Abstract
The iron regulatory hormone hepcidin is transcriptionally up-regulated in response to iron loading, but the mechanisms by which iron levels are sensed are not well understood. Large-scale genetic screens in the zebrafish have resulted in the identification of hypochromic anemia mutants with a range of mutations affecting conserved pathways in iron metabolism and heme synthesis. We hypothesized that transferrin plays a critical role both in iron transport and in regulating hepcidin expression in zebrafish embryos. Here we report the identification and characterization of the zebrafish hypochromic anemia mutant, gavi, which exhibits transferrin deficiency due to mutations in transferrin-a. Morpholino knockdown of transferrin-a in wild-type embryos reproduced the anemia phenotype and decreased somite and terminal gut iron staining, while coinjection of transferrin-a cRNA partially restored these defects. Embryos with transferrin-a or transferrin receptor 2 (TfR2) deficiency exhibited low levels of hepcidin expression, however anemia, in the absence of a defect in the transferrin pathway, failed to impair hepcidin expression. These data indicate that transferrin-a transports iron and that hepcidin expression is regulated by a transferrin-a–dependent pathway in the zebrafish embryo.
Introduction
Transferrin, a serum protein with 2 iron binding sites, is the major carrier of ferric iron in human plasma and is required to deliver iron to developing erythroid and other cells. Iron-bound holotransferrin binds transferrin receptor 1 (TfR1) on the surface of the cell, resulting in endocytosis of an endosome containing both transferrin and TfR1 (reviewed in Aisen1). After acidification of the endosome, ferrous iron exits the endosome via the divalent metal transporter (DMT1).2,3 Transferrin receptor 2 (TfR2), a homolog of TfR1, with restricted expression to hepatocytes and erythroid cells,4 binds transferrin at lower affinity than TfR15,6 and is involved in the regulation of iron homeostasis
As humans have no active means of iron excretion, maintaining iron homeostasis requires adjustment of iron absorption to reflect the body's iron stores. Hepcidin is a peptide hormone, primarily expressed in the liver,7 which modulates iron absorption and iron delivery to erythrocytes by binding the iron transporter ferroportin (fpn), resulting in internalization and degradation of fpn.8 In mammalian models, hepcidin is transcriptionally up-regulated in response to inflammation9,10 or iron overload11 and down-regulated in response to anemia, iron deficiency, or hypoxia.9 Although mutations in HFE,12 transferrin receptor 2 (TFR2),13 hepcidin (HAMP),14 or hemojuvelin (HJV or HFE2)15 have each been associated with hemochromatosis and impaired hepcidin expression,16 the mechanisms by which iron overload is sensed and hepcidin is regulated are not completely understood.
The zebrafish, Danio rerio, provides an excellent system for the identification and analysis of genes involved in iron metabolism and erythroid development. Large-scale genetic screens for embryonic hypochromic anemia have identified recessive mutations in genes affecting iron acquisition and use including DMT1,17 transferrin receptor 1a (TfR1a),18 and ferroportin1 (fpn1).19 Zebrafish embryos at 48 to 72 hours postfertilization (hpf), provide an excellent in vivo model for the study of hepcidin gene expression. At this stage, zebrafish embryos do not ingest food and are kept free of inflammatory stimuli, reducing the variables that may affect hepcidin expression. Our previous work demonstrated that fpn1 is required for normal cycling of iron through the reticuloendothelial system in the zebrafish and that induction of hepcidin expression in response to iron loading occurs 18 hours after injection of zebrafish embryos at 48 hpf with iron dextran.20 The zebrafish hepcidin peptide is 52% identical to human hepcidin and has been shown to mediate degradation of ferroportin and cellular iron retention in vitro.21
In this report, we identify a zebrafish strain, gavi (gav), with mutations in zebrafish transferrin (transferrin-a), which exhibits reduced transferrin-a expression and impaired hemoglobin production. We demonstrate that morpholino knockdown of transferrin-a reproduces the anemia phenotype and reduces nonheme iron stores in the somites of the embryo, while overexpression of transferrin-a partially restores hemoglobin production and iron stores in the somites and terminal gut. We also compare the effects of DMT1, TfR1a, TfR1b, TfR2, and transferrin-a deficiency on hepcidin expression in zebrafish embryos. We demonstrate that both transferrin-a and TfR2 are required for hepcidin expression in zebrafish embryos, while TfR1a and DMT1 are required for increased hepcidin expression in response to iron loading. These data support the hypothesis that transferrin-a is required for iron transport from the yolk sac to the embryo and that transferrin-a participates with TfR2 in the regulation of hepcidin expression in zebrafish embryos.
Methods
Zebrafish strains, maintenance, and determination of genotype
Zebrafish were maintained as described.22 Ethical approval was obtained from the Institutional Animal Care and Use Committees of Children's Hospital and Beth Israel Deaconess Medical Center. GavIT029 and gavHE067 were identified as mutants with hypochromic anemia in a large-scale ENU-mutagenesis screen conducted by the Tübingen 2000 consortium. Genetic mapping strains were created by mating gav/Tü heterozygotes to the polymorphic WIK strain. Embryos were collected from pairwise matings of mapping strain gav heterozygotes and scored at 72 hpf for hypochromic anemia. Genomic DNA extraction from individual embryos and bulk segregant analysis were performed as described23 using primers designed to amplify SSLP markers from the Massachusetts General Hospital Zebrafish Server website (http://zebrafish.mgh.harvard.edu). The gavIT029 and gavHE067mutations were amplified from genomic DNA using the following primer pairs: IT029 E4F2 5′-CCCCAATCTGATTCAGTTCC-3′ and IT029 E4R1 5′-TGCCCTGGTGACAACTCTTA-3′, and HE067 F9 5′-CAGATGCTTCCAAAATGCACGACTCC-3′ and HE067 R6 5′-ACCATGCAATCCCAGGTCGGTCAA-3′. For amplification of the aberrant splice forms of transferrin-a in the mutants, RNA was extracted from pools of mutants using the RNeasy kit (QIAGEN, Valencia, CA), cDNA was generated as described,20 and PCR was performed using the primers trf F 5′-GCCAGAAATCGCTGGTGAAG-3′ and trf R 5′-GAGCTAATACAGTTGCCAAAAGTGC-3′. PCR products were purified and sequenced directly. For characterization of gene expression, the following zebrafish anemia alleles were used: ciaTu25f,18 cdyTe216,17 and gavIT029.
