs of SALL4B transgenic and control mice. Percentage of different types of colonies found in CFU assays of SALL4B transgenic and control mice (i). CFUs from SALL4B transgenic mice compared with control mice showed a statistically significant increase in CFU-GM (ii) (transgenic: 53.6 ± 10.3 [n = 13] vs WT: 38.1 ± 3.1 [n = 8]; P = .002) and decrease in BFU-E (transgenic: 7.8 ± 3.8 [n = 13] vs WT: 14.1 ± 2.7 [n = 8]; P = .001).
Ineffective hematopoiesis in SALL4B transgenic mice. (A) Comparison of bone marrow of SALL4B transgenic (i) and control mice (ii). SALL4B transgenic mouse bone marrow showed increased cellularity, myeloid population (Gr-1/Mac-1 double positive), immature population (c-kit positive), and apoptosis (annexin V positive, PI negative), compared with control WT mice. (B) Increased number of immature cells and apoptosis in CFUs from SALL4B transgenic mice. On day 7 of culture, a greater number of immature cells (ii-iv, red arrows) and apoptotic cells (ii-iv, double red arrows) were observed in transgenic mouse CFUs than in control CFUs (i). Consistent with this morphologic observation, there was increased apoptosis (annexin V positive, PI negative; v) and more CD34+ immature cells (vi). (C) Comparison of bone marrow CFUs of SALL4B transgenic and control mice. Percentage of different types of colonies found in CFU assays of SALL4B transgenic and control mice (i). CFUs from SALL4B transgenic mice compared with control mice showed a statistically significant increase in CFU-GM (ii) (transgenic: 53.6 ± 10.3 [n = 13] vs WT: 38.1 ± 3.1 [n = 8]; P = .002) and decrease in BFU-E (transgenic: 7.8 ± 3.8 [n = 13] vs WT: 14.1 ± 2.7 [n = 8]; P = .001).
Similar expression patterns of β-catenin and SALL4 at different phases of CML
Dysregulated Wnt/β-catenin signaling is known to be involved in the development of LSCs. The best evidence for β-catenin's involvement in LSC self-renewal comes from the study of CML blast transformation. Jamieson et al11 demonstrated that Wnt signaling was activated in the blast phase of CML but not the chronic phase, concluding that dysregulated Wnt signaling, such as activation of β-catenin, could confer the property of self-renewal on the GMPs of CML and lead to their blastic transformation. Given the potential interaction between SALL4 and β-catenin and spalt's position as a downstream target of Wnt signaling in Drosophila, we examined SALL4 protein expression in CMLs in different phases. SALL4 expression was present in blast-phase CML (n = 12, 75%) but not the chronic phase (n = 11 100%). In the accelerated phase (n = 6, 10%), in which blast counts are increased, immature blasts expressing SALL4 were observed with a background of nonstaining more mature myeloid cells (data not shown).
Discussion
Homeobox and homeotic genes play important roles in normal development. Some homeobox genes, such as Hox and Pax, also function as oncogenes or as tumor suppressors in tumorigenesis or leukemogenesis. The important role of SALL4, a homeotic gene and a transcriptional factor, in human development was recognized because heterozygous SALL4 mutations lead to Duane-radial ray syndrome. SALL4′s oncogenic role in leukemogenesis is described here for the first time.
SALL4 and the Wnt/β-catenin signaling pathway. (A) Both SALL4A and SALL4B can interact with β-catenin. Nuclear extracts (lysates) prepared from Cos-7 cells were transiently transfected with HA-SALL4A or HA-SALL4B. (i) Anti-HA antibody recognized both SALL4A (165 kDa) and SALL4B (95 kDa). (ii) β-Catenin was detected in the lysates. (iii) Immunoprecipitation was performed with the use of an HA affinity resin and detected with an anti-β-catenin antibody. β-Catenin was readily detected in both HA-SALL4A and HA-SALL4B pull-downs. Untransfected cells subjected to the same immunoprecipitation condition as the transfected cells were used as a control. (B) Activation of the Wnt/β-catenin signaling pathway by both SALL4A and SALL4B. HEK-293 cells were transfected with 1.0 μg of either mock alone, or SALL4A or SALL4B plasmid, with or without Wnt1 (including Wnt1, and its coactivators: LRP6, MESD, and F25), and TOPflash (TOP) or FOPflash (FOP) reporter plasmid (Upstate USA, Chicago, IL). After 24 hours, luciferase activity was measured. SALL4A or SALL4B alone showed more potent activation of Wnt signaling pathway when compared with the positive control Wnt1. In addition, both SALL4 isoforms demonstrated a significantly synergistic activation of the Wnt signaling pathway with Wnt1. Data represent mean ± SD of 3 independent experiments. (C) Up-regulation of c-Myc and Cyclin D1 expression in SALL4B transgenic mice. RT-PCR analysis was performed on total bone marrow cells from 2 wild-type control mice (lanes 1-2), 2 preleukemic transgenic mice (lanes 3-4), and leukemic bone marrow cells from 2 leukemic transgenic SALL4B mice (lanes 5-6). Both c-Myc and Cyclin D1 expression were significantly up-regulated in SALL4B transgenic mice at both preleukemia MDS and leukemic stages. Beta actin was used as an internal standard.
