TLX1 is a homeodomain transcription factor generally associated with a favorable outcome in T-cell acute lymphoblastic leukemia (T-ALL). However, the molecular mechanisms of TLX1 deregulation remain unclear and various transcript levels in the absence of 10q24 abnormalities have been reported. A reproducible and accurate delineation of TLX1+ T-ALL will be necessary for proper therapeutic stratification. We have studied 264 unselected T-ALLs (171 adults and 93 children) and show that T-ALLs expressing high levels of TLX1 (n = 35, 13%), defined as a real-time quantitative polymerase chain reaction (RQ-PCR) level of TLX1 greater than 1.00 ABL, form a homogeneous oncogenic group, based on their uniform stage of maturation arrest and oncogenetic and transcriptional profiles. Furthermore, TLX1-high T-ALLs harbor molecular TLX1 locus abnormalities in the majority (31/33), a proportion largely underestimated by standard karyotypic screening. T-ALLs expressing TLX1 at lower levels (n = 57, 22%) do not share these characteristics. Prognostic analysis within the adult LALA94 and GRAALL03 prospective protocols demonstrate a better event-free survival (P = .035) and a marked trend for longer overall survival (P = .059) for TLX1-high T-ALLs, while the expression of lower levels of TLX1 does not impact on prognosis. We propose that TLX1+ T-ALLs be defined as cases expressing TLX1/ABL ratios greater than 1 and/or demonstrating TLX1 rearrangement. Therapeutic modification should be considered for those patients.
Introduction
A large proportion of T-cell acute lymphoblastic leukemias (T-ALLs) show a normal (30%-40%) or failed (15%-20%) karyotype.1,2 Molecular cytogenetic approaches have allowed identification an increasing number of oncogenetic lesions in this disease.3,4 Oncogenes such as TLX1/HOX11, TLX3/HOX11L2, TAL1, and the CALM-AF10 and MLL fusion transcripts appear lineage and stage-of-maturation-arrest specific and as such are likely to represent key leukemogenic features.5,,–8 More recently, evidence of NOTCH1 mutations in about half of T-ALLs has further enhanced the biologic heterogeneity of T-ALLs.9 However, few of these known oncogenetic markers have demonstrated clear prognostic significance. Conflicting outcomes have been associated with HOX11L2, TAL1 deregulation, and NOTCH1 mutations.7,10,,,–14 TLX1 overexpression and/or translocation generally confer a better prognosis, but this association, when found, varies between series.11,12,15,,–18
Deregulated expression of the TLX1 gene, situated on chromosome 10q24, is reported in 20% to 30% of T-ALLs, but only 14% of adults and 4% to 7% of childhood T-ALLs show a 10q24 translocation when evaluated by standard karyotypic methods.15,17,19 The stringency of the association between TLX1 expression and the presence of a translocation has therefore been questioned.18 Observed levels of expression, assessed by real-time quantitative polymerase chain reaction (RQ-PCR), vary greatly among samples.11,18 The reference genes used and RQ-PCR–defined thresholds for “high-level” TLX1 expression differ among authors, which makes data comparison difficult. It has been proposed that adults with a T-ALL expressing elevated levels of TLX1 should not undergo bone marrow transplantation during first remission.17 A stringent and standardized definition of TLX1+ T-ALL is therefore important if it is to be used for therapeutic stratification.
We undertook this study to clarify the biologic and clinical significance of TLX1 levels of expression in 264 (93 pediatric and 171 adult) T-ALLs that have undergone extensive conventional and molecular cytogenetic, immunophenotypic and oncogenetic analysis.
Patients, materials, and methods
Patients and diagnostic analysis
Diagnostic peripheral blood or bone marrow samples were analyzed from the following: 264 T-ALLs, defined by expression of cytoplasmic and/or surface CD3 and CD7, and negativity for CD19 and MPO; 30 B-cell acute lymphoblastic leukemias (B-ALLs); and 21 acute myeloid leukemias (AMLs). Patients provided informed consent in accordance with the Declaration of Helsinki, and approval for these studies was obtained from the Comite Consultatif de Protection des Personnes dans la Recherche Biomedicale Lyon B (CCPPRB) institutional review board. Patients were considered adults when older than 15 years. Ninety-eight adult T-ALLs were treated within the LALA-94 multicenter trial and 33 within the GRAALL-2003 trial. Details of patient classification, DNA and RNA extraction, immunophenotype, and TCR analysis were described previously.5,20,21
cDNA synthesis was performed centrally at the Necker facility, and RNA quality assessed and normalized by quantification of ABL on an ABI PRISM 7700 or 7000 (Perkin-Elmer Applied Biosystems, Branchburg, NJ), using guidelines from the Europe Against Cancer program.22 Samples with an ABL cycle threshold (Ct) more than 32 were excluded from analysis. Each experiment included 2 nontemplate controls for contamination, and all RQ-PCRs were performed in duplicate. All primers spanned an intron and absence of genomic amplification was confirmed by RQ-PCR from peripheral blood lymphocyte (PBL) DNA. Transcript quantification was performed after normalization by the ABL housekeeping gene from the standard curves using the delta of delta Ct method. Primers and RQ-PCR probes have been previously reported.8,23
Assessment of RQ-PCR amplification efficiency of TLX1 and ABL
Logarithmic TLX1 plasmid dilutions were conducted and quantified by RQ-PCR. Efficiency slopes for TLX1 and ABL dilutions were comparable with respective slope values of − 3.53 and − 3.45. In reproducibility experiments (n = 10), the Ct of detection for a dilution corresponding to one TLX1 copy per well was inconstant and varied between 38.04 and 41.1 (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Hence, we considered that Cts greater than 38 were not quantitative and were classified as TLX1 negative.