Morpholino injection and cRNA injection
Antisense morpholino oligonucleotides,24,25 obtained from Gene Tools (Philomath, OR), were designed either to interfere with translation or to impair appropriate splicing of transcripts (morpholino sequences shown in Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Morpholino sequences for TfR1a, TfR1b, and GATA1 were as described.18,26 For each transferrin-a morpholino (MO), a mismatch control morpholino was synthesized with identical sequence, except for 5 nucleotide substitutions. Morpholinos that did not produce an anemia phenotype were tagged with 3′-fluorescein to indicate adequacy of injection. Embryos were injected at the one-cell stage with 3 nanoliters in 1× Danieau medium, supplemented with phenol red. For single morpholino injections, the concentration was 0.25 mM, or 0.2 mM for GATA1 MO, while for combination morpholino injections, the injection solution contained 0.125 mM of each morpholino. For cRNA injections, zebrafish transferrin-a was cloned into the pCS2 + expression vector. The vector was digested with NotI, and the sense cRNA was synthesized using the SP6 mMachine RNA transcription kit (Ambion, Austin, TX). TfR1a cRNA was prepared as described.18 The cRNA was injected at a concentration of 1600 ng/microliter, with or without Trf-MO2 (0.0625 mM).
Whole mount in situ hybridization
Whole mount in situ hybridizations were performed as previously described.27 The development of endogenous pigments was inhibited by supplementing the embryo medium with 1-phenyl-2-thiourea (PTU) at a final concentration of 0.2 mM. The following antisense riboprobes were generated for use in the in situ hybridizations: transferrin-a, TfR2, hepcidin, and liver fatty acid binding protein (LFABP). Transferrin-a and partial TfR2 were each cloned into pCS2 +. The vectors were digested with EcoRI, and antisense riboprobes were synthesized from the T7 promoter. A full-length hepcidin clone in PCRII-Topo (Invitrogen, Carlsbad, CA) was digested with NotI, and the antisense riboprobe was synthesized from the SP6 promoter. The LFABP plasmid was a gift from W. Goessling. The antisense riboprobe was generated by digesting with SalI and transcribing from the T7 promoter. Photomicrographs of in situ hybridizations were obtained using a BX51 compound microscope (Olympus, Center Valley, PA) at 100× magnification using Nomarski optics and a Q-capture 5 digital camera (QImaging, Surrey, BC).
Whole mount embryo staining for hemoglobin and iron
Live anesthetized embryos were stained for hemoglobin with o-dianisidine, as described.28 Diaminobenzidine (DAB)–enhanced staining for ferric iron was performed as described.29 After fixation in 4% paraformaldehyde-phosphate buffered saline (PBS) and dehydration in methanol, the embryos were incubated in 2.5% potassium ferrocyanide/0.25 M HCl (no. HT20-1KT; Sigma-Aldrich, St Louis, MO) for 30 minutes at room temperature. The embryos were rinsed 3 times in phosphate buffered Tween (PBT), followed by incubation in 0.3% H2O2 in methanol for 20 minutes at room temperature. The embryos were rinsed twice in PBT and then incubated in DAB substrate for 5 to 7 minutes at room temperature. The DAB substrate was generated by dissolving one tablet of Sigma Fast 3,3′-DAB tetrahydrochloride with cobalt chloride enhancer (no. D-0426, Sigma-Aldrich) in 5 mL of water. This DAB-substrate preparation results in a dark blue or black stain. Photomicrographs of iron-stained embryos were obtained using an SZX51 zoom stereomicroscope (Olympus) at 40× magnification with a DP-71 camera (Olympus).
Iron injection and phenylhydrazine treatment
Embryos in the iron-injected cohorts were injected with iron dextran 100 mg/mL (Sigma-Aldrich) at 52 hpf, as described.19 For induction of hemolysis, embryos were maintained in phenylhydrazine 2 mg/L from 48 to 72 hpf.
Quantitative analysis of gene expression
At specified time points, embryos were pooled in groups of 18 to 20, anesthetized, and placed in RNAlater (Ambion). RNA extraction, generation of cDNA, and multiplex quantitative PCR assay for hepcidin, ferroportin1, and transferrin-a, normalized to β-actin expression, were performed as previously described.20 Number (N) equals 2 to 4 pools per condition or time point. Detection and analysis were performed on an ABI 7000. Transcript abundance was expressed as a fold increase over calibrator, according to the method as described.30 For each experiment, the calibrator pool consisted of sibling controls, at the same developmental stage, which were neither treated with iron, nor heat shock, nor morpholinos. Data presented are the means and standard errors.
Biostatistical analysis
Biostatistical analysis was performed using the method previously developed for analysis of zebrafish embryonic transcript levels.20 Heterogeneity among cohorts was analyzed by ANOVA using Instat 3.0 (GraphPad Software, LaJolla, CA). Tests for heterogeneity used the natural log for assessment of transcript levels. All estimates and standard errors presented have been converted back to the original units. When the global P value obtained from the ANOVA analysis was statistically significant, pairwise comparisons between the cohorts were performed using a 2-tailed student t test. P values less than or equal to .05 were deemed statistically significant. Multiple sequence alignments were constructed using the ClustalV algorithm with the MegAlign program (DNASTAR, Madison, WI). Residues that matched by 2 distance units in the PAM-250 weight table were considered homologous.
