Abstract
Posttranslationally modified histones and DNA hypermethylation frequently interplay to deregulate gene expression in cancer. We report that acute myeloid leukemia (AML) with an aberrant histone methyltransferase, the mixed lineage leukemia partial tandem duplication (MLL-PTD), exhibits increased global DNA methylation versus AML with MLL-wildtype (MLL-WT; P = .02). Among the differentially methylated genes, the SLC5A8 tumor suppressor gene (TSG) was more frequently hypermethylated (P = .003). In MLL-PTD+ cell lines having SLC5A8 promoter hypermethylation, incubation with decitabine activated SLC5A8 expression. Ectopic SLC5A8 expression enhanced histones H3 and H4 acetylation in response to the histone deacetylase inhibitor, valproate, consistent with the encoded protein—SMCT1—short-chain fatty acid transport function. In addition, enhanced cell death was observed in SMCT1-expressing MLL-PTD+ AML cells treated with valproate. Within the majority of MLL-PTD AML is a mechanism in which DNA hypermethylation silences a TSG that, together with MLL-PTD, can contribute further to aberrant chromatin remodeling and altered gene expression.
Introduction
The mixed lineage leukemia partial tandem duplication (MLL-PTD), present in 4% to 7% of adults with cytogenetically normal acute myeloid leukemia (CN-AML), is a cryptic gene rearrangement that most commonly duplicates introns 5 through 11 or introns 5 through 12 giving rise to an in-frame fusion transcript having exons 11 or 12 fused upstream of exon 5 within a full-length transcript.1,2 Like MLL-WT, the MLL-PTD has the transcriptional activating histone H3 lysine 4 methytransferase activity within the C-terminal SET domain.3,4 The Mll-PTD mouse exhibits enhanced histone H3K4 methylation activity supporting a gain-of-function role for the mutant rather than a loss of MLL WT function.5 However, MLL-PTD expression is concurrent with absence of MLL-WT expression in MLLPTD/WT primary human AML cells.6 The mechanism underlying this monoallelic silencing is not yet clear but may involve aberrant epigenetic mechanisms, that is, DNA methylation and histone modifications associated with gene inactivation.6 In contrast to initial studies,2,7-9 we reported improved disease-free survival in adult patients with MLL-PTD CN-AML treated with autologous transplantation during first remission.10 Even so, the majority of MLL-PTD+ patients relapsed early.10
In addition to structural genetic alterations, epigenetic gene silencing contributes to malignant transformation in AML and other cancers.11-13 We report here increased global DNA methylation in MLL-PTD+ compared with MLL-PTD− (MLL-WT) CN-AML. Among the hypermethylated and silenced genes associated with MLL-PTD was SLC5A8, a tumor suppressor gene (TSG)14-18 that encodes SMCT1, a plasma membrane transporter of endogenous monocarboxylates such as butyrate and pyruvate, both of which have histone deacetylase (HDAC) inhibitory activity.19,20 We showed that forced expression of SMCT1 enhanced histone acetylation and apoptosis in MLL-PTD AML cell lines treated with the HDAC inhibitor, valproic acid (VPA), a synthetic butyrate analog.
Methods
Bone marrow (BM) and blood samples were Ficoll-enriched and cryopreserved with institutional review board approval from consenting adults, in accordance with the Declaration of Helsinki, who were treated on Cancer and Leukemia Group B (CALGB) 9621 or CALGB 19808. Pretreatment cytogenetics were centrally reviewed under CALGB 8461.21 NotI/EcoRV/HinfI restriction landmark genomic scanning (RLGS) and bisulfite-PCR/sequencing were described previously.15,22 The EOL-1 (ATCC, Manassas, VA) and MUTZ-11 (gift from Dr Drexler, DSMZ [German Collection of Microorganisms and Cell Culture], Braunschweig, Germany) cell lines have been described.23,24 MLL-PTD fusion transcripts were detected by nested reverse transcription–polymerase chain reaction (RT-PCR)/sequencing and real time RT-PCR.6,25 Cells were transfected with SLC5A8-expressing pcDNA3.1/V5-His-TOPO vector15 or empty vector (Amaxa Biosystems, Gaithersburg, MD) and immunoblotting performed for SMCT1, the V5-epitope, or total acetylation of histones H3 and H4. Decitabine and VPA were purchased (Sigma-Aldrich, St Louis, MO). The difference in global DNA methylation between RLGS profiles from MLL-WT and MLL-PTD AML groups, measured as the number of events of 321 evaluable, was assessed using the Wilcoxon rank sum test. The association between the methylation of each RLGS spot and MLL groups was assessed using Fisher exact test. Hierarchical cluster analysis of samples was performed by applying the Jaccard binary metric26 similarity coefficient with compact linkage method, using all the spots with at least one methylation. This study's laboratory use of cryopreserved primary AML samples was approved by the institutional review board of the Ohio State University.
