Abstract
The purpose of this study is to correlate the presence of TP53 gene mutations with the clinical outcome of a cohort of patients with diffuse large B-cell lymphoma (DLBCL) assembled from 12 medical centers. TP53 mutations were identified in 102 of 477 patients, and the overall survival (OS) of patients with TP53 mutations was significantly worse than those with wild-type TP53 (P < .001). However, subsets of TP53 mutations were found to have different effects on OS. Mutations in the TP53 DNA-binding domains were the strongest predictors of poor OS (P < .001). Mutations in the Loop-Sheet-Helix and Loop-L3 were associated with significantly decreased OS (P = .002), but OS was not significantly affected by mutations in Loop-L2. A subset of missense mutations (His158, His175, Ser245, Gln248, His273, Arg280, and Arg282) in the DNA-binding domains had the worst prognosis. Multivariate analysis confirmed that the International Prognostic Index and mutations in the DNA-binding domains were independent predictors of OS. TP53 mutations also stratified patients with germinal center B cell–like DLBCL, but not nongerminal center B cell–like DLBCL, into molecularly distinct subsets with different survivals. This study shows the prognostic importance of mutations in the TP53 DNA-binding domains in patients with DLBCL.
Introduction
The TP53 tumor suppressor gene plays an important role in regulation of the cell cycle, cell proliferation, apoptosis, and genomic integrity. The p53 protein mediates cell-cycle arrest when cells experience stressful challenges such as DNA damage, hypoxia, or oncogene activation, whereas mutant p53 protein results in cell-cycle dysregulation, genomic instability, and uncontrolled proliferation of damaged cells.1,2 The TP53 gene also functions as an important mediator of tumor sensitivity to radiation therapy and chemotherapy in patients with cancer, and TP53 gene defects are frequently seen in almost every type of human cancer.3-8
Diffuse large B-cell lymphoma (DLBCL) is the most common type of non-Hodgkin lymphoma and is characterized by an aggressive clinical course.9-11 However, DLBCL exhibits considerable heterogeneity in terms of clinical, morphologic, molecular, and cytogenetic features. Recent gene expression profiling (GEP) studies have identified 3 distinct molecular subgroups of DLBCL: germinal center B cell–like (GCB), activated B cell–like (ABC), and primary mediastinal DLBCL.12-15 The GCB subgroup has a germinal center molecular signature and a better survival than does the ABC subgroup.13,16 In contrast, the molecular signature of the ABC subgroup is similar to that of mitogen-activated peripheral blood B cells. However, the molecular mechanisms underlying the differences in survival of the various subgroups of DLBCL are unclear.
The presence of TP53 mutation has been correlated with a poor response to treatment, rapid disease progression, and short survival in several cancers. A poor prognosis was found to be associated with specific mutations in the DNA-binding domain of TP53 in various cancers,17-22 and recently in DLBCL.23 In vitro studies have also shown that missense mutations in TP53 have different functional consequences and exhibit great variability in transactivation activity, with mutations in the DNA-binding domain usually resulting in loss of specific transactivation activity.24-26
In comparison to other cancers, the incidence of TP53 mutations is much lower in the hematologic malignancies.27,28 In DLBCL, most studies have reported that TP53 mutations are associated with poor overall survival (OS).23,29-37 However, those findings have not been consistent, probably reflecting the insufficient statistical power of individual studies as well as the variable classification of the mutations.32,35 Importantly, it is unclear whether some mutations carry a worse prognosis than others and whether TP53 mutations can be used as a prognostic factor to stratify the different molecular subgroups of DLBCL.12-15 To better understand the clinical consequences of TP53 mutations, we assembled clinical and molecular data from a number of reported studies to (1) correlate the structural profiles of TP53 mutations with survival in DLBCL, (2) analyze the structural profiles of TP53 mutations in the molecular subsets of DLBCL as defined by the GEP and immunohistochemical studies, and (3) delineate differences in the TP53 mutation profile in DLBCL compared with other types of human cancer. Our findings show that mutations in the TP53 DNA-binding domains are important and independent predictors of survival in DLBCL.
Methods
Establishment of collaborative groups
A total of 7 groups, representing 12 medical centers in 9 countries, agreed to participate in the study. Five groups were able to retrieve all of the necessary clinical data,23,31,33,34,36 whereas only partial data were available from 2 groups.29,30,32,35 The communicating authors from each group and the number of patients contributed to the study are shown in Table 1. The study was reviewed and approved by the Institutional Review Board at the University of Wisconsin School of Medicine and Public Health.