Results
Mutations in transferrin-a produce the gavi phenotype
The zebrafish mutant, gavi (gav), was identified as part of a large-scale screen for mutations resulting in hypochromic anemia. Two alleles were identified, gavHE067 and gavIT029, both of which exhibit a recessive pattern of inheritance, hypochromic anemia at 48 hpf, as evidenced by o-dianisidine staining for hemoglobin (Figure 1A,B), and lethality by 14 days of development. Using bulk segregant mapping, gavHE067 was localized to chromosome 2. Intermediate resolution mapping with a panel of 88 mutant embryos identified flanking microsatellite markers z9354 (3 recombinants/88) to the north and z26250 and z4300 (3 recombinants/88) to the south. This placed the gav gene near the transferrin-a locus (Figure 1C) that encodes the full-length zebrafish transferrin cDNA, which had been cloned previously and sequenced (GI:62204609).31,32 The transferrin-a protein sequence is 49% identical to human transferrin, but 67% identical to carp transferrin (Figure 1D). The gavHE067 and gavIT029 cDNA's for transferrin were sequenced. Each was found to have a single alternative splice form of transferrin-a. GavHE067 was found to have an A to G mutation (zebrafish assembly version 7 chromosome 2, nucleotide 12509998), creating a new exon donor leading to inclusion of 19 nucleotides from the intron following exon 7 (GenBank, accession number EU360800). This would result in truncation of the protein at residue 298. On the other hand, gavIT029 was found to have a G to A mutation (zebrafish assembly version 7 chromosome 2, nucleotide 12507046), resulting in a new exon donor and elimination of the last 89 nucleotides of exon 4 (GenBank, accession number EU360801). This splicing error would lead to truncation of the transferrin-a protein at residue 170 (Figure 1D).
The zebrafish mutant gavi. At 48 hpf, staining for hemoglobin with o-dianisidine revealed that the homozygous gavi (gav) mutant embryo (A) has decreased hemoglobin staining compared with a wild-type sibling (B). The scale bar represents 200 μm. Intermediate resolution mapping of the gavi mutation, using bulk segregant analysis, revealed that the mutation was on linkage group 2, near the transferrin-a locus (C). The green box represents the centromere. A protein sequence alignment (D) illustrates the relationship of the zebrafish transferrin-a protein sequence to transferrins from other species. Conserved residues are shaded light blue. The predicted truncation sites in the zebrafish mutants are indicated by asterisks: gavIT029 (red) and gavHE067 (blue).
The zebrafish mutant gavi. At 48 hpf, staining for hemoglobin with o-dianisidine revealed that the homozygous gavi (gav) mutant embryo (A) has decreased hemoglobin staining compared with a wild-type sibling (B). The scale bar represents 200 μm. Intermediate resolution mapping of the gavi mutation, using bulk segregant analysis, revealed that the mutation was on linkage group 2, near the transferrin-a locus (C). The green box represents the centromere. A protein sequence alignment (D) illustrates the relationship of the zebrafish transferrin-a protein sequence to transferrins from other species. Conserved residues are shaded light blue. The predicted truncation sites in the zebrafish mutants are indicated by asterisks: gavIT029 (red) and gavHE067 (blue).
Expression analysis of gavi mutants
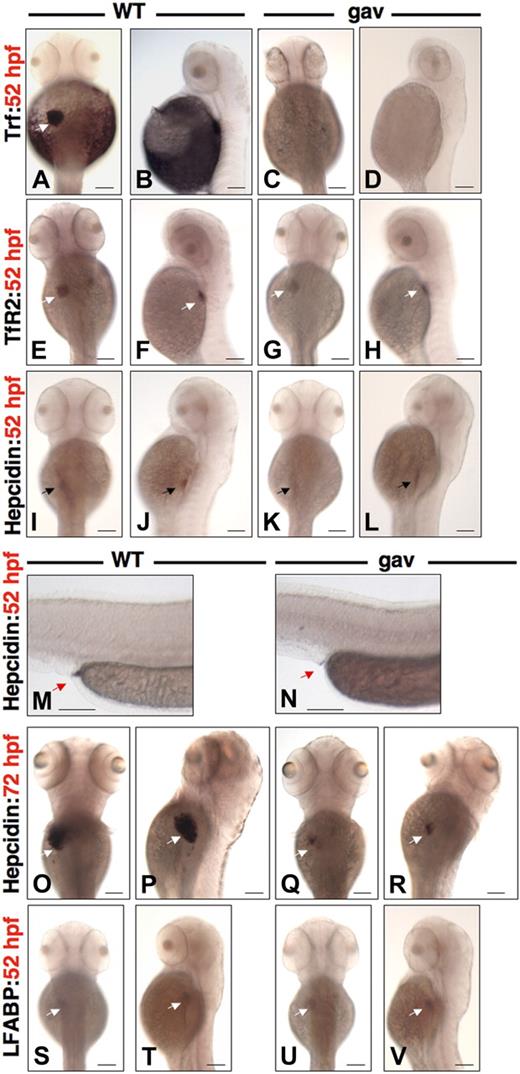
To evaluate transferrin-a expression in gavIT029, we performed whole mount in situ hybridization at 52 hpf in gavIT029 mutants, compared with WT siblings. In WT embryos, transferrin-a was expressed in the liver and yolk sac (Figure 2A,B), as previously described,32 however, expression was virtually absent in gavIT029 mutants (Figure 2C,D), likely due to nonsense-mediated decay. TfR2 expression was apparent at equivalent levels in the liver of both WT (Figure 2E,F) and gavIT029 mutants (Figure 2G,H). At 52 hpf, hepcidin expression was present at low levels in the foregut and liver of WT embryos (Figure 2I,J), but barely visible in gavIT029 mutants (Figure 2K,L). Hepcidin was expressed strongly, however, in a small area of the terminal gut (proctodeum) in WT (Figure 2M), but only weakly in gavIT029 (Figure 2N). At 72 hpf, hepcidin was expressed intensely in the liver of WT embryos (Figure 2O,P), but was expressed faintly in the liver of gavIT029 mutants (Figure 2Q,R). Expression in the foregut was no longer evident in either WT or gavIT029 mutants at 72 hpf, although expression persisted in the proctodeum (data not shown). Liver fatty acid binding protein (LFABP), a specific marker for zebrafish liver development,33 exhibited equivalent intensity of expression in WT (Figure 2S,T) and mutants (Figure 2U,V) at 52 hpf, consistent with normal liver development.