FISH analysis
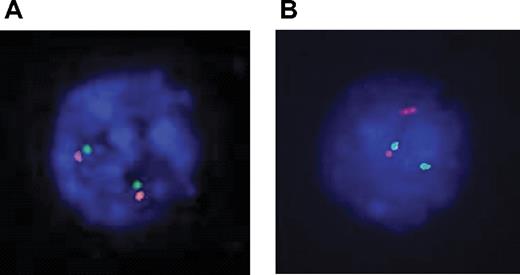
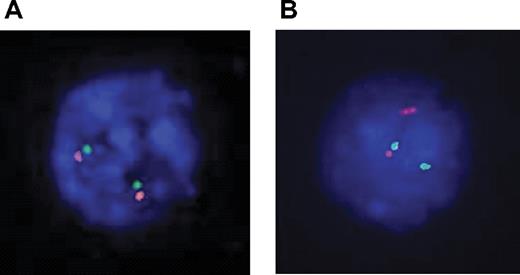
Dual-color fluorescent in situ hybridization (FISH) experiments were performed with 2 sets of overlapping probes flanking the TLX1 locus on each side of the breakpoint: 5′ RP11-179B2 and RP11-1031N22 labeled with FITC-dUTP and 3′ RP11-324L3 and RP11-119018 labeled with rhodamine-dUTP (Vysis, Downers Grove, IL; Figure S2). FISH images were read with a Leica DMRXA microscope (Leica, Solms, Germany) with 100×/1.30-0.6 objective, captured with a DDC-1300DS camera (Applied Spectral Imaging, Migdal Haemek, Israel), and processed by FISHView software version 3.0.0.14 (Applied Spectral Imaging). A normal TLX1 locus yields a fusion signal (Figure 2A). A TLX1 translocation yields a split signal (Figure 2B).
Ligation-mediated PCR
Ligation-mediated PCR (LM-PCR) was performed using a Jδ1 or Dδ3 probe, as described.24 Briefly, 1 μg DNA was digested with 2 blunt-end restriction enzymes, DraI and PvuII. Ligation of 50 pmol of an adaptor to both ends of the restriction fragments was followed by 2 rounds of PCR using nested adaptor–specific primers and the Jδ1- or Dδ3-specific primers 5′-gTTCCACAgTCACgggTTC-3′ and 5′-TgggACCCAgggTgAggATAT-3′, respectively. The LM-PCR products were sequenced in both directions using the Jδ1 primer and the nested adaptor–specific primer. The sequences were blasted in the NCBI nucleotide database.25 and on the Ensembl genome browser tool.26 Results obtained by LM-PCR were confirmed with a designed primer set.
TLX1-TCRD junction screen
A multiplex PCR reaction was performed using the 10q24 5′-gACATCCCTTCCTCAgACgC-3′ and the TCRD 3′ Jδ1 and Dδ3 aforementioned primers. Briefly, 100 ng DNA was amplified for 40 cycles in the presence of 0.2 nM each primer, 2 mM MgCl2, 200 μM dNTP, ABI Buffer II, and 1 U Taq Gold (Perkin Elmer Applied Biosystems, Branchburg, NJ). Cycling parameters included the following: preactivation for 7 minutes at 94°C followed by denaturation for 45 seconds at 94°C, annealing for 1 minute at 57°C, and extension for 90 seconds at 72°C for 40 cycles and a final extension for 10 minutes at 72°C.
Alleleic expression analysis
PCR amplification and sequencing of genomic DNA identified polymorphic markers in the 3′ untranslated region (3′UTR) of TLX1, as described.27 DNAse-treated mRNA from heterozygous samples was reverse transcribed, amplified, and sequenced. The monoallelic or biallelic expression pattern was determined by sequence analysis.