Results and discussion
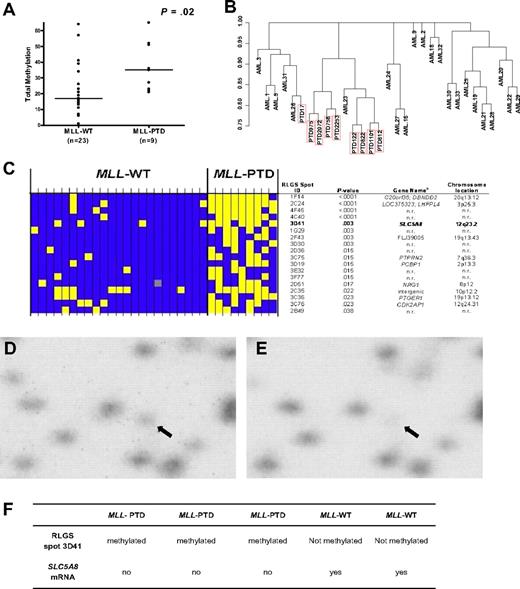
Global methylation profiles in CN-AML patients with MLL-WT (n = 23) and patients with MLL-PTD (n = 9) were obtained by RLGS. A significant difference in global DNA methylation (ie, the number of methylation events of 321 evaluable events) between patients with MLL-PTD and MLL-WT AML was observed (P = .02; Figure 1A). Unsupervised hierarchical clustering resulted in 8 of 9 MLL-PTD cases clustering together (Figure 1B). The methylation status for individual patients of the top 18 differentially expressed RLGS spots between the 2 MLL groups is shown in Figure 1C.
Differential global DNA methylation between CN-AML with and without MLL-PTD. (A) Each RLGS autoradiograph was viewed independently by 3 trained individuals who catalogued each spot loss or gain in comparison to a reference RLGS autoradiograph generated using normal donor BM DNA. RLGS data were compiled for all patients and 321 spots were evaluable across all profiles. Global DNA methylation, that is, the number of methylation events observed, was determined for each patient and a comparison of global DNA methylation between MLL-WT and MLL-PTD AML groups, was made using the Wilcoxon rank sum test. Horizontal bars represent the medians. (B) Unsupervised hierarchical clustering of patients was carried out based on RLGS spots that were methylated in at least one patient (265 spots). Jaccard binary similarity metric was used. (C) The 18 RLGS spots with the strongest association, measured by Fisher exact test, between methylation and MLL AML groups are shown. Yellow squares represent a methylated locus; blue squares represent an unmethylated locus; gray squares indicate spot was not evaluable. Columns represent individual patients and rows represent RLGS spots. RLGS spot names are shown on right. Row 5 depicts results for RLGS spot 3D41, that is, a DNA fragment corresponding to a region of the SLC5A8 promoter and exon 1. The corresponding gene names and chromosomal locations, if known and as reported in Supplemental Table 5 from Smiraglia et al27 are also shown. (D) RLGS was carried out as described in “Methods.” The area of the autoradiographs containing spot 3D41 (arrow) were scanned using a Storm 860 phosphorimager (Molecular Dynamics, Amersham Biosciences, Piscataway, NJ) and area with 3D41 enlarged (Photoshop v.8.0, Adobe Systems, San Jose, CA). Representative results are shown for the presence of 3D41 in a primary MLL-WT AML patient sample. (E) Representative RLGS results showing nearly complete loss of 3D41 in a primary MLL-PTD AML patient sample. (F) SLC5A8 mRNA detection in primary AML patient samples that exhibited loss or presence of RLGS spot 3D41.
