Abstract
BAALC expression is considered an independent prognostic factor in cytogenetically normal acute myeloid leukemia (CN-AML), but has yet to be investigated together with multiple other established prognostic molecular markers in CN-AML. We analyzed BAALC expression in 172 primary CN-AML patients younger than 60 years of age, treated similarly on CALGB protocols. High BAALC expression was associated with FLT3-ITD (P = .04), wild-type NPM1 (P < .001), mutated CEBPA (P = .003), MLL-PTD (P = .009), absent FLT3-TKD (P = .005), and high ERG expression (P = .05). In multivariable analysis, high BAALC expression independently predicted lower complete remission rates (P = .04) when adjusting for ERG expression and age, and shorter survival (P = .04) when adjusting for FLT3-ITD, NPM1, CEBPA, and white blood cell count. A gene-expression signature of 312 probe sets differentiating high from low BAALC expressers was identified. High BAALC expression was associated with overexpression of genes involved in drug resistance (MDR1) and stem cell markers (CD133, CD34, KIT). Global microRNA-expression analysis did not reveal significant differences between BAALC expression groups. However, an analysis of microRNAs that putatively target BAALC revealed a potentially interesting inverse association between expression of miR-148a and BAALC. We conclude that high BAALC expression is an independent adverse prognostic factor and is associated with a specific gene-expression profile.
Introduction
Clonal cytogenetic abnormalities have been shown to represent one of the most important factors for predicting clinical outcome in acute myeloid leukemia (AML).1-3 However, approximately 45% of adults younger than 60 years old with primary AML have normal cytogenetics (CN) at diagnosis and therefore lack informative chromosome markers. In this group, molecular genetic markers such as mutations in the FLT3 gene,4,5 the NPM1 gene,6-9 the CEBPA gene,10-12 and the MLL gene,13,14 and high expression levels of the ERG gene15,16 have been found to predict outcome
In addition, our group was first to report high expression levels of the brain and acute leukemia, cytoplasmic (BAALC) gene to be a poor prognostic factor in CN-AML.17,18 This gene, located on human chromosome 8 at q22.3, encodes at least 8 alternatively spliced transcripts in humans, and the protein has no homology to other known proteins.17,19,20 With regard to normal hematopoiesis, in vitro studies have shown that BAALC is up-regulated in CD34+ bone marrow (BM) cells, and that its down-regulation correlates with subsequent cell differentiation leading to the hypothesis that BAALC represents a marker of early hematopoietic progenitor cells.21
Although the prognostic value of high BAALC expression has been confirmed in 2 different studies,22,23 to date it has not been investigated together with a wider set of molecular markers predictive for outcome in CN-AML. Here we present a comprehensive study that evaluates the predictive value of BAALC expression in the context of the currently best-established molecular markers in CN-AML.24 Furthermore, to gain insights into the biologic function of this gene and its potential role in normal and malignant hematopoiesis, we conducted global gene expression and microRNA (miRNA) expression profiling.
Methods
Patients
A total of 172 adult patients younger than 60 years old with untreated primary CN-AML were included in this study. Patients were treated similarly on the Cancer and Leukemia Group B (CALGB) protocols 9621 (n = 83) and 19808 (n = 89). The clinical trials and companion protocols were approved by the Ohio State University Institutional Review Board and the Cancer and Leukemia Group B. All patients gave informed consent for the research use of their specimens, in accordance with the Declaration of Helsinki.
Treatment
Treatment on CALGB 9621 and 19808 has been detailed elsewhere.25,26 Briefly, patients received induction chemotherapy with cytarabine, daunorubicin, and etoposide with (ADEP) or without (ADE) PSC-833, a multidrug resistance protein inhibitor, also called valspodar. Upon achievement of complete remission (CR), patients received high-dose cytarabine and etoposide for stem cell mobilization followed by myeloablative treatment with busulfan and etoposide supported by autologous peripheral blood stem cell transplantation (APBSCT). Patients unable to undergo APBSCT received 2 additional cycles of high-dose cytarabine.
Clinical end points
CR was defined as recovery of morphologically normal BM and blood counts (ie, neutrophil count ≥ 1.5 × 109/L and platelet count ≥ 100 × 109/L), and no circulating leukemic blasts or evidence of extramedullary leukemia. Relapse was defined by 5% or more BM blasts, circulating leukemic blasts, or development of extramedullary leukemia. Overall survival (OS) was measured from the date on study until the date of death, censoring for patients alive at last follow-up. Disease-free survival (DFS) was measured from the date of CR until date of relapse or death, regardless of cause, censoring for patients alive at last follow-up. In our dataset, there were no patients who died in CR without first relapsing; hence, DFS is equivalent to relapse-free survival.