Large-scale expression analysis
An independent series of 92 T-ALL samples from Saint-Louis Hospital (Paris, France), including 56 children (median age, 9 years; range, 1 to 15 years) and 36 adults (median age, 27 years; range, 17 to 66 years) was previously analyzed by large-scale expression analysis using Affymetrix U133A arrays (Santa Clara, CA). Clinical, immunologic, and oncogenic groups of these cases have been described.8
LALA-94 and GRAALL-2003 trials
Ninety-eight adults from the LALA-9421 and 33 from the GRAALL-2003 clinical protocols could be classified into TLX1-high, -low, and -negative based on RQ-PCR–defined criteria. The LALA-94 multicenter prospective randomized trial was reported and discussed previously. The complete remission (CR) rate (86%), survival outcome (median, 28 months), and follow-up (median, 43 months) of the 98 LALA-94 T-ALLs with available cDNA did not differ significantly from the 236 T-ALLs included in the LALA94 protocol. The GRAALL-2003 protocol was a pediatric-inspired phase 2 trial that enrolled 224 adults with Ph-negative ALL between November 2003 and November 2005. Preliminary results have been presented recently, with a median follow-up of 18 months.28 We report here on 33 patients with available cDNA for our analysis. The outcome for these 33 patients did not differ from the overall 74 T-ALLs included in the GRAALL-2003.
Statistical analysis
Patient characteristics and CR rates were compared using the Fisher exact test, while median comparisons were performed with the Mann-Whitney test. Overall survival (OS) was calculated from the date of randomization until the date of death or last contact. Event-free survival (EFS) was calculated from the date of randomization until the date of induction failure, first relapse, death, or last contact. OS and EFS were estimated by the Kaplan-Meier method,29 then compared by the log-rank test.30 For OS and EFS estimations and comparisons, all patients who received an allogeneic stem-cell transplantation (SCT) were censored at SCT time. Adjustments were performed using the Cox model and tested by the log likelihood ratio test. All calculations were performed using the STATA software, version 9.0 (Stata, College Station, TX).
Results
RQ-PCR TLX1 quantification defines 3 groups of T-ALL
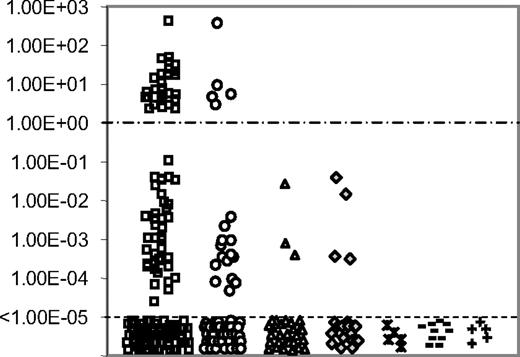
RQ-PCR quantification of TLX1 from 264 T-ALLs (171 adults and 93 children) was performed centrally and expressed as a TLX1/ABL ratio (Figure 1). Results range from 4.0 × 102 to undetectable. Thirty-five samples expressed TLX1 at high level with ratios of TLX1 over ABL greater than 1 (range, 2.0-4.0 × 102). For the purpose of this paper, these T-ALLs are designated as the “TLX1-high” group. Fifty-seven cases expressed TLX1 at a lower level, with a median TLX1 value of 8.0 × 10−4 (range, 1.0 × 10−5 to 1.0 × 10−1) and are designated as the “TLX1-low” group. One-hundred and seventy-two samples were classified in the TLX1-negative group. Strikingly, the TLX1-high and TLX1-low group do not overlap and are separated by at least 1 log of TLX1 level of expression (Figure 1). As reported, no B-cell ALL nor AML expressed high levels of TLX1, but low levels were observed for 3 of 30 B-ALLs and 4 of 21 AMLs. Low TLX1 expression was also found in the B-ALL (RS 4;11, RAJI, REH), myeloblastic (Kasumi and K562), and T-ALL (RPMI, MOLT4, CEM, MOLT13, MKB1, Jurkat, HBP-ALL) cell lines (data not shown). TLX1 was not expressed in 5 control bone marrows, 10 normal peripheral blood lymphocytes, and 5 normal neonatal thymi.
TLX1 level of expression. RQ-PCR quantification of TLX1 transcripts normalized for ABL in 171 adults (□) and 93 children (○) T-ALL samples, 30 B-ALLs (▵), 21 AMLs (♢), 5 normal bone marrows ( ), 10 normal peripheral blood lymphocytes (−) and 5 normal thymi (+). Results are expressed as TLX1/ABL ratio and displayed on a logarithmic scale. A clear difference of a minimum of 1 log-fold expression separates the TLX1-high from the TLX1-low samples. The cut-off value of 1.00 ABL is displayed (----). Samples with detectable TLX1 signal but inferior to 1.00× 10−5 were considered negative, based on plasmid dilutions experiments.
), 10 normal peripheral blood lymphocytes (−) and 5 normal thymi (+). Results are expressed as TLX1/ABL ratio and displayed on a logarithmic scale. A clear difference of a minimum of 1 log-fold expression separates the TLX1-high from the TLX1-low samples. The cut-off value of 1.00 ABL is displayed (----). Samples with detectable TLX1 signal but inferior to 1.00× 10−5 were considered negative, based on plasmid dilutions experiments.