Patient characteristics
This study includes a total of 477 patients with DLBCL treated with CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone) or CHOP-like chemotherapy with data on TP53 gene mutation status. In 329 cases, it was possible to retrieve all or part of the clinical data, and these cases were included in the analyses of clinicopathologic features. The clinical data included the age, sex, Ann Arbor stage, tumor size, date of diagnosis, performance score, sites of extranodal involvement, serum lactate dehydrogenase (LDH) level, presence of B symptoms, International Prognostic Index (IPI), initial chemotherapy, adjuvant radiation therapy, treatment response, disease progression, survival, and the date of death or last follow-up. The patients were divided into 2 groups according to TP53 gene mutation status: mutated (21.4%) and wild type (WT; 78.6%).
TP53 mutation detection methods
Although the various TP53 mutational analyses spanned exons 2 through 11, most of the studies focused on exons 5 to 9 (Table 1). Two groups used frozen tissue from a total of 131 specimens (27%), and 5 groups used paraffin-embedded specimens from 346 specimens (73%). A total of 199 patients were screened by single-stranded conformation polymorphism (SSCP) analysis followed by DNA sequencing, 103 patients were screened by denaturing gradient gel electrophoresis (DGGE) followed by sequencing, 62 patients were screened directly by DNA sequencing, and 113 patients were screened by denaturing high-performance liquid chromatography (dHPLC) followed by sequencing. The performance of each method was validated on cell lines with known TP53 mutations before use of the method in the study of DLBCL tumors.33,38-40 Several studies have shown equivalent performance (90%-100% concordance) in mutation detection accuracy between DGGE and SSCP and between DGGE and dHPLC.38-40 No significant false-positive rate has been observed (eg, disproportionate G-A change), and confirmatory sequencing after each screening method would mitigate any false-positive results. The direct sequencing method has been shown to have similar sensitivity in cases with tumor cells constituting greater than 40% of the tissue cellularity (M.S.-B. and M.A.P., oral communication, June 2008). The types of TP53 gene mutation (point or frameshift) and the sites of mutation (codon, exon, functional domain, or conserved area) were identified. For tumors with more than one mutation, the data for each mutation was recorded as a separate entry but analyzed as a single event for survival.
Classification of TP53 mutations for survival analysis
Mutations in exons 5 to 9 and the intervening introns were classified and analyzed according to their location, nature, and potential effect on protein structure and activity.41-43 Silent mutations or single nucleotide polymorphisms (5 cases) that did not change the amino acid sequence were not included in the analysis. TP53 mutations were categorized in several ways for survival analysis in comparison to the WT-TP53 group as follows: (1) Missense mutations were defined as mutations resulting in a single amino acid change. These included any TP53 missense mutation responsible for specific structural or functional characteristics, missense mutations affecting residues involved in direct DNA interaction (Ala119, Lys120, Ser121, Asn239, Ser241, Met243, Asn247, Arg248, Arg273, Cys275, Ala276, Cys277, Arg280, Arg282, and Arg283) and those at hot spot codons (Arg175, Arg196, Arg213, Gly245, Arg248, Arg273, and Arg282). (2) Analysis was also performed for missense mutations according to their positions and DNA-binding domain structures in a 3-dimensional crystal model. The DNA-binding domain included Loop-L2 (codons 164-194), which is believed to enhance the binding affinity of TP53 with the DNA helix under physiologic conditions; Loop-L3 (codons 237-250), which is involved in interaction with the DNA minor groove; and the LSH motif (Loop-Sheet-Helix; codons 119-135 and 272-287), which is involved in DNA binding in the major groove. The survival of patients with mutations in the DNA-binding domains was compared with patients with mutations in non–DNA-binding domains. In addition, missense mutations affecting residues involved in direct DNA contact (Asn239, Asn247, Arg248, Arg273, Arg280, and Arg283) and the zinc-binding site (Cys176, His179, Cys238, and Cys242) were evaluated by comparison to the WT-TP53 group. (3) Conserved and nonconserved missense mutations were defined as those with a change in an amino acid residue that is conserved or not conserved, respectively, during evolution (conservation based on CLUSTALW alignment of 35 p53 protein sequences from vertebrate species). These include amino acids in highly conserved areas II (codons 117-142), III (codons 171-181), IV (codons 234-258), and V (codons 270-286).41,42 (4) Nonmissense mutations were defined as any mutation other than a missense mutation, including mutations introducing a stop codon, deletions, or insertions (in-frame or frame-shift), and substitutions at splice sites.5 Missense mutations were also grouped into functional classes, based on yeast functional assays, according to their capacity to transactivate the promoters of several p53 target genes (WAF1, MDM2, BAX, GADD45, p53-AIP1, and Noxa genes) using a published database.26 Three functional groups were defined as follows: inactive, partially active, and active, with their functionality compared with the WT-TP53 group. (5) Finally, structural classes were classified based on the effect of the mutation on the nature of amino acid charge: polar neutral, apolar neutral, basic, and acidic, as well as the type of amino acid change according to the lateral group.