Differential global DNA methylation between CN-AML with and without MLL-PTD. (A) Each RLGS autoradiograph was viewed independently by 3 trained individuals who catalogued each spot loss or gain in comparison to a reference RLGS autoradiograph generated using normal donor BM DNA. RLGS data were compiled for all patients and 321 spots were evaluable across all profiles. Global DNA methylation, that is, the number of methylation events observed, was determined for each patient and a comparison of global DNA methylation between MLL-WT and MLL-PTD AML groups, was made using the Wilcoxon rank sum test. Horizontal bars represent the medians. (B) Unsupervised hierarchical clustering of patients was carried out based on RLGS spots that were methylated in at least one patient (265 spots). Jaccard binary similarity metric was used. (C) The 18 RLGS spots with the strongest association, measured by Fisher exact test, between methylation and MLL AML groups are shown. Yellow squares represent a methylated locus; blue squares represent an unmethylated locus; gray squares indicate spot was not evaluable. Columns represent individual patients and rows represent RLGS spots. RLGS spot names are shown on right. Row 5 depicts results for RLGS spot 3D41, that is, a DNA fragment corresponding to a region of the SLC5A8 promoter and exon 1. The corresponding gene names and chromosomal locations, if known and as reported in Supplemental Table 5 from Smiraglia et al27 are also shown. (D) RLGS was carried out as described in “Methods.” The area of the autoradiographs containing spot 3D41 (arrow) were scanned using a Storm 860 phosphorimager (Molecular Dynamics, Amersham Biosciences, Piscataway, NJ) and area with 3D41 enlarged (Photoshop v.8.0, Adobe Systems, San Jose, CA). Representative results are shown for the presence of 3D41 in a primary MLL-WT AML patient sample. (E) Representative RLGS results showing nearly complete loss of 3D41 in a primary MLL-PTD AML patient sample. (F) SLC5A8 mRNA detection in primary AML patient samples that exhibited loss or presence of RLGS spot 3D41.
We focused on RLGS spot “3D41” that corresponded to exon 1 of the TSG SLC5A8 and that was absent in 78% of MLL-PTD AMLs compared with only 17% of MLL-WT AMLs (P = .003; Figure 1C-E). Of the patient samples with material available for further analyses, 3 (AMLs 812, 9923 and 122) with complete absence (methylation) of the RLGS spot 3D41, had 24% to 78% CpG-methylation in the SLC5A8 CpG-island region tested by bisulfite-PCR/sequencing and no detectable gene expression (Figure 1F). In contrast, 2 MLL-WT patients (AMLs 1 and 22) with presence (low/absent methylation) of the RLGS spot 3D41 and consistent bisulfite-PCR/sequencing results (0%-12% CpG island methylation), expressed SLC5A8 (Figure 1F). SLC5A8 CpG island methylation was either an absent or rare event (0%-4%) in CD34+ cells from disease-free, normal donor BM samples (n = 5; not shown).
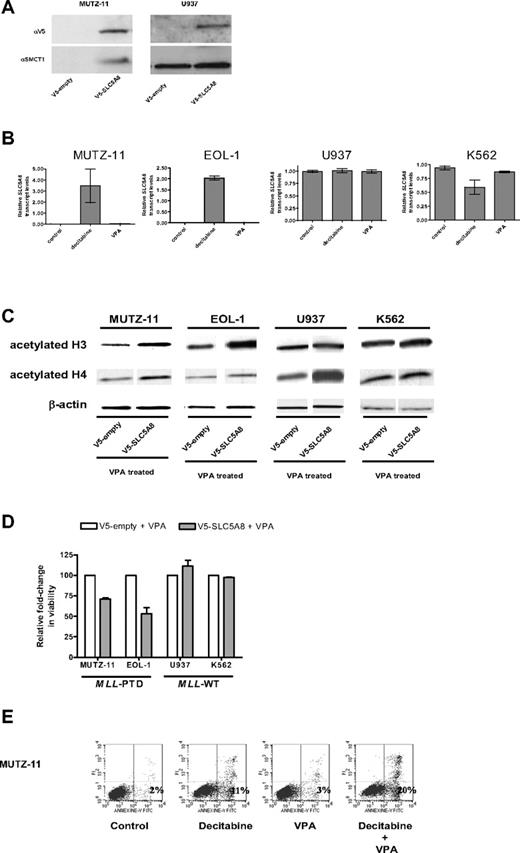
Similarly, the MLL-PTD+ MUTZ-11 and EOL-1 cell lines exhibited 92% (± 4%) and 91% (± 6%) CpG hypermethylation in the tested SLC5A8 promoter region, respectively, whereas the MLL-PTD− K562 and U937 cell lines exhibited only 43% (± 9%) and 28% (± 5%), respectively (not shown). Consistent with these results, SLC5A8 mRNA (not shown) and protein were detected only in the MLL-PTD− cells (Figure 2A). Incubation with the hypomethylating agent decitabine and not with the histone deacetylase inhibitor VPA, used here as a control, reversed the silencing of the SLC5A8 gene in MLL-PTD+ MUTZ-11 and EOL-1 cells and not in the MLL-WT K562 and U937 cells (Figure 2B).