SALL4 and the Wnt/β-catenin signaling pathway. (A) Both SALL4A and SALL4B can interact with β-catenin. Nuclear extracts (lysates) prepared from Cos-7 cells were transiently transfected with HA-SALL4A or HA-SALL4B. (i) Anti-HA antibody recognized both SALL4A (165 kDa) and SALL4B (95 kDa). (ii) β-Catenin was detected in the lysates. (iii) Immunoprecipitation was performed with the use of an HA affinity resin and detected with an anti-β-catenin antibody. β-Catenin was readily detected in both HA-SALL4A and HA-SALL4B pull-downs. Untransfected cells subjected to the same immunoprecipitation condition as the transfected cells were used as a control. (B) Activation of the Wnt/β-catenin signaling pathway by both SALL4A and SALL4B. HEK-293 cells were transfected with 1.0 μg of either mock alone, or SALL4A or SALL4B plasmid, with or without Wnt1 (including Wnt1, and its coactivators: LRP6, MESD, and F25), and TOPflash (TOP) or FOPflash (FOP) reporter plasmid (Upstate USA, Chicago, IL). After 24 hours, luciferase activity was measured. SALL4A or SALL4B alone showed more potent activation of Wnt signaling pathway when compared with the positive control Wnt1. In addition, both SALL4 isoforms demonstrated a significantly synergistic activation of the Wnt signaling pathway with Wnt1. Data represent mean ± SD of 3 independent experiments. (C) Up-regulation of c-Myc and Cyclin D1 expression in SALL4B transgenic mice. RT-PCR analysis was performed on total bone marrow cells from 2 wild-type control mice (lanes 1-2), 2 preleukemic transgenic mice (lanes 3-4), and leukemic bone marrow cells from 2 leukemic transgenic SALL4B mice (lanes 5-6). Both c-Myc and Cyclin D1 expression were significantly up-regulated in SALL4B transgenic mice at both preleukemia MDS and leukemic stages. Beta actin was used as an internal standard.
We identified the 2 SALL4 isoforms, SALL4A and SALL4B. During normal hematopoiesis, SALL4 isoforms are expressed in the CD34+ HSC/HPC population and rapidly turned off (SALL4B) or down-regulated (SALL4A) in normal human bone marrow and peripheral blood. In contrast, SALL4 was constitutively expressed in all AML samples (n = 81) that we examined, and failed to turn off in human primary AML and myeloid leukemia cell lines. To directly test the leukemogenic potential of constitutive expression of SALL4 in vivo, we generated SALL4B transgenic mice. The transgenic mice exhibited dysregulated hematopoiesis, much like that of human MDS, and AML that was transplantable. The MDS-like features in these SALL4B transgenic mice apparently did not require cooperating mutations and were observed as early as 2 months of age. The ineffective hematopoiesis observed in these mice was characterized, as it is in human MDS, by hypercellular bone marrow and paradoxic peripheral blood cytopenias (neutropenia and anemia) and dysplasia, which were probably secondary to the increased apoptosis noted in the bone marrow. The reason for the late onset of leukemia development in these transgenic mice may be the accumulation of additional genetic damage during the 8 or more months of replicative stress. Late onset of disease may also be a consequence of SALL4-induced genomic instability.