Immunohistochemistry
Immunohistochemical studies were performed to classify DLBCL into GCB and non-GCB types in 85 cases with the use of the following antibody panel (CD20, CD10, MUM-1, BCL-6) and the algorithm of Hans et al.44 GEP analysis was previously described, and the results were used for molecular classification of 113 additional cases.13,23
Statistical and survival analyses
Associations between categorical variables and TP53 mutation status were assessed using the chi-square analysis or Fisher exact test. A nonparametric Wilcoxon rank sum test was used to compare continuous variables between groups. OS was defined as the time from diagnosis to death resulting from any cause or, for patients remaining alive, the time from diagnosis to last contact. Patients who were alive or lost in follow-up were classified as censored observations. The Kaplan-Meier method was used for univariate survival analysis, and the log-rank test was used to assess the difference between survival curves. Multivariate Cox proportional hazard models were used to evaluate which variables were independent prognostic factors for OS.45,46 Predictive variables were selected by forward stepwise selection with a P value less than .05. The proportional hazard assumption was verified by examining the Schoenfeld residuals. All P values are 2-sided. Statistical analyses were performed with SAS software version 8.2 (SAS Institute, Cary, NC).
Results
Mutation profile of the TP53 gene
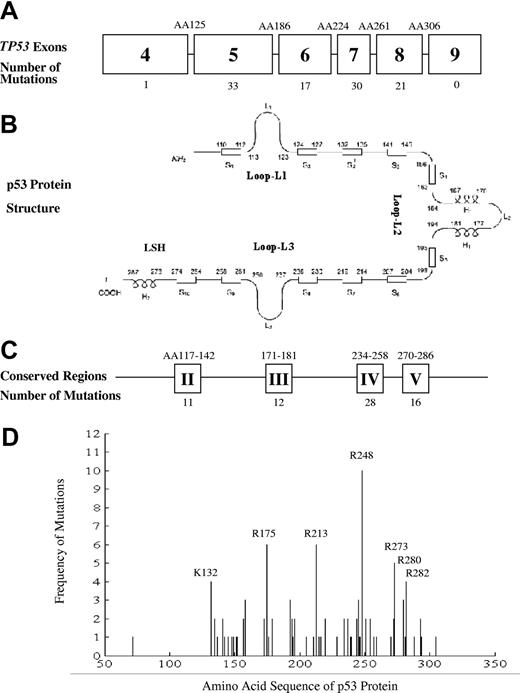
The frequency and types of TP53 mutations in DLBCL are shown in Table 1 and Figure 1. The TP53 gene was screened or sequenced at exons 5 to 8 in 244 tumors, exons 5 to 9 in 199 tumors, and exons 2 to 11 in 34 tumors. Of the 477 cases, 102 (21.4%) had TP53 mutations, including 92 single nucleotide missense mutations and 10 other nonmissense mutations (5 single nucleotide deletions, 4 multiple nucleotide deletions, and one 10 nucleotide insertion before Arg283). Two cases showed missense mutations with single amino acid changes occurring in both alleles. Of these 2 cases, the primary mutation was identified in exon 5, with the other allele having a missense mutation in exon 6 or 7 (His193 and Ile254, respectively). The distribution of the 102 mutations was 1 in exon 4, 33 in exon 5, 17 in exon 6, 30 in exon 7, and 21 in exon 8 (Table 1; Figure 1A). No mutations were identified in exon 9. Silent mutations were identified in 5 cases at Ser149, Arg213, Gly244, and Arg248.
Schematic representation of the TP53 gene and its mutations in diffuse large B-cell lymphoma. (A) The distribution of TP53 mutations in exons 4 to 9, (B) their relation to p53 protein structure, (C) the mutations in conserved regions, and (D) the distribution and frequency of TP53 mutations with peaks at known hot spot exons depicted.