TLX1 level of expression. RQ-PCR quantification of TLX1 transcripts normalized for ABL in 171 adults (□) and 93 children (○) T-ALL samples, 30 B-ALLs (▵), 21 AMLs (♢), 5 normal bone marrows ( ), 10 normal peripheral blood lymphocytes (−) and 5 normal thymi (+). Results are expressed as TLX1/ABL ratio and displayed on a logarithmic scale. A clear difference of a minimum of 1 log-fold expression separates the TLX1-high from the TLX1-low samples. The cut-off value of 1.00 ABL is displayed (----). Samples with detectable TLX1 signal but inferior to 1.00× 10−5 were considered negative, based on plasmid dilutions experiments.
), 10 normal peripheral blood lymphocytes (−) and 5 normal thymi (+). Results are expressed as TLX1/ABL ratio and displayed on a logarithmic scale. A clear difference of a minimum of 1 log-fold expression separates the TLX1-high from the TLX1-low samples. The cut-off value of 1.00 ABL is displayed (----). Samples with detectable TLX1 signal but inferior to 1.00× 10−5 were considered negative, based on plasmid dilutions experiments.
More than 90% of TLX1-high T-ALLs have an abnormal 10q24 locus and show monoallelic TLX1 expression
Karyotype results were available for 175 cases (22 failed and 67 unavailable). Eleven (6%) show clonal 10q24 abnormalities; all 11 cases belonged to the TLX1-high group. However, 17 TLX1-high T-ALLs with successful karyotypes did not demonstrate 10q24 rearrangement (Table 1). The remaining 7 TLX1-high samples did not have karyotypic data available. Among these 24 TLX1-high samples without evident karyotypic 10q24 abnormality, 2 could not be explored by molecular techniques due to the absence of appropriate material. Of the 22 evaluable samples, 20 demonstrated either a split 10q24 locus by FISH (Figure 2A), suggestive of a translocation (n = 16), or a TLX1-TCRD junction by LM-PCR (n = 4). Two samples showed a normal (fusion) FISH pattern (Figure 2B), with no evidence of a TCR-TLX1 junction by LM-PCR. Overall, of 33 TLX1-high T-ALLs with cytogenetic and/or DNA available for appropriate analysis, 31 (94%) demonstrated a TLX1 cytogenetic aberration.
Interphasic dual color 10q24 FISH. Probes were labeled as follows: TLX1-5′ FITC, green signal; TLX1-3′ rhodamine, red signal. (A) Two fusion signals, indicative of 2 intact 10q24 loci. (B) A fusion signal and a split signal, indicative respectivley of a normal 10q24 locus and a 10q24 locus rupture, suggesting a translocation.
Interphasic dual color 10q24 FISH. Probes were labeled as follows: TLX1-5′ FITC, green signal; TLX1-3′ rhodamine, red signal. (A) Two fusion signals, indicative of 2 intact 10q24 loci. (B) A fusion signal and a split signal, indicative respectivley of a normal 10q24 locus and a 10q24 locus rupture, suggesting a translocation.
Twelve of 15 TLX1-high samples tested were informative (heterozygous) for a single nucleotide polymorphism in the 3′UTR region of TLX1. Monoallelic expression of TLX1 was observed in all 12. Of note, 1 of the 2 TLX1-high samples for whom no TLX1 cytogenetic lesion was evidenced by FISH or LM-PCR was evaluable for allele-specific expression and showed a monoallelic pattern of TLX1 expression.
TLX1-low T-ALLs have an intact 10q24 locus
Among the 42 TLX1-low T-ALLs with an available karyotype, none showed karyotypic 10q24 abnormalities (Table 1). FISH analysis was performed for 14 TLX1-low T-ALLs. The TLX1 locus appeared intact in all of these cases, including the 3 patients expressing the highest TLX1 levels of the TLX1-low group. FISH was also performed on 11 cell lines expressing low levels of TLX1 and all showed normal TLX1 loci. Among TLX1-high samples, the sequenced translocation breakpoints on chromosome 10, identified by LM-PCR, were relatively clustered (full details of this cluster will be published in detail elsewhere). One particular set of primers successfully amplified 30% of the TLX1-TCR junctions among the 21 TLX1-high samples tested. The sensitivity of this primer set was evaluated at 10 × 10−3 to 10 × 10−4 log dilution (Figure 3). To verify whether we could evidence a minor TCRD-TLX1 subclone causing the low TLX1 expression in the TLX1-low T-ALLs, we searched the TLX1-low samples for TCRD-TLX1 rearrangements with this designed PCR primer set. A total of 30 TLX1-low T-ALLs was tested, but no TLX1-TCRD junctions were amplified. For allele-specific expression analysis, it was not possible to amplify sufficient cDNA from the 3′UTR region of a significant number of TLX1-low samples to reliably study their allelic expression.