Epigenetic silencing of SLC5A8 and functional consequences of forced expression of SLC5A8 in MLL-PTD+ cell lines. (A) Whole cell lysates were prepared and immunoblotting performed as described in “Methods.” Anti-SLC5A8 antibody (A-15) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Nucleofections with V5-empty and V5-SLC5A8 expression vectors and immunoblotting to detect the V5-epitope were performed as described in “Methods.” (B) The demethylating agent, decitabine, but not the histone deacetylase inhibitor, VPA activates SLC5A8 transcription in MLL-PTD+ AML cell lines. Cell lines were incubated in the absence or presence of the hypomethylating reagent decitabine (2.5 μM) or VPA (1 mM) for 48 or 24 hours, respectively. SLC5A8 mRNA levels were measured by real time RT-PCR using SYBR Green dye for detection (Prism 7700 SDS, Applied Biosystems, Foster City, CA). Primers were SLC5A8RT-for, 5′-TCCGAGGTCTACCGTTTTG-3′ and SLC5A8RT-rev, 5′-GGGCAGGGCATAAA-TAAC-3′. The ΔΔCt method of relative quantification was carried out. SLC5A8 mRNA levels were normalized to 18S rRNA levels. Results are presented as relative SLC5A8 transcript levels (means ± SD). (C) Forced SMCT1 expression enhances VPA-induced acetylation of histone H3 and H4. Immunoblot analyses for total acetylated histones H3 and H4 were carried out on empty vector or V5-SLC5A8 vector transfected cells. Twenty-four hours after nucleofection, 1 mM VPA was added to the cultures for an additional 24 hours. Immunoblot detection of β-actin was used as a loading control. (D) Overexpression of SMCT1 sensitizes MLL-PTD+ cells to the growth inhibitory effects of VPA. Cell lines were incubated for an additional 24 hours with or without VPA (1 mM) beginning 24 hours posttransfection. The effect on viable cell numbers was measured using the trypan blue exclusion assay and is depicted as a fold-change in relation to the appropriate VPA-treated, empty-vector transfected cells. Error bars represent SD. (E) The sequential combination of decitabine followed by VPA results in enhanced apoptosis in the MLL-PTD+ cell lines. Cells were treated as described above, except with the inclusion of a sequential combination of decitabine (48 hours) followed by VPA (24 hours), and then harvested for staining with annexin V/propidium iodide and fluorescence-activated cell sorting analysis. The percentage of cells undergoing early apoptosis (lower right quadrant) is indicated.
Epigenetic silencing of SLC5A8 and functional consequences of forced expression of SLC5A8 in MLL-PTD+ cell lines. (A) Whole cell lysates were prepared and immunoblotting performed as described in “Methods.” Anti-SLC5A8 antibody (A-15) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Nucleofections with V5-empty and V5-SLC5A8 expression vectors and immunoblotting to detect the V5-epitope were performed as described in “Methods.” (B) The demethylating agent, decitabine, but not the histone deacetylase inhibitor, VPA activates SLC5A8 transcription in MLL-PTD+ AML cell lines. Cell lines were incubated in the absence or presence of the hypomethylating reagent decitabine (2.5 μM) or VPA (1 mM) for 48 or 24 hours, respectively. SLC5A8 mRNA levels were measured by real time RT-PCR using SYBR Green dye for detection (Prism 7700 SDS, Applied Biosystems, Foster City, CA). Primers were SLC5A8RT-for, 5′-TCCGAGGTCTACCGTTTTG-3′ and SLC5A8RT-rev, 5′-GGGCAGGGCATAAA-TAAC-3′. The ΔΔCt method of relative quantification was carried out. SLC5A8 mRNA levels were normalized to 18S rRNA levels. Results are presented as relative SLC5A8 transcript levels (means ± SD). (C) Forced SMCT1 expression enhances VPA-induced acetylation of histone H3 and H4. Immunoblot analyses for total acetylated histones H3 and H4 were carried out on empty vector or V5-SLC5A8 vector transfected cells. Twenty-four hours after nucleofection, 1 mM VPA was added to the cultures for an additional 24 hours. Immunoblot detection of β-actin was used as a loading control. (D) Overexpression of SMCT1 sensitizes MLL-PTD+ cells to the growth inhibitory effects of VPA. Cell lines were incubated for an additional 24 hours with or without VPA (1 mM) beginning 24 hours posttransfection. The effect on viable cell numbers was measured using the trypan blue exclusion assay and is depicted as a fold-change in relation to the appropriate VPA-treated, empty-vector transfected cells. Error bars represent SD. (E) The sequential combination of decitabine followed by VPA results in enhanced apoptosis in the MLL-PTD+ cell lines. Cells were treated as described above, except with the inclusion of a sequential combination of decitabine (48 hours) followed by VPA (24 hours), and then harvested for staining with annexin V/propidium iodide and fluorescence-activated cell sorting analysis. The percentage of cells undergoing early apoptosis (lower right quadrant) is indicated.