Schematic representation of the TP53 gene and its mutations in diffuse large B-cell lymphoma. (A) The distribution of TP53 mutations in exons 4 to 9, (B) their relation to p53 protein structure, (C) the mutations in conserved regions, and (D) the distribution and frequency of TP53 mutations with peaks at known hot spot exons depicted.
When the TP53 mutation distribution pattern was analyzed, 68 (66%) of 102 mutations were found in codons involved in DNA-binding motifs of the central core domain. These included 23 mutations in Loop-L3 (codons 237-250) that interacts with the DNA minor groove, 26 mutations in the LSH helix motif (codons 119-135 and 272-287) that interacts with the DNA major groove, and 19 mutations in Loop-L2 that enhances the binding affinity of TP53 with the DNA helix under physiologic conditions (Figure 1B).41,42 In 37 cases, the mutations were localized to codons that have been previously described as TP53 hot spots in non-Hodgkin lymphoma and other cancers (Arg175, Arg196, Arg213, Gly245 Arg248, Arg273, and Arg282).47-51 Amino acid residues in the hot spot exons were mutated at higher frequencies (2- to 10-fold) than were other exons (Figure 1D). Ninety-two missense mutations could be classified according to their charge and side-chain structures, and these included 8 polar neutral, 42 apolar neutral, 20 basic, and 22 acidic mutations. In 3 cases, the mutations were localized to codons involved in the zinc-binding site (Cys176, His179, Cys238, and Cys242).
Mutations in 67 cases were localized to highly conserved areas (Figure 1C): 11 mutations in area II (codons 117-142), 12 mutations in area III (codons 171-181), 28 mutations in area IV (codons 234-258), and 16 mutations in area V (codons 270-286). Eighty-three of the 92 missense mutations could be classified by functionality based on the capacity to transactivate promoters of several p53 target genes, and these included 68 inactive mutations, 9 partially active mutations, and 6 active mutations.
Correlation between TP53 mutations and clinical features
In 329 of 477 patients, clinical data were available for analysis (292 with complete data and 37 with partial data). The patients were treated between 1983 and 2002 and were followed for a median of 78 months (range, 7-229 months). Of the 329 cases, only 16 cases had a follow-up period of less than 2 years (range, 7-22 months), and these constituted only 4.9% of the analyzed dataset. Table 2 shows the clinical and pathologic features of the patients according to TP53 gene mutation status. The median ages at diagnosis were 51 years (range, 13-88 years) and 64 years (range, 14-91 years) for the mutant and WT groups, respectively. Of the 292 cases with complete clinical data, 98 cases had a single nucleotide mutation or deletion in exons 5 to 8, one case had a deletion in exon 4, another had an insertional mutation in exon 8, and 2 cases had mutations in introns.
TP53 mutations were significantly associated with a younger age at diagnosis (P = .003), high serum LDH level (P < .001), tumor at least 10 cm (P = .014), and a high IPI risk group (P = .035), and these patients were less likely to have received radiation therapy (P = .004; Table 2). Thirty-four (57%) of 60 TP53 mutated cases with treatment response data achieved a complete remission (CR), 10 (17%) had a partial response (PR), 6 (10%) had no response, and 10 (17%) had progressive disease (PD) after initial chemotherapy. The patients with mutations exhibited poorer response to this treatment than did patients with WT-TP53 (Table 2).
Prognostic value of TP53 gene status
Patients with a TP53 mutation had a 1.9-fold increased risk of DLBCL-specific death after treatment than did patients with WT-TP53 in univariate analysis (Table 3). The 5 components of the IPI and B symptoms were also found to be predictive. In multivariate analysis, TP53 mutation and a high IPI score were found to be independent predictors of poor survival (Table 4). Mutation analysis also showed that patients with a TP53 mutation in Loop-L3 and the LSH motif had hazard ratios for DLBCL-specific death of 2.3-fold and 3.4-fold, respectively (Table 4).