Multiplex-PCR products of TCRD-TLX1 junctions in T-ALLs. Top lanes: TCRD-TLX1 junctions among 8 TLX1-high T-ALLs. Next to these are loaded 10 representative products of the same PCR experiment, performed on 30 TLX1-low samples. No TCRD-TLX1 junction was amplified among the 30 TLX1-low T-ALLs tested. Bottom lanes: Logarithmic dilutions estimated the PCR sensitivity to reach 10−3/−4. The DNA ladder appears on the left.
Multiplex-PCR products of TCRD-TLX1 junctions in T-ALLs. Top lanes: TCRD-TLX1 junctions among 8 TLX1-high T-ALLs. Next to these are loaded 10 representative products of the same PCR experiment, performed on 30 TLX1-low samples. No TCRD-TLX1 junction was amplified among the 30 TLX1-low T-ALLs tested. Bottom lanes: Logarithmic dilutions estimated the PCR sensitivity to reach 10−3/−4. The DNA ladder appears on the left.
TLX1-high but not TLX1-low T-ALL share common immunogenotypic and transcriptional features
Of the 264 T-ALLs, 241 cases have undergone TCR expression and rearrangement analysis and have been TCR classified as described.20 All 32 TLX1-high tested demonstrated TCRB V(D)J rearrangement on at least one allele and a uniform CD34− (31/32, 97%), CD1a+ (32/32, 100%), CD4/8 DP (28/32, 87%) cortical phenotype. They were closely correlated (90%) to an Immature bêta (IMB)/pre-AB stage of maturation arrest (Table 2).20 TLX1-low and TLX1-neg T-ALLs were not tightly linked to any specific stage of maturation arrest, although the TLX1-low group harbors fewer TCR-expressing cases compared with the TLX1-neg group (P = .01).
TLX1-high T-ALLs expressed TLX1 by definition and were uniformly negative for TLX3, CALM-AF10, or SIL-TAL1 (Table 3). In contrast, TLX1-low T-ALLs constituted a heterogeneous oncogenic subgroup. Interestingly, more “HOX”-expressing cases such as TLX3+ or CALM-AF10+ samples31 were found among the TLX1-low samples (P < .05) compared with the TLX1-negative T-ALLs.
TLX1 and array analysis of an independent series of 92 T-ALLS
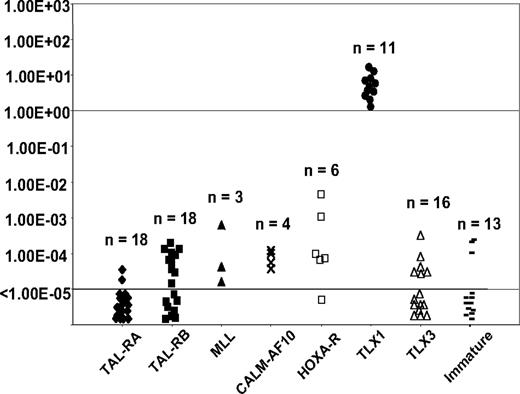
Homogeneous T-ALL oncogenic subgroups (TAL-RA, TAL-RB, MLL, CALM-AF10, HOXA-t, TLX1, TLX3, and immature) have been defined by gene expression profiling from an independent series of 92 T-ALL samples, using unsupervised clustering (U133A Affymetrix microarray) and correlation with immunologic and oncogenic transcript expression data.8 We measured TLX1 expression levels in these cases by RQ-PCR using identical conditions and classified cases as TLX1-high, -low, and -negative as described in “RQ-PCR TLX1 quantification defines 3 groups of T-ALL.” Distribution of the cases within the oncogenic subgroups was analyzed with respect to the TLX1 status. As expected, all TLX1-high cases shared a homogeneous gene expression profile and clustered in the TLX1 subgroup. In contrast, TLX1-low cases were distributed among the other subgroups (Figure 4), as were TLX1-negative cases. Moreover, there was no case in the so-called HOX-R branch that had low TLX1 expression as sole homeobox gene expression (ie, all cases in this unsupervised branch expressed HOXA, TLX1-high, or TLX3), suggesting that low TLX1 expression cannot trigger the biologic profile defining this branch. These data reinforce the view that whereas TLX1-high cases represent a homogeneous oncogenic subgroup with biologic significance, low TLX1 expression does not trigger oncogenic pathways.
TLX1 level of expression measured by RQ-PCR among the different oncogenic groups as defined by transcriptional profile. Levels of TLX1 were normalized for ABL and displayed on a logarithmic scale. The total number of cases in each group is indicated. All TLX1-high samples co-cluster.
TLX1 level of expression measured by RQ-PCR among the different oncogenic groups as defined by transcriptional profile. Levels of TLX1 were normalized for ABL and displayed on a logarithmic scale. The total number of cases in each group is indicated. All TLX1-high samples co-cluster.