Cytogenetics and additional molecular markers
Pretreatment cytogenetic analyses of BM were performed by CALGB-approved institutional cytogenetic laboratories as part of CALGB 8461, a prospective cytogenetic companion, and were centrally reviewed, as previously reported.1 To be considered cytogenetically normal, at least 20 metaphase cells had to be analyzed and the karyotype found to be normal in each case. The presence or absence of additional molecular markers, that is, FLT3 internal tandem duplication (FLT3-ITD),5,27 mutations in the NPM17 and CEBPA10 genes, MLL partial tandem duplication (MLL-PTD),13,28 and FLT3 tyrosine kinase domain (FLT3-TKD) mutations29,30 as well as ERG expression levels,15,16 were assessed centrally as reported previously.
RNA extraction and real-time RT-PCR for BAALC
Mononuclear cells from pretreatment blood were enriched by Ficoll-Hypaque gradient and cryopreserved until they were thawed for this analysis. cDNA was synthesized from total RNA extracted using Trizol reagent (Invitrogen, Carlsbad, CA) and used for the TaqMan real-time reverse transcription–polymerase chain reaction (RT-PCR) assay as previously reported,15,18 including the previously described primers and probes.18,23 Positive controls (cDNA from the BAALC-expressing cell line KG1a), negative controls (water control of the cDNA synthesis), and standard curves (serial dilutions of plasmids containing BAALC or GPI cloned fragments) were included in each run.
mRNA and miRNA gene-expression profiling
RNA was extracted from thawed tissue samples using Trizol reagent and processed for Affymetrix U133 plus 2.0 GeneChip (Affymetrix, Santa Clara, CA) hybridizations. Briefly, from 5 μg total RNA, double-stranded cDNA was prepared with the use of the T7-Oligo(dT) primer (Affymetrix). In vitro transcription was performed with the BioArray HighYield RNA Transcript Labeling Kit (T7; Enzo Life Science, Farmingdale, NY). Biotin-cRNA (10 μg) was fragmented and hybridized to the U133 plus 2.0 GeneChip for 16 hours at 45°C. Scanned images were converted to CEL files using GCOS software (Affymetrix).
For the miRNA microarray chips, biotinylated first-strand cDNA was synthesized in reverse transcription from 2.5 to 5.0 μg total RNA using biotin-labeled random octamer oligo primer from pretreatment BM and blood mononuclear cell samples and hybridized to miRNA microarray chips, as previously reported.31 Images of the miRNA microarrays were acquired as reported previously.31
The quantitative real-time PCR assay for the miRNA expression analysis was performed as described in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Statistical methods
BAALC expression levels represented a continuum ranging from less than 0.001 to approximately 5.5 (when expressed using the comparative cycle threshold [CT] method18 ; Figure S1). To combine expression levels from patients on each of the 2 protocols and evaluate the impact of BAALC expression levels on clinical outcome without seeking an optimal cut point, patients were divided into low and high expressers using the median BAALC expression level within each protocol. This cut point was chosen to make our results comparable with those of previously published studies,21,23 and also based on the trends observed in the OS and DFS of patients divided into quartiles by BAALC expression levels; for both end points, patients in quartile 1 had better outcome than patients in quartile 2, followed by patients in quartiles 3 and 4 (OS, P < .001 and DFS, P = .02, test for trend32 ; Figure S2).
Associations between the high and low BAALC groups and baseline clinical, demographic, and molecular features were analyzed using Fisher exact and Wilcoxon rank sum tests for categoric and continuous variables, respectively.
Estimated probabilities of OS and DFS were calculated using the Kaplan-Meier method, and the log-rank test evaluated differences between survival distributions. Proportional hazards models were constructed to determine whether BAALC was associated with outcome when adjusting for other prognostic variables. Full models were fit with the variables significant in univariable analyses (P < .20). If a particular variable failed to meet the proportional hazards assumption, then an artificial time-dependent covariate was included in all models containing that variable. Using backward selection, the least significant variable was removed one at a time until all variables remaining in the final model were significant at α = .05. Any variables that were initially excluded were added back into the model to confirm that they were not statistically significant. In a similar fashion, logistic regression was used to model the impact of BAALC on the achievement of CR.