Prognostic value of the DNA-binding domain mutations
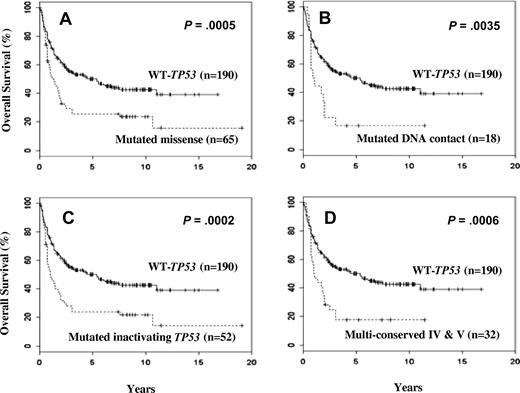
Kaplan-Meier analysis showed that patients with any TP53 mutation had a poor OS (median survival, 1.3 year) compared with patients without mutation (median survival, 4.5 years; P < .001; Figure 2A,B). Based on location, mutations in the DNA-binding domains were the strongest predictors of poor OS (median survival, 1.0 year; P < .001; Figure 2C). In contrast, patients with mutations in the non–DNA-binding domains had an OS similar to patients with WT-TP53 (P = .158; Figure 2D). The 5-year OS was only 19% for patients with mutations of the DNA-binding domain compared with 45% for patients with WT-TP53. The complete remission rate was only 54% for patients with mutations of the DNA-binding domain compared with 69% for patients with WT-TP53 (Table 5).
Kaplan-Meier survival analysis of patients with diffuse large B-cell lymphoma stratified by TP53 mutation status. (A) Distribution pattern of TP53 mutations in the central core domain model designed from published crystal structure41 (red indicates patients with poor survival; green, patients with survival similar to WT group; blue, survival data unavailable). (B) Overall survival of patients with TP53 mutations versus those with WT-TP53. (C) Overall survival of patients with DNA-binding domain mutations versus those with WT-TP53. (D) Overall survival of patients with non–DNA-binding domain mutations versus those with WT-TP53.
Kaplan-Meier survival analysis of patients with diffuse large B-cell lymphoma stratified by TP53 mutation status. (A) Distribution pattern of TP53 mutations in the central core domain model designed from published crystal structure41 (red indicates patients with poor survival; green, patients with survival similar to WT group; blue, survival data unavailable). (B) Overall survival of patients with TP53 mutations versus those with WT-TP53. (C) Overall survival of patients with DNA-binding domain mutations versus those with WT-TP53. (D) Overall survival of patients with non–DNA-binding domain mutations versus those with WT-TP53.
In Table 6, the clinical features of the patients with DNA-binding mutations and non–DNA-binding mutations are compared. Univariate analysis showed that the 2 groups did not differ significantly in clinical features except that patients with DNA-binding mutations tended to present with a larger tumor size. Twenty-one (54%) of 41 patients with DNA-binding mutations achieved a CR, 8 (21%) had a PR, 2 (5%) had no response, and 10 (17%) had PD after initial treatment.
When the subsets of mutations in the DNA-binding domains were examined, patients with mutations in the Loop-L3 domain or LSH motif had a significantly decreased OS (median survival, 1.0 year for each of the groups, P = .012 and P = .002), compared with patients with WT-TP53 (median survival, 4.5 years; Table 7). However, the OS was not significantly decreased for patients with mutations in the Loop-L2 domain (Figure 3A-C; Table 7).
Prognostic significance of TP53 mutations in specific DNA-binding motifs. (A) Overall survival of patients with mutations in the Loop-L2 domain. (B) Overall survival of patients with mutations in the Loop-L3 motif. (C) Overall survival of patients with mutations in the LSH domain.
Prognostic significance of TP53 mutations in specific DNA-binding motifs. (A) Overall survival of patients with mutations in the Loop-L2 domain. (B) Overall survival of patients with mutations in the Loop-L3 motif. (C) Overall survival of patients with mutations in the LSH domain.
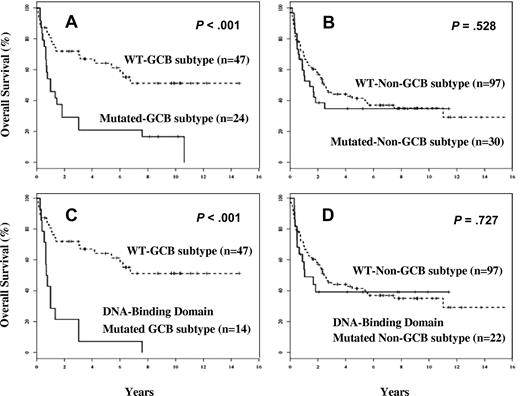
A total of 198 cases were classified into GCB or non-GCB subgroups according to GEP (113 cases)13,23 or by immunohistochemical stains (85 cases).44 In the GCB subgroup, mutations were found in 24 (34%) of 71 cases, whereas 30 (24%) of 127 cases were mutated in the non-GCB subgroup (P = .18). The TP53 mutation profile stratified GCB-DLBCL into molecularly distinct subsets and predicted for poor OS in those with TP53 mutations (P < .001; Figure 4A). However, TP53 mutations did not predict for survival in the non-GCB subgroup (Figure 4B). The predictive value of TP53 mutations in GCB-DLBCL was mainly due to the mutations in the DNA-binding domains (Figure 4C), but these mutations were not predictive in the non-GCB subgroups (Figure 4D).