Prognostic value of TLX1-high versus TLX1-low expression within the adult LALA-94 and GRAALL-2003 therapeutic protocols
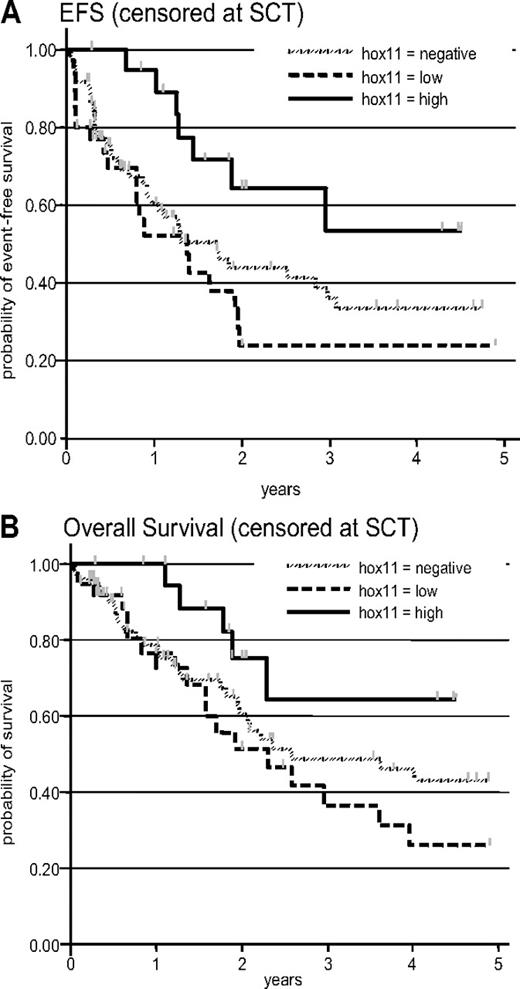
Ninety-eight patients from the LALA-94 and 33 from the GRAALL-2003 protocol were included. The median age of the included patients was 29 years (range, 16-58 years). The median white blood cell count at diagnosis of all included patients was 21.4 × 109/L (range, 0.2-759 × 109/L). There were 20 TLX1-high, 37 TLX1-low, and 74 TLX1-neg samples. Patient characteristics among the 3 subgroups are shown in Table 4. Overall, 118 (90%) of 131 patients achieved CR including 20 of 20 patients in the TLX1-high, 30 of 37 in the TLX1-low, and 68 of 74 in the TLX1-neg subgroup. However, the achievement of CR at first induction was greater in the TLX1-high subgroup compared with combined TLX1-low and TLX1-neg subgroups (Table 4). Median EFS of all 131 patients was 23 months. EFS events were induction death in 5 patients (3, 2, and 0 in the TLX1-neg, -low, and -high subset, respectively), resistance to induction in 8 patients (3, 5, and 0 in the TLX1-neg, -low, and -high subset, respectively), relapse in 54 patients (32, 15, and 7 in the TLX1-neg, -low, and -high subset, respectively; including 1 relapse after SCT), and death in first CR in 6 patients (3, 2, and 1 in the TLX1-neg, -low, and -high subset, respectively; including 4 deaths after SCT). After censoring at stem-cell transplantation (SCT) time, median EFS of TLX1-neg, -low, and -high patients was 20 months, 16 months, and not reached, respectively (Figure 5A, P = .04 by log-rank test). After adjustment on trial and age, the difference between TLX1-high patients and TLX1-low/-neg patients was statistically significant (hazard ratio in the TLX1-low/-neg subgroup, 1.53 [95% CI: 1.03-2.27; P = .035]). Median OS of all 131 patients was 43 months. After censoring at SCT time, median OS of TLX1-neg, -low, and -high patients was 31 months, 28 months, and not reached, respectively (Figure 5B, P = .08 by log-rank test). After adjustment on trial and age, there was a marked trend for longer OS in TLX1-high patients compared with TLX1-low/-neg patients (hazard ratio in the TLX1-low/-neg subgroup, 1.56 [95% CI: 0.98-2.48; P = .059]).
Prognostic analysis. (A) Event-free survival, censored at the stem-cell transplantation time (P = .04 by log-rank test); after adjustment for trial and age, the P value for the TLX1-high versus TLX1-low/-neg comparison was .035. (B) Overall survival, censored at stem-cell transplantation time (P = .08 by log-rank test); after adjustment for trial and age, the P value for the TLX1-high versus TLX1-low/-neg comparison was .059.
Prognostic analysis. (A) Event-free survival, censored at the stem-cell transplantation time (P = .04 by log-rank test); after adjustment for trial and age, the P value for the TLX1-high versus TLX1-low/-neg comparison was .035. (B) Overall survival, censored at stem-cell transplantation time (P = .08 by log-rank test); after adjustment for trial and age, the P value for the TLX1-high versus TLX1-low/-neg comparison was .059.