To study the functional consequences of SLC5A8 reactivation, cell lines were transfected with empty vector or V5-tagged SLC5A8 expression vector and treated with VPA. As SMCT1 is a transporter of VPA into cells, we hypothesized that forced expression of SMCT1 would increase VPA pharmacologic activity. Consistent with the restored and/or enhanced function of the SMCT1, histones H3 and H4 acetylation increased with SMCT1 forced expression in all cells, including MUTZ-11 and EOL-1 cells in which the endogenous gene was constitutively silenced (Figure 2C).
While VPA was cytotoxic to both MLL-WT cell lines, MUTZ-11 and EOL-1 cell viabilities were not significantly affected by VPA under similar treatment conditions (data not shown). However, MUTZ-11 and EOL-1 cells expressing SLC5A8/SMCT1 showed reduced cell viability with VPA in comparison to the VPA treated empty-vector transfected cells (Figure 2D). Cell viability remained unchanged in similarly treated and V5-SLC5A8-transfected K562 and U937 cell lines (Figure 2D). We speculate that the enhanced VPA-induced cytotoxicity in the MLL-PTD+ MUTZ-11 and EOL-1 may be due to reexpression of the MLL-WT allele that sensitizes MLL-PTD+ cells to HDAC inhibitors.6 Consistent with this, and our previous report,6 the fraction of MLL-PTD+ cells undergoing early apoptosis was higher after incubation with the combination of decitabine followed by VPA compared with either drug alone and untreated controls (Figure 2E).
We demonstrate that MLL-PTD presence is associated with global DNA hypermethylation relative to MLL-WT AML and provide evidence that the TSG, SLC5A8, is epigenetically silenced in this molecular subset of AML. DNA methylation-induced silencing of TSGs, such as SLC5A8, may represent a second “hit” in myeloid blasts harboring MLL-PTD that itself contributes to leukemogenesis via H3K4 methylation-induced transcriptional up-regulation of genes involved in self-renewal and proliferation of hematopoietic precursors. As in colon cancer,15,18-20 SLC5A8 silencing may contribute to an aggressive phenotype in subsets of CN-AML. Indeed, although a high proportion of MLL-PTD CN-AML patients treated on newer intensive regimens exhibited long term disease-free survival, the majority of MLL-PTD patients still relapsed within 1.7 years after remission induction.10 The underlying reasons for this remain largely unknown but may also include other molecular and epigenetic defects present in these AMLs. Finally, a recent clinical strategy in AML is to overcome aberrant epigenetic events, that is, DNA methylation and histone deacetylation, both of which frequently cooperate to silence TSGs.12 Based on the data provided in this report, one could envision a sequential treatment for MLL-PTD AML consisting of the hypomethylator, decitabine, followed by the HDAC inhibitor VPA as a rational attempt to improve clinical outcome in this subset of patients.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to Hans G. Drexler for generously providing us with the MUTZ-11 cell line. The authors gratefully acknowledge sample processing and storage services provided by Ms Donna Bucci of the CALGB Leukemia Tissue Bank at The Ohio State University Comprehensive Cancer Center, Columbus.
This work was supported in part by National Cancer Institute (Frederick, MD) grants CA089341, CA096887, CA101140, CA114725, CA016058, CA077658, CA089317 and CA101956 and the Coleman Leukemia Research Foundation.
National Institutes of Health
Authorship
Contribution: S.P.W., C.P., G.M., and M.A.C. helped design this study; S.P.W., S.L., G.M., and M.A.C. helped write this report, and all authors agreed on the final version; K.M., D.M., and S.L. performed statistical analyses; C.P. and R.D. contributed reagents and intellectual expertise; S.P.W., B.H., L.Y., C.Y., L.J.R., J.W., S.L., and T.W. carried out laboratory-based research and raw data analyses; and G.M., M.A.C., and C.D.B. were directly or indirectly involved in patient care and/or sample procurement.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael A. Caligiuri, MD, 458A Starling-Loving Hall, West 10th Avenue, Columbus, Ohio, 43210; e-mail: Michael.caligiuri@osumc.edu.