Kaplan-Meier survival analysis by TP53 mutations in the molecular subtypes of diffuse large B-cell lymphoma. (A) Overall survival of patients with GCB-DLBCL with TP53 mutations. (B) Overall survival of patients with non–GCB-DLBCL with TP53 mutations. (C) Overall survival of patients with GCB-DLBCL with DNA-binding domain mutations. (D) Overall survival of patients with non–GCB-DLBCL with DNA-binding domain mutations.
Kaplan-Meier survival analysis by TP53 mutations in the molecular subtypes of diffuse large B-cell lymphoma. (A) Overall survival of patients with GCB-DLBCL with TP53 mutations. (B) Overall survival of patients with non–GCB-DLBCL with TP53 mutations. (C) Overall survival of patients with GCB-DLBCL with DNA-binding domain mutations. (D) Overall survival of patients with non–GCB-DLBCL with DNA-binding domain mutations.
Prognostic values of structural subsets of TP53 mutations
Mutations in exons 5 to 9 were classified into different subsets according to the effect or location of the mutation in the primary sequence or tertiary structure of the p53 protein. Cases with missense mutations as a group had a poor OS compared with the WT-TP53 cases (P < .001; Figure 5A) but not other mutations (nonmissense, P = .418).
Kaplan-Meier survival analysis of patients with diffuse large B-cell lymphoma defined by functional or structural subsets of TP53 mutations. (A) Overall survival of patients with missense TP53 mutations. (B) Overall survival of patients with TP53 mutations in direct DNA-contact codons. (C) Overall survival of patients with inactivating TP53 mutations defined by yeast functional assays. (D) Overall survival of patients with TP53 mutations in the conserved regions IV and V.
Kaplan-Meier survival analysis of patients with diffuse large B-cell lymphoma defined by functional or structural subsets of TP53 mutations. (A) Overall survival of patients with missense TP53 mutations. (B) Overall survival of patients with TP53 mutations in direct DNA-contact codons. (C) Overall survival of patients with inactivating TP53 mutations defined by yeast functional assays. (D) Overall survival of patients with TP53 mutations in the conserved regions IV and V.
The presence of TP53 mutations at direct DNA contact sites was also associated with poor median survival and OS compared with the WT-TP53 cases (P = .004; Figure 5B). Nonfunctional inactivating mutations as defined by yeast-based assays also predicted for poor OS compared with the WT-TP53 (P < .001; Figure 5C).
The mutations were also analyzed based on the conserved or nonconserved status. TP53 mutations in conserved regions IV and V, but not II and III, were associated with poor OS compared with mutations in the nonconserved regions or WT-TP53 (P < .001; Figure 5D). The structural distribution of mutations in these 2 regions overlapped significantly with those in the DNA-binding domain; thus, the survival curve for these 2 regions is similar to that seen for patients with the DNA-binding domain mutations (Figure 2C).
Approximately 36% of these TP53 mutations affect only 6 hot spot residues (His175, Ser245, Gln248, Arg249, His273, and Arg282), and survival analysis showed that the presence of any TP53 mutation at hot spots correlated with poor OS. These patients had a 5-year OS of only 21% (P = .002) and a median survival of less than 1 year. Some mutations (Tyr163 and Arg249), which are hot spots in other cancers, were not identified in DLBCL. A small group of missense mutants in the DNA-binding domains (His175, Ser245, Gln248, His273, Arg280, and Arg282) overlapped with the hot spot codons, and they were also associated with significantly decreased survival. The effect of these mutations was relatively homogeneous with OS of less than 5 years in the majority of the patients and a median survival of less than 1 year.