Our investigation of the potential mechanism of SALL4 involvement in leukemogenesis demonstrated that both SALL4A and SALL4B interacted with β-catenin, an essential component of the Wnt signaling pathway involving self-renewal of HSCs. In addition, both were able to activate the Wnt/β-catenin pathway in a reporter gene assay, consistent with SALL family function in Drosophila20,44 and humans.30 Furthermore, similar to the situation with β-catenin, SALL4 expression in CML varied at different phases of the disease: SALL4 expression was absent in the chronic phase, became detectable in the accelerated phase only in immature blasts, and was strongly positive in the blast phase. The down-stream target genes of Wnt/β-catenin, such as c-Myc and Cyclin D1, were up-regulated in SALL4B transgenic mice at both preleukemic and leukemic stages. On the basis of these studies, we propose a working hypothesis: constitutive expression of SALL4 in AML may enable leukemic blasts to gain stem cell properties, such as self-renewal and/or lack of differentiation, and thus become LSCs.
In summary, the novel oncogene SALL4 plays an important role in normal hematopoiesis and leukemogenesis. SALL4B transgenic mice exhibit MDS-like phenotype with subsequently AML transformation that is transplantable. Few animal models are currently available for the study of human MDS. The SALL4B transgenic mice that we generated provide a suitable animal model for understanding and treating human MDS and its subsequent transformation to AML. The interaction between SALL4 and the Wnt/β-catenin signaling pathway not only provides a plausible mechanism for SALL4 involvement in leukemogenesis but also advances our understanding of the activation of the Wnt/β-catenin signaling pathway in CML blastic transformation.
Prepublished online as Blood First Edition Paper, June 8, 2006; DOI 10.1182/blood-2006-02-001594.
Supported in part by the National Institutes of Health under grants K08 CA097185 and P20 RR016464 (Y.M.) and K08 DK063220, and by the National Blood Foundation (L.C.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We gratefully acknowledge Dr Stephen Blacklow for making available Wnt plasmid constructs, Nikki Kong for immunohistochemical staining, and Dr Daniel G. Tenen and Dr David Ward for reviewing the paper.


![Figure 2. SALL4 expression in human primary AML and myeloid leukemia cell lines. (A) SALL4 mRNA expression in AML. Real-time PCR quantification of SALL4A and SALL4B normalized to GAPDH showed that both SALL4A and SALL4B were expressed in purified CD34+ cells, but SALL4A was rapidly down-regulated and SALL4B turned off in normal bone marrow (n = 3) and normal peripheral blood (n = 3) cells. In contrast, in 15 primary AML samples and 3 myeloid leukemia cell lines (Kasumi-1, THP-1, and KG.1), the expression of SALL4A or SALL4B, or both, failed to be down-regulated. The results were calibrated against the expression of SALL4A or SALL4B in purified CD34+ cells. Y-axis: Log scale on the relative quantification. (B) Constitutive expression of SALL4 protein in human AML (FAB M1-M5, n = 81) is demonstrated by immunohistochemical staining. No SALL4 expression was detected in normal bone marrow (i), normal thymus (ii), or normal spleen (iii). All cell nuclei remained blue. Nuclei of CD34+ HSCs/HPCs showed brown staining indicating SALL4 expression (iv); acute myeloid leukemia blasts showed similar staining (v, low power [× 4]) in microarray leukemia tissue samples. Each circle represents one leukemia sample. (vi) High-power (× 400) view of one leukemia sample shown in panel vii. The red arrows indicate positive nuclear staining.