For DNA microarrays, summary measures of gene expression were computed for each probe set using the robust multichip average (RMA) method, which incorporates quantile normalization of arrays.33 Expression values were logged (base 2) before analysis. A filtering step was performed to remove probe sets that did not display significant variation in expression across arrays. In this procedure, a chi-square test was used to test whether the observed variance in expression of a probe set was significantly larger than the median observed variance in expression for all probe sets using α = .01 as the significance level. A total of 24 183 probe sets passed the filtering criterion and were included in subsequent analyses. A comparison between the gene-expression profiles of patients with low (n = 24) and high (n = 26) expression of BAALC was performed by univariable 2-sample t tests using α = .001 as the significance level (for the definition of the low and high BAALC groups, see Document S1). A global test of differences in expression profiles between the 2 classes was also performed. This test is based on a permutation distribution of the number of significant genes in the 2-sample comparison.
For miRNA microarrays, calculation, normalization, and filtering of signal intensity for each microarray spot and batch effect adjustment were performed as described in Document S1. Of the human miRNA probes, 305 passed the filtering criteria for the training set and were included in subsequent analyses. A comparison of low and high BAALC expressers was made just as for the DNA microarray data, although a univariable significance level of α = .005 was used.
A more focused investigation of the relationship between expression of BAALC and miRNAs that potentially target BAALC was also undertaken. Prediction of putative miRNA target sites in BAALC was based on the following publicly available algorithms: http://www.microrna.org/,34 http://www.targetscan.org/,35 and http://microrna.sanger.ac.uk/.36 Among the 305 miRNA probes studied, 14 probes, representing 11 unique miRNAs, were identified as putatively targeting BAALC. Correlations between expression of these miRNA probes and BAALC were measured using the Spearman rank correlation coefficient for all 75 patients for whom miRNA array analysis was performed.
Analyses were performed using BRB-ArrayTools version 3.4.0 (National Cancer Institute, Bethesda, MD) developed by Dr Richard Simon and Amy Peng Lam and using the R version 2.3.1 (R Foundation for Statistical Computing, Vienna, Austria). All analyses were performed by the CALGB Statistical Center.
Results
Associations of BAALC expression with molecular markers and clinical characteristics
At diagnosis, patients with high BAALC expression were more likely than patients with low BAALC expression to harbor FLT3-ITD (44% vs 28%, P = .04), have NPM1 wild-type alleles (48% vs 17%, P < .001), carry a CEBPA mutation (32% vs 10%, P = .003), present with MLL-PTD (15% vs 3%, P = .009), and have high expression of the ERG gene (45% vs 29%, P = .05; Table 1). Low BAALC expressers were more likely to harbor FLT3-TKD than high BAALC expressers (15% vs 3%, P = .005).
With respect to clinical characteristics, the BAALC expression groups differed with regard to FAB subtype distribution (P = .006), with high BAALC expressers more often presenting with AML FAB M1 and M2 and less often with AML FAB M4 and M5 (Table 1).
BAALC expression as a prognostic marker
Patients with high BAALC expression less frequently achieved CR (79% vs 90%; P = .09). In a multivariable model for CR, BAALC expression provided independent prognostic information (P = .04). When controlling for ERG expression (P = .007) and age (P = .02), the odds of achieving CR were almost 4 times less for high BAALC expressers than for low BAALC expressers (odds ratio = .27; Table 2).
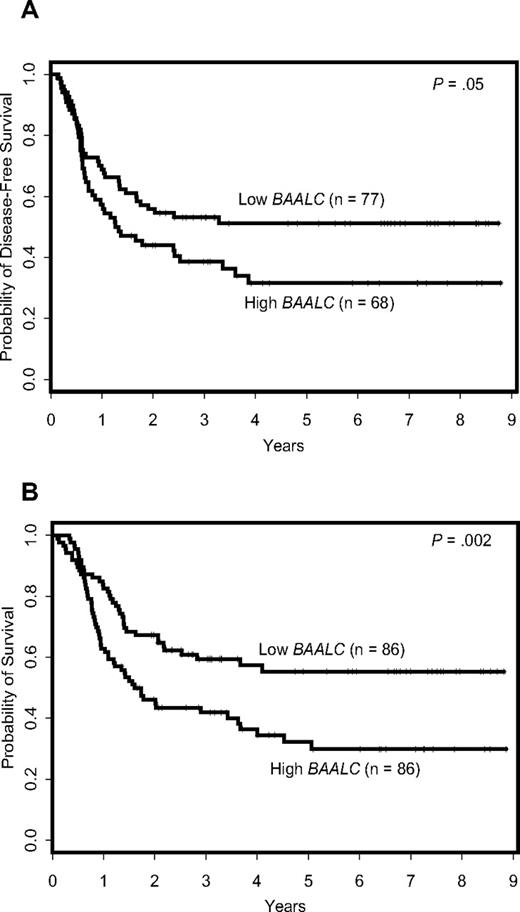
Patients expressing high BAALC levels had a shorter DFS (P = .05) and OS (P = .002; Figure 1) compared with patients expressing low BAALC levels; the estimated 3-year DFS rates were 39% and 53% and the estimated 3-year OS rates 42% and 59% for the 2 groups (Table 1).