Discussion
In this study, we correlated the clinical and mutation data on 477 cases from several reported studies to investigate the prognostic significance of somatic TP53 mutations in DLBCL.23,29-37 Most studies of TP53 mutations in DLBCL have focused on exons 5 to 9, and these exons have been shown to contain greater than 90% of the mutations reported in different types of lymphomas.47-51 The previous analysis of exons 2 to 11 in 34 cases in this series showed the absence of mutations outside of exons 5 to 9, indicating that analysis of exons 5 to 9 and their splice junctions appears adequate in DLBCL.31
This study extends our initial observations in DLBCL23 and confirms that the presence of TP53 mutations is an independent predictor of OS in patients with DLBCL. The relative risk of death from DLBCL after standard CHOP or CHOP-like chemotherapy for patients with TP53 mutations in exons 5 to 8 was 2-fold higher than for patients with WT-TP53. Interestingly, TP53 mutations in the DNA-binding domains were associated with a significantly worse OS than mutations in non–DNA-binding domains. This finding suggests that these mutations result in loss of function, either because of direct loss of DNA contact or because of the conformation change that does not allow p53 to recognize its DNA targets. Therefore, the domain location of a TP53 mutation plays a critical role in determining the clinical outcome in DLBCL.
Biologic studies have shown that TP53 mutations are heterogeneous in their function; therefore, the different functions may lead to different clinical outcomes. Functional studies of some TP53 mutants in tumor cell lines have shown loss of transcriptional activity (p21Waf1 up-regulation) and defects in the capacity to induce cell-cycle arrest or apoptosis.52-54 Our study shows that mutations in the Loop-L3 and LSH motifs are the strongest predictors of poor survival in DLBCL. Similarly, mutations affecting the Loop-L3 and LSH motifs have been reported to predict a poor outcome, with doxorubicin resistance, in breast and ovarian tumors.17-22,55 Colorectal cancers with TP53 mutations in conserved regions or affecting Loop-L3 are poorly differentiated and more aggressive than those with other mutations.18,21 In head and neck squamous cell carcinoma, TP53 mutations in direct DNA contact areas resulted in accelerated tumor progression and reduced therapeutic responsiveness.56,57 Mutations in the Loop-L2 motif do not appear to be prognostically important in DLBCL; however, this observation has not been reported in other human cancers.17-22 This finding may not be representative of DLBCL because of the low number cases with the Loop-L2 mutations in this series. Therefore, further study is needed to evaluate the role and the predictive value of Loop-L2 mutations in DLBCL.
Besides a loss of function of mutated p53, several studies suggest that a gain of function of mutant p53 also confers tumorigenetic potential by enhancing tumor cell growth or resistance to drug-induced apoptosis, thereby decreasing patient survival.58-61 In DLBCL, we found that several missense mutants in the DNA-binding domains (His158, His175, Ser245, Gln248, His273, Arg280, and Arg282) correlated with the highest mortality rate (median survival, 0.67 year). Blandino et al60 have shown that particular TP53 mutants (eg, His175 and His179) may give tumor cells a selective survival advantage during chemotherapy.60 A recent study by Song et al61 using a humanized TP53 knock-in mouse model showed that TP53-Gln248 mice rapidly developed certain types of cancers not commonly seen in TP53−/− mice, indicating that some TP53 gain-of-function mutants can promote tumorigenesis.
The gain-of-function of some TP53 mutations is also believed to result in poor patient survival through direct or indirect effects on the expression of genes involved in cancer progression, such as growth regulation, metabolism, angiogenesis, drug resistance, and genomic instability.58,59 Some TP53 mutants seen in our cases of DLBCL (eg, His175, Trp248, His273, and Gly281) have previously been found to activate the expression of proliferating cell nuclear antigen62-64 and to negatively regulate CD95/Fas/Apol (a death receptor pathway) resulting in antiapoptotic effects.65 Several gain-of-function mutants (eg, Ala143, His175, Trp248, His273, and Gly281) observed in our cases of DLBCL can also result in mitogenic effects by stimulating the expression of growth factors or growth factor receptors, such as insulin-like growth factor 1 receptor and epidermal growth factor receptor.63,66-69 In transfection assays, Ludes-Meyers et al66 found that some mutants (His175, Trp248, His273, and Gly281) induce vascular growth factor receptor expression and play a pivotal role during neoangiogenesis and tumor growth. Interestingly, several mutants (His175, Trp248, His273, and Gly281) were also found to up-regulate c-MYC in vitro, in contrast to WT-p53, which repressed c-MYC expression.70 These mutants have also been found to induce MDR-1 gene expression, thereby contributing to chemotherapy resistance in tumor cell lines.62,70-72 It appears that individual TP53 mutants share some but not all transcriptional targets and, therefore, may result in distinct tumor phenotypes. Of particular interest, Scian et al73 recently found that mutant p53 can increase NF-κB activity and protect cells against chemotherapy-induced death. A significant correlation was also shown between the presence of endogenous mutated p53 protein and constitutive NF-κB activation in human tumors.73 These observations suggest that particular mutations in the DNA-binding domain can exhibit either the dominant-negative effects or gain-of-function effects or both that are ultimately responsible for the survival of lymphoma cells and the poor survival of patients with DLBCL.