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/8/10.1182_blood-2006-02-001594/4/m_zh80200602550002.jpeg?Expires=1767808501&Signature=m07PV3a4Wpl-7efbhbHdS1U~T0bog0Go1v84RrInpF~FhuM-gUHFU8SIpBUf9ZQlWnkWEcEvxBolbfvEhBF2efGdJgh~L3z-2pRrwc39EKveJ7yNoajfL9d2BXusl0~W-L-o3HIaq4xWZLPGbd9z6gHd-jixPIo~YJH5IG2yfzaWo9osfCls9-wTIe~qxBihBU4H2pzGmQ~vodO2GZoi3meYrJCQfKbAu6szT8-7lsNqALQtHJkzRLebNdzqNq9ZKC4rR6qaNawMGgCEMS8br5o-RGzj~lVGrieKlCaIYdEMwPrlVWENESxUH~0hUrHEvsMwSt0kggCeKrkfMpVm~g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 4. SALL4B transgenic mice have an MDS-like/AML phenotype. (A) MDS-like changes in SALL4B transgenic mice. Giemsa staining of peripheral blood from normal, age-matched WT littermates showed normal neutrophils (i), and normal red blood cells and platelets (ii, black arrow). In transgenic mice, neutrophils were hypersegmented (v), and pseudo-Pelger-Huet-like atypical white cells were present (vi-viii), together with increased numbers of immature cells (ix-xi). Nucleate red cells (xii, red arrow), giant platelets (xiii, red arrow), and polychromasia (xiv) were also observed in the transgenic mice. A binucleate dysplastic erythrocyte (xv, red arrow) and a dysplastic megakaryocyte with a hypolobulated nucleus (xvi, red arrow) were found in the cytospin from transgenic mouse bone marrow. An erythroid precursor (iii) and a megakaryocyte (iv) from WT control animals are shown for comparison. (B) AML (AML is defined as blast count more than 20% in peripheral blood and/or bone marrow with multiple organ involvements) observed in SALL4B transgenic mice (mouse 25). Blasts were present in the peripheral blood (i, × 600), bone marrow biopsy specimen (ii, × 100; iii, × 400), bone marrow smear (iv, × 600), liver (v, × 100), lymph node (vi, × 400), and spleen (vii-viii, gross view; ix, × 100; and the inset, × 400). (C) Flow cytometric analysis of leukemia in SALL4B transgenic mice. Leukemia cells from bone marrow, spleen, and lymph nodes were positive for CD45 and myeloid markers such as c-kit, Gr-1, and Mac-1, and negative for B cells (B220), T cells (CD3), and erythrocytes (Ter119). Numbers in quadrants indicate the percentage of total cells. (D) Serial transplantation of SALL4B-induced AML to NOD/SCID mice. Gross picture (i-ii) and histology (iii-vi, × 200) on splenomegaly (i [black arrow], iii), hepatomegaly (i [double black arrows], iv), lymph node enlargement (ii [black arrow], v), and pale kidney (ii [double black arrows], vi) caused by leukemia infiltration in a NOD/SCID mouse 6 weeks after leukemia transplantation.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/8/10.1182_blood-2006-02-001594/4/m_zh80200602550004.jpeg?Expires=1767808501&Signature=FuQQ3c~Gt8E7IMVplPXs4-1Bcw3RecbWDX1zr2I3QzwMdiE4cAc43BjjaAl9b-dk2wNp3PYMro2MYvu~CmSxvG7Tttc9xUoxV3W2QtxUtrciO1gqhd563t0JE5TeAXwMsC3NgNRIEKIqraAViFHiK1XP7eyl5l8KrbLc~uLpF8NkKgmAMG1NzUNoM-a4TUpMA9DEsA~gmrnBW-QwsnQqD~7nPORCpuUmCsZOhxbud1lvA1aHFz78d5gBEEoByiafQ~1wDPUBLjfEsvb~oW2bXFQ6LwpF3JeQlCZghyBgIUbtCqHTdw5O~pXNLHSqg0sO8RS5~2qQo3wxUUqP5Iikfw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Ineffective hematopoiesis in SALL4B transgenic mice. (A) Comparison of bone marrow of SALL4B transgenic (i) and control mice (ii). SALL4B transgenic mouse bone marrow showed increased cellularity, myeloid population (Gr-1/Mac-1 double positive), immature population (c-kit positive), and apoptosis (annexin V positive, PI negative), compared with control WT mice. (B) Increased number of immature cells and apoptosis in CFUs from SALL4B transgenic mice. On day 7 of culture, a greater number of immature cells (ii-iv, red arrows) and apoptotic cells (ii-iv, double red arrows) were observed in transgenic mouse CFUs than