Outcome of cytogenetically normal AML patients according to BAALC expression levels. (A) Disease-free survival. (B) Overall survival.
Outcome of cytogenetically normal AML patients according to BAALC expression levels. (A) Disease-free survival. (B) Overall survival.
In a multivariable model for DFS, BAALC expression was no longer significant (P > .20) when controlling for the FLT3-ITD (P < .001) and ERG expression (P = .006). In contrast, in a multivariable model for OS, BAALC expression provided additional independent prognostic information (P = .04), when controlling for the FLT3-ITD (P = .004), NPM1 mutation status (P = .02), CEBPA mutation status (P = .02), and white blood cell (WBC) count (P = .01). Patients with high BAALC expression were almost twice as likely to die as patients with low BAALC expression (Table 2).
Gene-expression profiling of AML patients with low and high BAALC expression
Gene-expression profiling (GEP) was performed to determine whether a unique signature was associated with the expression levels of BAALC in CN-AML. A comparison of GEP of patients with low (n = 24) and high (n = 26) expression levels of BAALC identified a gene-expression signature composed of 312 differentially expressed (P < .001) probe sets corresponding to 233 unique genes, hypothetical genes/proteins, and open reading frames (ORFs; Figure 2). With regard to the expression of the BAALC gene, itself, the GEP results were consistent with those obtained by real-time RT-PCR, because the 2 probe sets representing BAALC on the Affymetrix U133 plus 2.0 GeneChip were part of the signature and were up-regulated 4.7- and 4.8-fold in the high BAALC group (Table S1).
Heat map of the BAALC signature. Columns represent samples and rows represent genes ordered by hierarchical cluster analysis using average linkage and one minus Pearson correlation as the distance metric. Shading indicates relative expression of each gene with respect to the gene's median expression (black equal to, red above, and blue below the median value).
Heat map of the BAALC signature. Columns represent samples and rows represent genes ordered by hierarchical cluster analysis using average linkage and one minus Pearson correlation as the distance metric. Shading indicates relative expression of each gene with respect to the gene's median expression (black equal to, red above, and blue below the median value).
Within the derived gene-expression signature, 48 individual genes showed at least a 2-fold change in expression between high and low BAALC expressers. Of the 48 genes, 29 were up-regulated and 19 down-regulated in the high BAALC expression group (Table 3; for a complete list of the 312 differentially expressed probe sets see Tables S1,S2). Several genes reported to be associated with hematopoietic stem cells were among the 29 genes up-regulated in high BAALC patients including PROM1 (also known as CD133), CD34, and KIT. Moreover, CD34, C5orf23, FZD6, JUP, KIT, B4GALT6, CRYGD, PLAGL1, and ITM2C that were up-regulated here were previously reported as being associated with CD133, a marker expressed on primitive cell populations such as hematopoietic stem and progenitor cells.37,38
Consistent with the adverse prognostic impact of high BAALC expression levels, genes previously reported to be predictive of poor outcome (ie, HGF, MN1, CD200)39-42 or genes involved in drug-resistance mechanisms in AML, such as ABCB1 (also known as MDR1),43-49 were up-regulated.