Another effect of some mutations (eg, His175, Gly245, Arg248, Arg273, and Arg283) is the production of mutant p53 proteins that inhibit the tumor-suppressive activities of p63 and p73 in vitro.74-76 These observations have also been confirmed in mouse models showing that mutant p53 proteins (eg, His175, Thr170, Phe270, Arg273) promote tumorigenesis through down-regulation of p63/p73.74,75 Therefore, efforts aimed at liberating p63/p73 from the blockade by mutant p53 might have therapeutic value in cancer treatment. Future studies will be needed to elucidate the importance of mutant TP53 on its family members, p63 and p73, in DLBCL.
The development of DNA microarray technology has provided the opportunity for a genome-wide approach to the study of DLBCL.12-16,77 Previously, 2 molecularly distinct subgroups of DLBCL were identified: GCB-DLBCL and ABC-DLBCL. In our previous study, we could not show that TP53 mutations stratified patients prognostically within the GCB and ABC subgroups.23 In the current series, however, we showed that the TP53 mutations do stratify GCB-DLBCL, but not non–GCB-DLBCL, into distinct subsets with a different OS. Therefore, TP53 mutation and microarray analysis both contribute to the risk stratification of patients with DLBCL. Our study strongly indicates that identification of TP53 mutations in the DNA-binding domains will be important for developing targeted therapy directed at this critical tumor suppressor pathway in patients with DLBCL.78,79
Recently, large randomized clinical trials have shown that the addition of rituximab to standard CHOP or CHOP-like therapy has improved the survival of patients with DLBCL.80,81 However, preliminary studies from Farinha et al82 and our group (K.H.Y. et al, unpublished data, August 2008) suggest that TP53 mutations will continue to be predictive of poor OS in the rituximab-CHOP era. If large studies confirm the adverse effect of TP53 mutations in rituximab-CHOP–treated patients with DLBCL, novel therapeutic strategies will be needed. Our work provides the framework to test hypotheses about the type and location of TP53 mutations in future studies of rituximab-CHOP–treated DLBCL.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Megali Olivier from the International Agency for Research on Cancer, World Health Organization, for advice on the functional classification of TP53 mutations; Dr Qiang Cui at the University of Wisconsin School of Medicine, Drs Leo Kinarsky and Simon Sherman at the University of Nebraska Medical Cener for p53 structural analysis; Dr Xiao Li at the University of Nebraska Medical Center for statistical consultation; and Drs Kazunori Kanehira and Korise Rasmusson at the University of Wisconsin School of Medicine for data assembly and manuscript preparation.
This work was supported by grants from the US Public Health Service (grants CA36727 and CA84967) awarded by the National Cancer Institute, Department of Health and Human Services and by awards from the Gundersen Medical Foundation and the University of Wisconsin Paul P. Carbone Comprehensive Cancer Center. T.C.G. is a Lymphoma Research Foundation Mantle Cell Lymphoma Program Research Grantee.
National Institutes of Health
Authorship
Contribution: K.H.Y. designed the study, performed research, collected and analyzed data, and wrote the paper; K.L., M.B.M., G.W.B.C., M.S.-B., F.R.K., C.H., P.G., M.A.P., E.C., J.D., R.D.G., A.R., L.R., J.H., R.M.B., E.S.J., W.H.W., L.M.S., and J.M.V. contributed vital new reagents or analytical tools and collected data; J.C.E., A.H.Y., W.M.R., and B.S.K., analyzed data; T.D.O. and J.S.M. analyzed data and assisted with the design of the study and the paper writing; and W.C.C., D.D.W., and T.C.G. contributed vital new reagents or analytical tools, collected and analyzed data, and assisted with the design of the study and the paper writing.
A list of the members of the TP53-DLBCL International Collaborative Study is available on the Blood website; see the Supplemental Appendix link at the top of the online article.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ken H. Young, Department of Pathology and Laboratory Medicine, the University of Wisconsin Paul P. Carbone Comprehensive Cancer Center, University of Wisconsin School of Medicine and Public Health, Madison, WI 53792-2472; e-mail: khyoung@wisc.edu.