Discussion
In this study, we sought to determine the oncobiologic and clinical significance of TLX1 expression in T-ALL, because published data on the subject were conflicting. We show that 2 independent groups of TLX1-expressing T-ALLs exist: a homogenous good-prognosis group with high-level TLX1 expression, due to TLX1-TCR juxtapositioning in the majority, and a heterogeneous group with low-level TLX1 expression and neutral prognostic impact. Therapeutic stratification of TLX1+T-ALL is increasingly envisaged; this will require reproducible distinction of TLX1-high and TLX1-low T-ALL within the different prospective adult and pediatric T-ALL trials.
High-level expression was defined as TLX1/ABL RQ-PCR ratios greater than 1 (TLX1 > 1.00 ABL), with a clear demarcation of at least one log from TLX1-low cases. Distinction of low-level expression from TLX1 negativity depends on the sensitivity of the technique. We considered samples with an absolute TLX1 Ct higher than 38 to be TLX1 negative, based on reproducibility experiments conducted with TLX1 plasmid dilutions. This ensured that the TLX1-low group expressed reasonably quantifiable TLX1 amounts. The fact that no TLX1 was detected in normal peripheral blood, bone marrow, thymic cDNA, or genomic DNA confirms the leukemic origin of these low-level TLX1 transcripts. This bimodal pattern of TLX1 expression in T-ALL has been reported, but its biologic significance was unexplored.11 Multicenter reproducibility of these RQ-PCR criteria needs to be evaluated.
TLX1-high T-ALLs corresponded to 18% of adult and 5% of pediatric cases, in keeping with similar estimates,11,23 but lower than reported by other authors.17,18 Only 39% demonstrated karyotypic 10q24 abnormalities, although the incidence of 10q24 abnormalities (1/56, 2% of pediatric T-ALLs; 10/119, 8% of adult T-ALLs) and the overall karyotype failure rate (22/197; 11%) approximated published prevalence.17,18 Combined interphase FISH and TCRD LM-PCR analyses demonstrated that the majority of TLX1-high cases without 10q24 karyotypic abnormalities were due to TLX1 rearrangement. Given that neither TCRA/D-TLX1 t(10;14) nor TCRB-TLX1 t(7;14) is cryptic, the low detection rate is likely to result from difficulty in obtaining representative mitosis from T-ALL blasts, although half of the TLX1-high cases without apparent 10q24 rearrangements did demonstrate clonal abnormalities. The 2 TLX1-high T-ALLs that demonstrated no TLX1 rearrangement by FISH or TCRD LM-PCR may bear 10q24 abnormalities that are beyond the scope of the molecular approaches used, such as intragenic insertion of a transcriptional deregulator other than TCRD. The monoallelic expression of TLX1 in one of these cases is in keeping with deregulation in cis. This 6% false-negative rate by FISH/LM-PCR (2/33 fully analyzed TLX1-high T-ALLs) suggests that standardized RQ-PCR is the most appropriate method for initial screening, if therapeutic stratification is to be envisaged. Such an approach is, however, entirely dependent on a reproducible capacity to distinguish TLX1-high from TLX1-low cases, and a combined RQ-PCR and FISH approach is optimal. Based on the data presented here, cases with a TLX1/ABL ratio greater than 1 are likely to be associated with a TLX1 rearrangement, those with TLX1/ABL ratios below 0.1 are unlikely to have TLX1 rearrangement if the sample is representative of the leukemia, and those with intermediate ratios should be analyzed by FISH with TLX1 and TCR probes.
TLX1-high T-ALLs demonstrated a homogenous stage of maturation arrest (IMB, pre-AB) characteristic of cortical thymocytes arrested around the TCRβ selection,32 a correspondingly distinct gene expression profile8 and were mutually exclusive of the other major T-ALL oncogenic markers, TLX3, CALM-AF10, and SIL-TAL1, in marked contrast to TLX1-low T-ALLs. Taken together, TLX1-high T-ALLs represent a distinct oncogenic group, biologically different from TLX1-low T-ALLs.