Phenotypic characterization of gavi. Whole mount in situ hybridization for transferrin-a (A-D), TfR2 (E-H), hepcidin (I-R), and LFABP (S-V) for WT siblings (A,B,E,F,I,J,M,O,P,S,T) versus gavIT029 (C,D,G,H,K,L,N,Q,R,U,V) in dorsal and lateral views at 52 hpf (A-N,S-V) and 72 hpf (O-R). White arrow indicates the liver. Black arrow indicates the foregut. Red arrow indicates the proctodeum (terminal gut). The scale bar represents 100 μm.
Phenotypic characterization of gavi. Whole mount in situ hybridization for transferrin-a (A-D), TfR2 (E-H), hepcidin (I-R), and LFABP (S-V) for WT siblings (A,B,E,F,I,J,M,O,P,S,T) versus gavIT029 (C,D,G,H,K,L,N,Q,R,U,V) in dorsal and lateral views at 52 hpf (A-N,S-V) and 72 hpf (O-R). White arrow indicates the liver. Black arrow indicates the foregut. Red arrow indicates the proctodeum (terminal gut). The scale bar represents 100 μm.
Morpholino knockdown of transferrin-a produces anemia and decreased somite iron stores
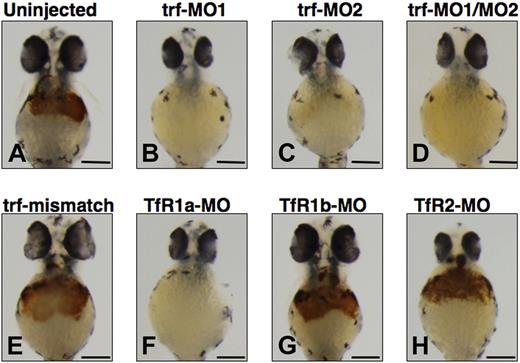
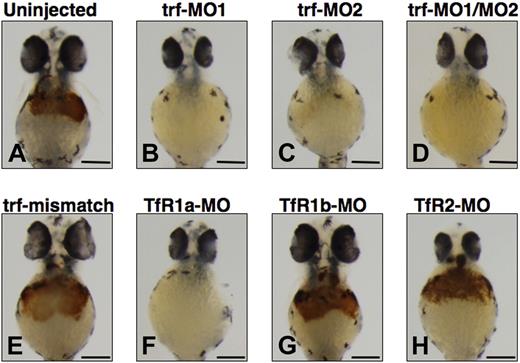
We used antisense morpholinos (MOs) designed to knock down transferrin-a and the transferrin receptors (TfRs) to confirm the requirement of transferrin-a for hemoglobin production. In comparison to uninjected WT embryos (Figure 3A), injection of trf-MO1, directed against the ATG of the transferrin-a gene, resulted in severe anemia at 48 hpf (Figure 3B) in 100% of the embryos injected (n = 177), compared with none in uninjected embryos (n = 3477). Injection of trf-MO2 (Figure 3C), which targets the exon 4 donor and thus replicates the effect of the gavIT029 mutation, resulted in anemia in 84% of the injected embryos (n = 99). Coinjection of trf-MO1 and trf-MO2 produced anemia (Figure 3D) in 96% of the injected embryos (n = 553). To confirm that the morpholinos exert a sequence-specific effect, mismatch control morpholinos for trf-MO1 and trf-MO2 were devised (Table S1), which were identical to the original morpholinos with the exception of 5 mismatched bases. Coinjection of the mismatch control morpholinos (trf-MO1M and trf-MO2M) failed to produce anemia (Figure 3E) in any embryos (n = 213).
Effect of morpholino knockdown of transferrin-a or TfRs on o-dianisidine staining for hemoglobin at 48 hpf. Uninjected embryos (A) exhibited normal levels of hemoglobin, while morpholino knockdown of transferrin-a via injection of trf-MO1 (B) targeting the ATG, or trf-MO2 (C) targeting the exon 4 donor, or coinjection of trf-MO1/trf-MO2 (D) produced anemia. In contrast, coinjection of mismatch controls trf-MO1M/trf-MO2M (E) failed to produce anemia. Morpholino knockdown of the erythroid-specific transferrin receptor (TfR1a; F) resulted in anemia, while knockdown of the ubiquitously expressed transferrin receptor (TfR1b; G) or TfR2 (H) did not cause anemia. The scale bar represents 200 μm.
Effect of morpholino knockdown of transferrin-a or TfRs on o-dianisidine staining for hemoglobin at 48 hpf. Uninjected embryos (A) exhibited normal levels of hemoglobin, while morpholino knockdown of transferrin-a via injection of trf-MO1 (B) targeting the ATG, or trf-MO2 (C) targeting the exon 4 donor, or coinjection of trf-MO1/trf-MO2 (D) produced anemia. In contrast, coinjection of mismatch controls trf-MO1M/trf-MO2M (E) failed to produce anemia. Morpholino knockdown of the erythroid-specific transferrin receptor (TfR1a; F) resulted in anemia, while knockdown of the ubiquitously expressed transferrin receptor (TfR1b; G) or TfR2 (H) did not cause anemia. The scale bar represents 200 μm.
Exploiting morpholino knockdowns, we evaluated which transferrin receptors are required for hemoglobin production in the zebrafish. In mammalian models, TfR1 is ubiquitously expressed and is the most important effector of iron uptake for erythrocytes and other cells, while TfR2 is expressed exclusively in hepatocytes and erythrocytes and is not a major iron importer for most mammalian cells.4,34 In the zebrafish there are 2 functional TfR1 orthologs (TfR1a and TfR1b) with different expression patterns, but only one TfR2 ortholog (TfR2). TfR1a is expressed in erythrocytes, while TfR1b is expressed ubiquitously.18 We show here that TfR2 is expressed specifically in the liver (Figure 2G,H). Consistent with prior experiments,18 morpholino knockdown of TfR1a resulted in anemia (Figure 3F) in 96% of the injected embryos (n = 260), while knockdown of TfR1b (Figure 3G) did not yield anemia (n = 59). We found that morpholino knockdown of TfR2 (Figure 3H) failed to produce anemia or morphologic defects in any of the injected embryos (n = 431).