Among the 19 most down-regulated genes in the high BAALC expression group, we found the group of cancer/testis antigen CT45 genes that are highly expressed in the testis and in solid tumor cell lines50 and appear to be identical to the Ki-A10 antigen expressed in Hodgkin lymphoma.51 Also down-regulated were members of the HOXB gene family (HOXB6 and HOXB7 by at least 2-fold; HOXB5 and HOXB9 to a lesser degree, 1.98-fold and 1.77-fold, respectively) and the histone cluster 1 and histone cluster 2 families, as well as the plasminogen activator, urokinase receptor gene (PLAUR, also known as CD87) and the coreceptor for plasminogen/tissue plasminogen activator annexin A2 (ANXA2).
miRNA expression profiling of AML patients with low and high BAALC expression
A group of 50 patients with high (n = 24) and low (n = 26) BAALC expression levels as determined by real-time RT-PCR was examined using microarray miRNA expression profiling to identify differences in expression of miRNAs. A global comparison of all miRNA probes revealed no significant miRNA expression signature that differentiated between the low and high BAALC expressers. However, we also performed an analysis that focused on the expression of miRNAs predicted in silico to have binding sites in BAALC. Data were available for analysis of 11 predicted miRNAs (represented by 14 probes). One probe, representing miR-148a (probe sequence TGAGTATGATAGAAGTCAGTGCACTACAGAACTTTGTCTC), had a rather striking inverse correlation with the expression of BAALC (Spearman correlation coefficient −.63; P < .001). In support of this observation, we used an independent set of patients and performed expression analysis of miR-148a by real-time PCR. Again, we observed an inverse correlation between the expression levels of miR-148a and BAALC (Figure S3). In addition, expression levels of miR-148a were lower (median, 0.19) within the AML patients compared with the expression levels in blood from healthy volunteers (median, 4.41; Figure S4).
Discussion
Recently, there has been substantial progress in using molecular markers such as gene mutations or changes in levels of gene expression to risk-stratify patients with CN-AML.24 With the number of molecular markers increasing, it is important to investigate each of these markers not only separately but also in relation to each other. In this report, we analyzed 172 patients with CN-AML for the expression levels of BAALC and tested the prognostic value of BAALC expression in the context of a comprehensive set of other established molecular markers in CN-AML.
In our set of patients, high BAALC expressers were less likely to achieve CR than patients with low BAALC expression (P = .04), when controlling for ERG expression and age. This observation is in accordance with recent results of a large German study.23 High BAALC expression also predicted poor OS independently from the FLT3-ITD, NPM1, and CEBPA mutation status, and WBC. Patients with high BAALC expression were almost twice as likely to die as those with low BAALC expression. Although associated with DFS as a single marker (P = .05), BAALC expression did not provide additional prognostic information in a multivariable analysis for DFS. This is in contrast to our initial report on BAALC as a prognostic marker.18 However, in this previous study, we did not include patients harboring the very unfavorable genotype of FLT3-ITD in the absence of a FLT3 wild-type allele,18 nor were NPM1 and CEBPA mutational status and ERG expression considered. Because patients undergoing allogeneic transplantation in first CR were not included in our current study, we were not able to address the previously reported observation that patients with high BAALC expression may benefit from this form of consolidation therapy.23
GEP revealed a unique signature associated with high BAALC expression levels. The multidrug resistance gene ABCB1 (MDR1) was identified as one of the most highly up-regulated genes in high BAALC expressers, which is consistent with the resistant disease associated with these patients. MDR1 protein (also called P-glycoprotein) is an ATP-dependent drug efflux pump and is responsible for decreased drug accumulation in multidrug resistant cells and often mediates the development of resistance to anticancer drugs.52 Nevertheless, the addition of valspodar (PSC-833), a P-glycoprotein inhibitor, to the induction therapy did not alter the CR rates within the group of high BAALC expressers (78% with PSC-833 vs 80% without PSC-833).
Other groups have sought to identify gene signatures associated with drug resistance in adult AML. Heuser et al53 reported a gene-expression profile that distinguished AML samples from patients with good response to induction chemotherapy from those of patients with poor response. Interestingly, we found that several genes overexpressed in the high BAALC expression group in our study were also overexpressed in the poor-response group in the study of Heuser et al.53 These include the CD34, CD133, MN1, and JUP genes and the ORF C5orf23 (FLJ14054).
Consistent with the worse outcome associated with high BAALC expression, the GEP signature included up-regulation of other genes whose overexpression has been found to be adversely associated with outcome in either CN-AML, such as the MN1 gene, or among cytogenetically heterogeneous AML patients, such as the HGF and CD200 genes.39-42 Overexpression of the MN1 gene was recently reported as being associated with inferior OS and a higher risk of relapse in adults aged 60 years or younger with primary or secondary CN-AML.41 In our study, MN1 was up-regulated by 3.1-fold in high BAALC expressers. Two other studies, performed on patient populations with various cytogenetic findings, reported that plasma hepatocyte growth factor (HGF) protein levels were significantly higher in AML patients compared with healthy controls and that higher plasma HGF levels correlated with shorter survival.39,40 Finally, in a recent report, Tonks et al42 investigated CD200 expression levels in AML and found an association of CD200 expression with worse survival in AML patients with karyotypes that did not contain t(8;21)(q22;q22) or inv(16)(p13q22).