The survival curves of the combined LALA-94 and GRAALL-2003 adult trials reported here clearly show that only high levels of TLX1 expression confer a better prognosis. TLX1 translocation or expression has generally been associated with a better prognosis, although the strength of the association varies between series and small patient numbers often preclude appropriate prognostic evaluation1,11,15 The absence of a standardized PCR definition of TLX1-positive T-ALLs also makes data comparison difficult. Notably, the trend toward better prognosis seems to be more pronounced when authors consider “high” TLX1 expressers or TLX1-translocated samples.11,17,18 A better leukemia-free survival in adult T-ALL was associated with high TLX1 expression in the study by Ferrando et al.17 A trend for a better outcome was observed in pediatric T-ALL by Kees and al.18 , when 19.7% of the cohort was classified as TLX1-high, compared with only 5% in the present series. It is possible that the use of a higher threshold to define TLX1-high T-ALL would have allowed even better prognosis discrimination, although performing such an analysis in pediatric protocols would require large patient numbers, given the relative rarity of pediatric TLX1-high T-ALL, as defined here. Because all T-ALLs with 10q24 abnormalities correspond to TLX1-high cases, the t(10;14) is associated with a trend toward better EFS even with limited patient numbers in both pediatric and adult T-ALL.11,15
TLX1-low T-ALLs are not a homogeneous group, because they demonstrated variable immunophenotypic, oncogenotypic and transcriptional features and a similar response to treatment as TLX1-negative cases. We cannot formally eliminate the possibility of minor TCR-TLX1 subclones in these cases but consider this to be unlikely because no TLX1-TCR junction could be identified among 30 TLX1-low samples tested with a sensitivity of 10 × 10−3 to 10 × 10−4. Given that most of the TLX1-low T-ALLs demonstrate other oncogenic markers, oncogenic cooperation involving low levels of TLX1 is possible, as proposed for NOTCH1 mutations.33 TLX1-low T-ALLs did, however, tend to correlate with an immature stage of maturation arrest and, more particularly, the “HOXA”-associated oncogenic markers CALM-AF10, MLL, and HOXA-TCR,8,31 compared with TCR+, SIL-TAL+, or TAL-RA T-ALLs. This low expression of TLX1 could reflect an underlying global homeodomain gene deregulation program34,35 with low-level transcription of TLX1 occurring as some kind of leakage effect. While such considerations are beyond the scope of this paper, if such a mechanisms occurs, it is unlikely to be restricted to T-ALL, because low TLX1 expression was also seen in B-lineage ALL and AML.
Taken together, the importance of a standardized definition of TLX1 positivity cannot be overstressed if, as suggested in adult T-ALL,17 TLX1 status is to be used to dictate therapeutic decisions such as bone marrow transplantation. As detailed above, we propose that TLX1-high T-ALLs should be defined by a molecular RQ-PCR approach, refined when necessary by FISH, although initial FISH screening will detect the majority of cases and will need to be considered if RQ-PCR classification does not prove reproducible in interlaboratory comparisons within prospective clinical trials. We also propose that within adult T-ALL, TLX1 positivity be restricted to cases with 10q24 rearrangement and/or TLX1/ABL RQ-PCR ratios greater than 1 and that such cases be stratified as low-risk acute leukemia. All adult cases with lower RQ-PCR ratios and absence of 10q24 rearrangements by FISH with appropriate probes should be considered TLX1 negative. It is probable that similar criteria will be appropriate in pediatric T-ALL, but this will require assessment within prospective large-scale trials.
The online version of this article contains a data supplement.
Partial results were presented in abstract form at the 48th Annual Meeting of the American Society of Hematology, Orlando, FL, December 10, 2006.36
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Fondation de France (UB: 032145; No. Engt: 2005006547) and the Association de Recherche contre le Cancer (No. 3136). J.B. is a Fellow of the National Cancer Institute of Canada (NCIC) through an award from the Terry Fox Foundation (TFF).
We thank all participants in the LALA94, GRAALL, and FRALLE trials for collecting and providing data and samples, especially Véronique Lhéritier, data manager, Christine Pérot, and Jacqueline Van Den Akker.
Authorship
Contribution: V.A. and E.A.M. designed and supervised the research; J.B., E.C., C.M., and P.B. performed molecular analyses; J.B., I.R., and G.S. performed cytogenetic analyses; H.D. and X.T. performed clinical data analyses; J.B., V.A., E.A.M., H.D., and J.S. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Vahid Asnafi, Hôpital Necker Enfants Malades, Tour Pasteur 2e étage, Laboratoire d'hématologie, 149 rue de Sèvres, 75015 Paris, France; e-mail:vahid.asnafi@nck-aphp.fr.





 ), 10 normal peripheral blood lymphocytes (−) and 5 normal thymi (+). Results are expressed as TLX1/ABL ratio and displayed on a logarithmic scale. A clear difference of a minimum of 1 log-fold expression separates the TLX1-high from the TLX1-low samples. The cut-off value of 1.00 ABL is displayed (----). Samples with detectable TLX1 signal but inferior to 1.00× 10−5 were considered negative, based on plasmid dilutions experiments.
), 10 normal peripheral blood lymphocytes (−) and 5 normal thymi (+). Results are expressed as TLX1/ABL ratio and displayed on a logarithmic scale. A clear difference of a minimum of 1 log-fold expression separates the TLX1-high from the TLX1-low samples. The cut-off value of 1.00 ABL is displayed (----). Samples with detectable TLX1 signal but inferior to 1.00× 10−5 were considered negative, based on plasmid dilutions experiments.