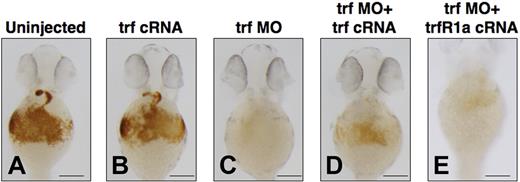
To explore the effects of transient overexpression of transferrin-a on zebrafish embryos, we injected transferrin-a cRNA into WT embryos at the 1-cell stage and stained for hemoglobin at 48 hpf. Compared with uninjected controls (Figure 4A), transferrin-a cRNA-injected embryos (Figure 4B) did not exhibit an increase in hemoglobin staining. In contrast, embryos injected with trf-MO2 to knock down transferrin-a expression exhibited severe anemia (Figure 4C), which was ameliorated (Figure 4D) in 86% (n = 81) by concomitant overexpression of transferrin-a cRNA. Coinjection of TfR1a cRNA with transferrin-a morpholino also partially reversed the anemia phenotype (Figure 4E), indicating that both genes act in the same pathway to mediate erythroid iron uptake. Rescue of the anemia phenotype was apparent as early as 36 hpf (Figure S1), before the onset of hepcidin expression by in situ hybridization.
Coinjection of transferrin-a or TfR1a cRNA rescues anemia in transferrin-a morphants. O-dianisidine staining for hemoglobin at 48 hpf comparing (A) uninjected WT, (B) overexpression of transferrin-a cRNA, (C) morpholino knockdown of transferrin-a, (D) coinjection of transferrin-a cRNA with transferrin-a morpholino, (E) coinjection of TfR1a cRNA with transferrin-a morpholino. The scale bar represents 200 μm.
Coinjection of transferrin-a or TfR1a cRNA rescues anemia in transferrin-a morphants. O-dianisidine staining for hemoglobin at 48 hpf comparing (A) uninjected WT, (B) overexpression of transferrin-a cRNA, (C) morpholino knockdown of transferrin-a, (D) coinjection of transferrin-a cRNA with transferrin-a morpholino, (E) coinjection of TfR1a cRNA with transferrin-a morpholino. The scale bar represents 200 μm.
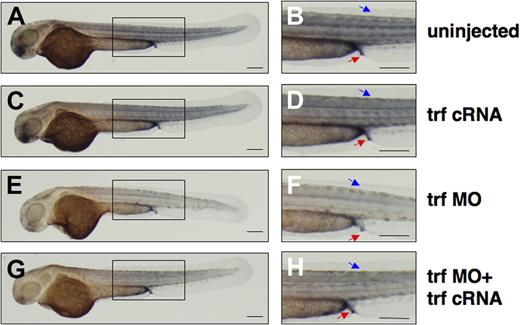
To elucidate the effect of transferrin-a deficiency on embryonic iron distribution, we performed DAB-enhanced staining for ferric iron at 52 hpf. In uninjected controls (Figure 5A,B) and transferrin-a cRNA injected embryos (Figure 5C,D), iron staining was concentrated in the somites, proctodeum, and yolk sac extension. On the other hand morpholino-knockdown of transferrin-a resulted in decreased iron staining in these areas (Figure 5E,F), including the proctodeum, an area of hepcidin expression at 52 hpf (Figure 2M). Coinjection of transferrin-a cRNA with trf-MO2 partially rescued iron staining in the somites and proctodeum (Figure 5G,H), in 72% (n = 47). These findings support the hypothesis that transferrin-a is required for iron transport from the yolk sac into the developing zebrafish embryo.
Coinjection of transferrin-a cRNA improves somite iron stores in transferrin-a morphants. DAB-enchanced Perl stain for ferric iron at 52 hpf in (A,B) uninjected WT, (C,D) transferrin-a cRNA overexpression, (E,F) morpholino knockdown of transferrin-a, (G,H) coinjection of transferrin-a cRNA with transferrin-a morpholino. Red arrow indicates the proctodeum. Blue arrow indicates the somites. The scale bar represents 200 μm.
Coinjection of transferrin-a cRNA improves somite iron stores in transferrin-a morphants. DAB-enchanced Perl stain for ferric iron at 52 hpf in (A,B) uninjected WT, (C,D) transferrin-a cRNA overexpression, (E,F) morpholino knockdown of transferrin-a, (G,H) coinjection of transferrin-a cRNA with transferrin-a morpholino. Red arrow indicates the proctodeum. Blue arrow indicates the somites. The scale bar represents 200 μm.
Quantitative analysis of gene expression in gavi
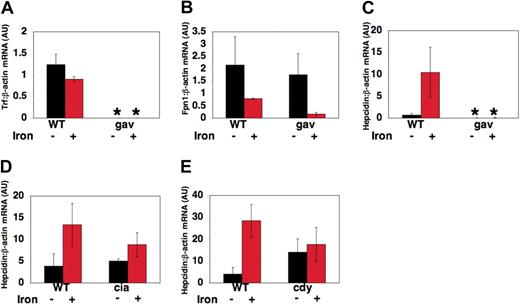
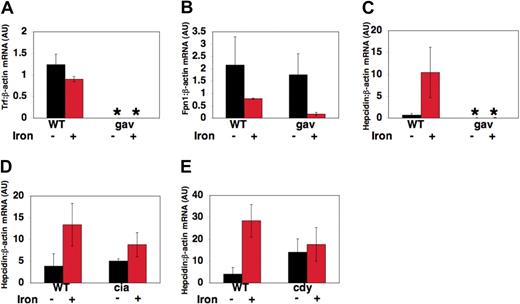
To explore the effects of transferrin-a deficiency on transcript levels of genes important in iron metabolism, RNA was obtained from pools of gavIT029 embryos and pools of WT siblings at 72 hpf, after injection with iron dextran to detect iron-induced transcript levels, or without injection to detect “basal” transcript levels. Quantitative realtime RT-PCR was used to assess the ratio of transferrin-a, fpn1, and hepcidin to β-actin expression in arbitrary units (AU), relative to a control sample. As expected, gavIT029 embryos expressed significantly lower levels of transferrin-a mRNA (Figure 6A) than their wild-type siblings (0.0008 ± 0.0001 AU vs 1.24 ± .24 AU, P = .0008) and the level of transferrin-a expression did not change significantly after iron dextran injection. On the other hand, fpn1 expression levels (Figure 6B) were not significantly different in uninjected gavi mutants compared with WT. The decrease in fpn1 mRNA transcript levels, which we observed in both gavi mutant and WT embryos after iron injection, resembles the decrease in fpn1 mRNA expression observed in duodenal enterocytes obtained from mice that were maintained on an iron-supplemented diet.35
Changes in gene expression in gavi and other hypochromic mutants. At 52 hpf, zebrafish embryos were either injected with iron dextran or anesthetized, but not injected. RNA was obtained from pools of anemic mutants and their wild-type siblings at 72 hours after fertilization, 18 hours after iron injection. Quantitative multiplex real-time RT-PCR, normalized to β-actin expression, was performed for transferrin-a (A), fpn1 (B), and hepcidin (C-E) in gavIT029 (A-C), TfR1a-deficient chianti (ciaTu25f) mutants (D) and DMT1-deficient chardonnay (cdyTe216) mutants (E). Gene expression is reported as fold increase over calibrator, a WT uninjected control pool. *P < .05 compared with WT uninjected controls.