The notion of BAALC as a marker of hematopoietic progenitor cells21 is further supported by our GEP results as high BAALC expression was associated with genes found up-regulated in undifferentiated hematopoietic cells (CD133, CD34, KIT, NPR3, JUP, C5orf23, FZD6, B4GALT6, CRYGD, PLAGL1, ITM2C).37,38,54,55 Notably, CD133, the most highly up-regulated gene in the high BAALC expresser group (7.1-fold), is considered to identify hematopoietic stem cells because CD133+/CD34+ cells have a higher clonogenic capacity than CD133−/CD34+ cells.56,57 In recent studies, BAALC has been found among the most highly up-regulated genes in CD133+ cells in both cord blood and peripheral blood.37,38 On the other hand, genes expressed in more differentiated subtypes of AML were down-regulated in the high BAALC expresser group. One example is the plasminogen activator, urokinase receptor CD87, whose expression in AML varies according to the FAB subtype, with the highest expression in M5 and the lowest in M0.58 This is consistent with our current results and previous reports18,23 showing that high BAALC expressers less often present with M5 and more often present with FAB M1 morphology than the lower BAALC expressers.
Finally, we tested whether high BAALC expression was associated with a specific miRNA expression profile. miRNAs are naturally occurring 19- to 25-nucleotide RNAs that hybridize to complementary mRNA targets and inhibit translation of the corresponding proteins.59 Because miRNAs have recently been shown to be predictive for more aggressive phenotypes in chronic lymphocytic leukemia and solid tumors,31,60,61 we performed miRNA-expression profiling in search of a distinctive miRNA signature that would differentiate patients with high BAALC expression levels from those with low BAALC expression. This analysis failed to reveal significant differences in global miRNA-expression between the BAALC expression groups. In a second step, we focused our search on miRNAs with predicted binding sites in BAALC. We observed the expression of one such miRNA, miR-148a, being inversely correlated with the expression of BAALC, thereby suggesting that miR-148a might act as a negative regulator for BAALC.
In summary, high BAALC expression levels predict lower rates of CR and shorter survival independent of other prognostic molecular markers. Furthermore, high BAALC expressers are characterized by a gene-expression signature consistent with less differentiated AML blasts. It is possible that this less differentiated phenotype and overexpression of genes previously reported associated with drug resistance contribute to the lower CR rate and worse survival observed in high BAALC expressers.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors gratefully acknowledge sample processing and storage services provided by Ms Donna Bucci of the CALGB Leukemia Tissue Bank at The Ohio State University Comprehensive Cancer Center (Columbus, OH).
This work was supported in part by National Institutes of Health (NIH) grants CA101140, CA114725, CA077658, CA016058, CA096887, CA089341, and CA098933 and the Coleman Leukemia Research Foundation.
National Institutes of Health
Authorship
Contribution: C.L., M.D.R., A.S.R., K.M., M.E.R., C.-G.L., A.d.l.C., G.M., and C.D. Bloomfield contributed to the design and analysis of this study; C.L., M.D.R., A.S.R., K.M., A.d.l.C., G.M., and C.D. Bloomfield contributed to the writing of this paper and all authors agreed on the final version; C.L., S.P.W., P.P., C.D. Baldus, and T.V. carried out laboratory-based research; A.S.R. and M.D.R. performed statistical analyses; B.L.P., J.E.K., R.A.L., G.M., and C.D. Bloomfield were involved directly or indirectly in care of patients and/or sample procurement.
A complete list of the Cancer and Leukemia Group B institutions, principal investigators, and cytogeneticists who participated in this study is provided in Document S2, available on the Blood website; see the Supplemental Materials link at the top of the online article.
Conflict-of-interest disclosure: A.d.l.C. has applied for a patent on BAALC expression as a diagnostic marker for acute leukemia and has a financial interest in the patent. The remaining authors declare no competing financial interests.
Correspondence: Clara D. Bloomfield, The Ohio State University, The Comprehensive Cancer Center, 519 James Cancer Hospital, 300 West 10th Ave, Columbus, OH 43210; e-mail: clara.bloomfield@osumc.edu.