in control CFUs (i). Consistent with this morphologic observation, there was increased apoptosis (annexin V positive, PI negative; v) and more CD34+ immature cells (vi). (C) Comparison of bone marrow CFUs of SALL4B transgenic and control mice. Percentage of different types of colonies found in CFU assays of SALL4B transgenic and control mice (i). CFUs from SALL4B transgenic mice compared with control mice showed a statistically significant increase in CFU-GM (ii) (transgenic: 53.6 ± 10.3 [n = 13] vs WT: 38.1 ± 3.1 [n = 8]; P = .002) and decrease in BFU-E (transgenic: 7.8 ± 3.8 [n = 13] vs WT: 14.1 ± 2.7 [n = 8]; P = .001).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/8/10.1182_blood-2006-02-001594/4/m_zh80200602550005.jpeg?Expires=1767808501&Signature=TeddbokKMspy1LzDHKy-Xegzwhfo92UKXVUmY7CPIHepmye9j8X-oR2BNPKpV3icfOz8Z3BldWBRv0whjBPMaql~6n~~ZXmrpw58m8kavsHkisl41xn6AhP-lOK6xy7rsk6o6uhljovHr2OsUSzVc7-66QA05hAgqdtCOaGLExmMhjx3yT4ttrK4hwnF1swRQvV3RJv8ZAsmhir6af2HXBvTWKe3vMxIDSJdyZgsOrpUhJkJ5Y3MFd-VQ7PbVcINSd1NHW7LX1I5BXBNSx8Gz1fJzfzpPgrYo30XrXFTnguffgqRxcpLLkvxC6aNbX-uBSG1-6G597V3XRpqLIl7Cw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 2. SALL4 expression in human primary AML and myeloid leukemia cell lines. (A) SALL4 mRNA expression in AML. Real-time PCR quantification of SALL4A and SALL4B normalized to GAPDH showed that both SALL4A and SALL4B were expressed in purified CD34+ cells, but SALL4A was rapidly down-regulated and SALL4B turned off in normal bone marrow (n = 3) and normal peripheral blood (n = 3) cells. In contrast, in 15 primary AML samples and 3 myeloid leukemia cell lines (Kasumi-1, THP-1, and KG.1), the expression of SALL4A or SALL4B, or both, failed to be down-regulated. The results were calibrated against the expression of SALL4A or SALL4B in purified CD34+ cells. Y-axis: Log scale on the relative quantification. (B) Constitutive expression of SALL4 protein in human AML (FAB M1-M5, n = 81) is demonstrated by immunohistochemical staining. No SALL4 expression was detected in normal bone marrow (i), normal thymus (ii), or normal spleen (iii). All cell nuclei remained blue. Nuclei of CD34+ HSCs/HPCs showed brown staining indicating SALL4 expression (iv); acute myeloid leukemia blasts showed similar staining (v, low power [× 4]) in microarray leukemia tissue samples. Each circle represents one leukemia sample. (vi) High-power (× 400) view of one leukemia sample shown in panel vii. The red arrows indicate positive nuclear staining.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/8/10.1182_blood-2006-02-001594/4/m_zh80200602550002.jpeg?Expires=1767808568&Signature=uqtXuAXRoaXf-qFP7glwUgBpDF-lI9IhcFYAbY2gRUTtRXSTQuY8Nd6WdmID67bS4nEbPwiwOG8XElfXnCo3~PPKl0D0TscvL6rFix6GuJKTSU5RY9jdSqqZMX~AP4XDt1RRPxDlbDDnVcrAdEg1JQQInRddB8-hOqR8gVqWy6AXyS2bL1Jl-s~SMkSW~UdfFAM0WpSVM7gaGKUQcX2RX1SWFYo0dmnNtFKb-Yxmtwho5-OVFe6T89Z5gVV1IBNclt3a8rIy7r77xJfHUdqcAGi4VoikQiQmB5aIpXD2AqBHAB4-n5wjldC0vKcbvdKrx3cReT-bTxzw3oxfoTko2A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 4. SALL4B transgenic mice have an MDS-like/AML phenotype. (A) MDS-like changes in SALL4B transgenic mice. Giemsa staining of peripheral blood from normal, age-matched WT littermates showed normal neutrophils (i), and normal red blood cells and platelets (ii, black arrow). In transgenic mice, neutrophils were hypersegmented (v), and pseudo-Pelger-Huet-like atypical white cells were present (vi-viii), together with increased numbers of immature cells (ix-xi). Nucleate red cells (xii, red arrow), giant platelets (xiii, red arrow), and polychromasia (xiv) were also observed in the transgenic mice. A binucleate dysplastic erythrocyte (xv, red arrow) and a dysplastic