Changes in gene expression in gavi and other hypochromic mutants. At 52 hpf, zebrafish embryos were either injected with iron dextran or anesthetized, but not injected. RNA was obtained from pools of anemic mutants and their wild-type siblings at 72 hours after fertilization, 18 hours after iron injection. Quantitative multiplex real-time RT-PCR, normalized to β-actin expression, was performed for transferrin-a (A), fpn1 (B), and hepcidin (C-E) in gavIT029 (A-C), TfR1a-deficient chianti (ciaTu25f) mutants (D) and DMT1-deficient chardonnay (cdyTe216) mutants (E). Gene expression is reported as fold increase over calibrator, a WT uninjected control pool. *P < .05 compared with WT uninjected controls.
In contrast, hepcidin transcript levels (Figure 6C), were 100-fold lower in uninjected gavi mutants than WT siblings (0.006 AU ± 0.003 AU vs 0.686 ± 0.386 AU, P = .04). After iron dextran injection, hepcidin expression was still significantly decreased in gavi mutants compared with iron-injected WT siblings (0.007 ± 0.004 AU vs 10.42 ± 5.75 AU, P = .01). This differed from chianti (Figure 6D) and chardonnay (Figure 6E) mutants. Chianti mutants, which have a defect in erythroid TfR1, exhibited equivalent basal hepcidin expression levels to those in wild-type sibling embryos (5.06 ± 0.52 AU vs 3.88 ± 2.88 AU, P = .56). Chardonnay mutants, which exhibit anemia due to impaired function of the iron transporter DMT1, did not exhibit significantly reduced basal levels of hepcidin expression compared with WT siblings (14.0 ± 6.13 AU vs 4.04 ± 3.07 AU, P = .39). In both chianti and chardonnay, however, the response to iron injection was blunted compared with WT.
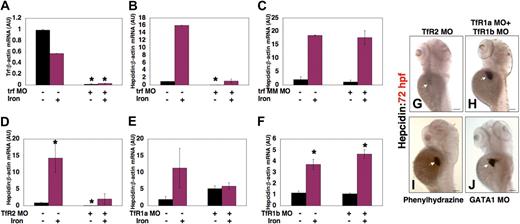
Morpholino knockdown of transferrin-a or TfR2 reduces hepcidin expression
Morpholino knockdown of transferrin-a produced a similar effect on hepcidin expression to that seen in gavIT029 mutants. After coinjection of trf-MO1 and trf-MO2, transferrin-a expression (Figure 7A) was significantly reduced (0.015 ± 0.007 AU vs 0.988 ± 0.012 AU, P = .02), and hepcidin expression (Figure 7B) was also significantly decreased (0.023 ± 0.016 AU vs 0.972 ± 0.028 AU, P = .04), compared with uninjected controls. Hepcidin expression increased in transferrin-a morphants after iron injection, but was not significantly increased compared with morphants that were not injected with iron (1.07 ± 0.53 vs 0.023 ± 0.016, P = .32). Injection of a mismatch control morpholino (Figure 7C) failed to reduce hepcidin expression, compared with uninjected controls (1.16 ± 0.67 AU vs 2.05 ± 1.05 AU, P = .55), or to impair the response to iron. Knockdown of TfR2 (Figure 7D), however, resulted in a significant reduction in the basal level of hepcidin expression (0.072 ± 0.006 AU vs 0.875 ± 0.125 AU, P = .007), while iron injection increased hepcidin transcript levels in the TfR2 knockdown embryos (2.02 ± 1.55 vs 0.072 ± 0.006, P = .10). In contrast, knockdown of TfR1a (Figure 7E) did not significantly lower the basal level of hepcidin expression (5.12 ± 0.88 AU vs 1.90 ± 0.90 AU, P = .18), although the effect of iron injection on hepcidin expression was blunted in TfR1a knockdown embryos (5.89 ± 1.03 AU vs 5.12 ± 0.88 AU, P = .63), as was seen in the TfR1a-deficient chianti mutant (Figure 6D). Furthermore, knockdown of TfR1b (Figure 7F) failed to decrease basal hepcidin expression levels (0.504 ± 0.226 AU vs 0.689 ± 0.311 AU, P = .69) or to impair the response to iron injection (9.32 ± 1.98 AU vs 0.504 ± 0.226 AU, P = .03). Whole mount in situ hybridization at 72 hpf confirmed that hepcidin expression was decreased in TfR2 morphants (Figure 7G), but normal in embryos injected with morpholinos to knockdown TfR1a and TfR1b simultaneously (Figure 7H). Taken together, these data indicate that transferrin-a and TfR2 are required for hepcidin expression in the zebrafish embryo, while TfR1a and DMT1 are needed for augmentation of hepcidin expression in response to iron loading.
Effect of morpholino knockdown of transferrin-a or TfRs on gene expression. Transferrin-a expression was reduced after morpholino knockdown of transferrin-a (A). Hepcidin expression was also reduced after knockdown of transferrin-a (B), but not after injection of a mismatch morpholino with 5 nucleotide substitutions (C). Knockdown of TfR2 (D) resulted in low hepcidin expression that failed to increase significantly after iron dextran injection. In contrast, knockdown of TfR1a (E) or TFR1b (F) did not impair baseline hepcidin expression, although the response to iron injection was blunted in TfR1a morphants. (G-J) Whole mount in situ hybridization at 72 hpf, in the absence of iron injection. Decreased hepcidin expression was observed in TfR2 morphant embryos (G), while knockdown of TfR1a and TfR1b in combination (H) failed to reduce hepcidin expression. Producing anemia by treatment of embryos with phenylhydrazine (I) or morpholino knockdown of GATA1 (J) resulted in strong hepcidin expression. White arrow marks the liver. *P < .05 compared with embryos that were neither injected with iron dextran nor morpholino.
Effect of morpholino knockdown of transferrin-a or TfRs on gene expression. Transferrin-a expression was reduced after morpholino knockdown of transferrin-a (A). Hepcidin expression was also reduced after knockdown of transferrin-a (B), but not after injection of a mismatch morpholino with 5 nucleotide substitutions (C). Knockdown of TfR2 (D) resulted in low hepcidin expression that failed to increase significantly after iron dextran injection. In contrast, knockdown of TfR1a (E) or TFR1b (F) did not impair baseline hepcidin expression, although the response to iron injection was blunted in TfR1a morphants. (G-J) Whole mount in situ hybridization at 72 hpf, in the absence of iron injection. Decreased hepcidin expression was observed in TfR2 morphant embryos (G), while knockdown of TfR1a and TfR1b in combination (H) failed to reduce hepcidin expression. Producing anemia by treatment of embryos with phenylhydrazine (I) or morpholino knockdown of GATA1 (J) resulted in strong hepcidin expression. White arrow marks the liver. *P < .05 compared with embryos that were neither injected with iron dextran nor morpholino.
Increased erythropoietic activity has been shown to suppress hepcidin expression in a mouse model.36 To evaluate whether increased erythropoietic activity plays a role in regulating hepcidin expression in zebrafish embryos, independent of the transferrin pathway, we induced hemolysis in zebrafish embryos by treating them with phenylhydrazine37 from 48 to 72 hpf. This treatment resulted in a complete absence of circulating erythrocytes, but hepcidin expression was, in fact, markedly increased at 72 hpf (Figure 7I). This is consistent with our observation of normal hepcidin expression in chianti and chardonnay mutant embryos, both of which have been shown to exhibit increased erythropoietic activity, manifest by increased numbers of erythroid precursor cells.17,18
To evaluate the effect of anemia on hepcidin expression in the absence of a defect in iron metabolism or enhanced erythropoietic activity, we produced anemia by an alternate method. We injected embryos with a GATA1 morpholino,26 which decreases the numbers of erythroid progenitors. Although GATA1 morphants had no circulating erythrocytes at 72 hpf, hepcidin expression was strongly present (Figure 7J). Taken together, these data indicate that anemia, in the absence of transferrin-a deficiency, fails to inhibit basal hepcidin expression in zebrafish embryos.
Discussion
Transferrin-a is required for hemoglobin production in the zebrafish
Gavi, the first transferrin-a–deficient mutant characterized in the zebrafish, exhibits severe hypochromic anemia and embryonic lethality, demonstrating that transferrin-a is required for hemoglobin production in the zebrafish. These findings complement studies in the zebrafish mutant chianti, which demonstrated that the erythroid transferrin receptor 1 (TfR1a) is required for hemoglobin production.18 Gavi serves as a zebrafish model for congenital atransferrinemia, a rare condition in humans resulting in severe hypochromic anemia and premature death,38,39 in the absence of intermittent plasma infusions, which partially restore serum transferrin levels.40
Both alleles of gav (gavIT029 and gavHE067) are due to mutations that cause aberrant splicing of the transferrin-a message. This resembles the gene defect in hypotransferrinemic (hpx/hpx) mice, which display low serum transferrin levels due to a mutation that results in aberrant splicing of transferrin mRNA.41 In the hpx/hpx mouse, provision of transferrin protein improves the anemia phenotype and reverses the lethality of the phenotype.42 Similarly, in our study, injection of normal transferrin-a cRNA into zebrafish embryos ameliorated anemia.