megakaryocyte with a hypolobulated nucleus (xvi, red arrow) were found in the cytospin from transgenic mouse bone marrow. An erythroid precursor (iii) and a megakaryocyte (iv) from WT control animals are shown for comparison. (B) AML (AML is defined as blast count more than 20% in peripheral blood and/or bone marrow with multiple organ involvements) observed in SALL4B transgenic mice (mouse 25). Blasts were present in the peripheral blood (i, × 600), bone marrow biopsy specimen (ii, × 100; iii, × 400), bone marrow smear (iv, × 600), liver (v, × 100), lymph node (vi, × 400), and spleen (vii-viii, gross view; ix, × 100; and the inset, × 400). (C) Flow cytometric analysis of leukemia in SALL4B transgenic mice. Leukemia cells from bone marrow, spleen, and lymph nodes were positive for CD45 and myeloid markers such as c-kit, Gr-1, and Mac-1, and negative for B cells (B220), T cells (CD3), and erythrocytes (Ter119). Numbers in quadrants indicate the percentage of total cells. (D) Serial transplantation of SALL4B-induced AML to NOD/SCID mice. Gross picture (i-ii) and histology (iii-vi, × 200) on splenomegaly (i [black arrow], iii), hepatomegaly (i [double black arrows], iv), lymph node enlargement (ii [black arrow], v), and pale kidney (ii [double black arrows], vi) caused by leukemia infiltration in a NOD/SCID mouse 6 weeks after leukemia transplantation.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/8/10.1182_blood-2006-02-001594/4/m_zh80200602550004.jpeg?Expires=1767808568&Signature=D-iKkJJanm3y7HPz1Wegz9yiDEYwqzX1fmFQAT1PmiLKLmPGs4o5fyF3uEPK4wQ2j0X7WyIn6~cAV6bIhEzFvvggeIECRxLs5ICem4DEXcQbEk7otXVoQ5f7TPKhIfox~QnAHaCHaFH4FJxLvMqA6g4Zoyi-cgCTSQPM21acJC2ACSKGd3ruq~xPEItyKe5lax-D1UoG5YVKmEISQqEkll14ORpQpt~GFrd51gqSBN16sEm-Q-v5SQ1WvP~YbAnR15CwqRvIov2QoWyJGy2lcHTK0uQEjxe8lTgQeh4X1x7iO7gemW5yYlChlC0zD-1O18BECVrIyhSU60evz5ybnw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Ineffective hematopoiesis in SALL4B transgenic mice. (A) Comparison of bone marrow of SALL4B transgenic (i) and control mice (ii). SALL4B transgenic mouse bone marrow showed increased cellularity, myeloid population (Gr-1/Mac-1 double positive), immature population (c-kit positive), and apoptosis (annexin V positive, PI negative), compared with control WT mice. (B) Increased number of immature cells and apoptosis in CFUs from SALL4B transgenic mice. On day 7 of culture, a greater number of immature cells (ii-iv, red arrows) and apoptotic cells (ii-iv, double red arrows) were observed in transgenic mouse CFUs than in control CFUs (i). Consistent with this morphologic observation, there was increased apoptosis (annexin V positive, PI negative; v) and more CD34+ immature cells (vi). (C) Comparison of bone marrow CFUs of SALL4B transgenic and control mice. Percentage of different types of colonies found in CFU assays of SALL4B transgenic and control mice (i). CFUs from SALL4B transgenic mice compared with control mice showed a statistically significant increase in CFU-GM (ii) (transgenic: 53.6 ± 10.3 [n = 13] vs WT: 38.1 ± 3.1 [n = 8]; P = .002) and decrease in BFU-E (transgenic: 7.8 ± 3.8 [n = 13] vs WT: 14.1 ± 2.7 [n = 8]; P = .001).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/8/10.1182_blood-2006-02-001594/4/m_zh80200602550005.jpeg?Expires=1767808568&Signature=JsP4uyXmD76CA1Y8abj~A56YCassUJQcxOKJA~ZCosbTmdHHd7EiZf27Qr5YQiwkMTlT3djfUP2s0i~Knc~mtdFpMuTvC27FRcvAh55Zkjyrv0cWFB~NwNa2R0~1OtAxY2yrWAD18NxpTPZlXIaARYSzalE74-xoZhPexlGMRpmJciIpdlinPwZD0HZufFLZrtNLdAKglrvBFTSUAxggN1EpL92jRCGgN8YtPbRcYqiBpJ8T8TQzgUulJmhMaUEcMQgKlXCD072v7mOQSGKhgV8SdfUiNu~twXwBJseXb5Jk~l8euiNQS~DVic1zesWXgg27fVhUWbaWasbr9z-hDA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)