Transferrin-a is required for normal iron distribution in the zebrafish embryo
Supporting a role for transferrin-a in iron transport from the yolk sac to the developing embryo, we demonstrated that transferrin-a deficiency resulted in a deficit of stainable iron stores in the somites and terminal gut of the zebrafish embryo, which improved after experimental overexpression of transferrin-a cRNA. Further supporting this hypothesis, transferrin-a (Figure 2A,B) is expressed in the yolk syncytial layer,32 as is fpn1, which is also required for transporting iron from the yolk sac to the embryo.19 Although histologic iron accumulation has been demonstrated in nonerythropoietic tissues in atransferrinemic patients42-44 and the hpx/hpx mouse at 45 days of age,42 the absence of stainable iron accumulation in the gavi mutant embryos is consistent with the newborn hpx/hpx mouse, in which iron deposits were not stainable.42
Transferrin-a and TfR2 are required for hepcidin expression in zebrafish embryos
In this report, we provide the first in situ hybridization expression data for hepcidin during embryonic development. We found that strong hepatic expression of transferrin-a and TfR2 at 52 hpf preceded strong hepatic expression of hepcidin at 72 hpf and that transferrin-a–deficient mutants and morphants exhibited low levels of hepcidin expression, which remained inappropriately low after iron dextran injection. Taken together, these data demonstrate that transferrin-a is required in the zebrafish embryo for normal hepcidin expression. These findings complement recent work, in animal models and human subjects, supporting the hypothesis that transferrin plays an important role in regulating postnatal hepcidin expression. Hpx/hpx mice exhibit low levels of hepcidin expression,45 while provision of holotransferrin to cultured primary hepatocytes stimulates hepcidin expression.46 Furthermore, administration of plasma to an atransferrinemic patient resulted in an increase in urinary hepcidin levels.40 Low levels of hepcidin expression in atransferrinemic patients may explain the increased intestinal iron absorption previously observed in these patients.43,47
Of the hypochromic anemia mutants that we have studied in the zebrafish (reflecting deficiencies in fpn1,20 TfR1a, DMT1, and transferrin-a), only the transferrin-a–deficient gavi exhibited decreased embryonic hepcidin expression. Furthermore, we demonstrated that elimination of circulating erythrocytes, by treatment with phenylhydrazine or by knocking down GATA1, failed to impair embryonic hepcidin expression. This differs from studies of mouse strains with hypochromic anemia, such as mk/mk and sla/sla mice, which have defects in DMT1 and hephaestin, respectively, or phenylhydrazine-treated mice, all of which exhibit low levels of hepcidin expression.9,45 This discrepancy could be due to differences in hepcidin regulation across species or to differences in regulation at different stages of development. The mk/mk and sla/sla mice were evaluated 4 to 10 weeks after birth (Cindy N. Roy, Johns Hopkins University, e-mail, April 29, 2008), while we evaluated zebrafish embryos. Supporting a role for developmental differences in the regulation of hepcidin expression, hepcidin expression was markedly reduced in the livers of severely anemic fpn1-deficient adult zebrafish, but not in fpn1-deficient embryos.20
The expression and function of TfR2 have not been evaluated previously in the zebrafish. We demonstrate here that TfR2 is coexpressed with transferrin-a in the liver of the zebrafish embryo and that knockdown of TfR2 fails to produce anemia or a morphologic defect. Instead, we found that morpholino knockdown of TfR2, but not TfR1a or TfR1b, abrogated hepcidin expression. This is consistent with experimental data in mouse models and human patients, which support a critical role for TfR2 in regulating hepcidin expression. Liver-specific knockout of TfR2 in mice results in decreased hepcidin expression and increased liver iron stores.48 In addition, patients who are homozygous for TfR2 mutations develop early onset hemochromatosis49 and exhibit low levels of urinary hepcidin.13
Transferrin-a, TfR1a, TfR2, and DMT1 facilitate increased hepcidin expression in response to iron loading in zebrafish embryos
Although transferrin-a and TfR2 were the only genes identified that were required for basal hepcidin expression in zebrafish embryos, we found that knockdown of transferrin-a, TfR2, TfR1a, or DMT1 impaired the enhancement of hepcidin expression in response to iron injection, while deficiency of TfR1b did not. As TfR1a and DMT1 are required for erythroid iron acquisition, while TfR1b is not,18 these data imply that erythroid iron acquisition via transferrin-a/TfR1a/DMT1 facilitates iron-induced increases in hepcidin expression in zebrafish embryos. In mammalian systems, DMT1 has previously been implicated in the release of iron from the endosome after endocytosis of holotransferrin/transferrin receptor.2,3
How transferrin and the transferrin receptors interact to regulate hepcidin expression is not yet understood. Recently, it has been shown, in mammalian cell culture, that TfR2 competes with TfR1 to bind HFE,50 an atypical MHC class I protein (reviewed in Pietrangelo51 ), and that holotransferrin competes with HFE to bind TfR1.52 Genomic analysis has failed to identify a close ortholog for human HFE in the zebrafish53 or in the other fish species for which sequence data are available (P.F. and Y.G., unpublished data, April 14, 2008). We have identified 9 orthologs of MHC class I in the zebrafish with low sequence homology to human HFE (25.9%-31.6% sequence identity). It remains to be determined if any of these zebrafish MHC class I orthologs interacts with TfR2. While many aspects of iron homeostasis appear to be conserved in the zebrafish, further dissection of the pathways involved in regulating iron metabolism may reveal important differences, which may provide insight into the evolution of iron homeostasis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We would like to thank the Tübingen 2000 Screen Consortium (see the Appendix for a list of members) for generating the gavi zebrafish mutants. We would also like to thank Dr Alan Davidson, Dr Tina Schmid, and Ms Jennifer Axe for assistance in identification and maintenance of anemia mutants identified in the Tübingen 2000 screen. We thank Dr Victoria Petkova of the Beth Israel Deaconess Medical Center Realtime PCR Core for assistance with the realtime PCR assay.
This work was supported by National Institutes of Health grants DK 61685-07 (P.G.F.) and DK053298-07 (L.I.Z.), Howard Hughes Medical Institute (L.I.Z.), and the American Society of Hematology Scholar Award (P.G.F.).
National Institutes of Health
Appendix
Tübingen 2000 screen consortium members: F. Bebber van, E. Busch-Nentwich, R. Dahm, H. G. Frohnhöfer, H. Geiger, D. Gilmour, S. Holley, J. Hooge, D. Jülich, H. Knaut, F. Maderspacher, C. Neumann, T. Nicolson, C. Nüsslein-Volhard, H. Roehl, U. Schönberger, C. Seiler, C. Söllner, M. Sonawane, A. Wehner, C. Weiler, and B. Schmidt, Max-Planck-Institut für Entwicklungsbiologie, Tübingen, Germany; U. Hagner, E. Hennen, C. Kaps, A. Kirchner, T. I. Koblizek, U. Langheinrich, C. Metzger, R. Nordin, M. Pezzuti, K. Schlombs, J. deSantana-Stamm, T. Trowe, G. Vacun, A. Walker, and C. Weiler, Artemis Pharmaceuticals/Exelixis Deutchland, Köln, Germany.
Authorship
Contribution: P.G.F., Y.G., J.L.H., V.J.L., S.F.B., K.A.D., and R.A.W. performed experiments; P.G.F. and L.I.Z. designed the research; and P.G.F. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paula Fraenkel, Division of Hematology/Oncology, Department of Medicine, Beth Israel Deaconess Medical Center, 330 Brookline Ave, Boston, MA 02215; e-mail: pfraenke@bidmc.harvard.edu.