Abstract
Platelet factor 4 (PF4) is a negative regulator of megakaryopoiesis, but its mechanism of action had not been addressed. Low-density lipoprotein (LDL) receptor–related protein-1 (LRP1) has been shown to mediate endothelial cell responses to PF4 and so we tested this receptor's importance in PF4's role in megakaryopoiesis. We found that LRP1 is absent from megakaryocyte-erythrocyte progenitor cells, is maximally present on large, polyploidy megakaryocytes, and near absent on platelets. Blocking LRP1 with either receptor-associated protein (RAP), an antagonist of LDL family member receptors, or specific anti-LRP1 antibodies reversed the inhibition of megakaryocyte colony growth by PF4. In addition, using shRNA to reduce LRP1 expression was able to restore megakaryocyte colony formation in bone marrow isolated from human PF4-overexpressing mice (hPF4High). Further, shRNA knockdown of LRP1 expression was able to limit the effects of PF4 on megakaryopoiesis. Finally, infusion of RAP into hPF4High mice was able to increase baseline platelet counts without affecting other lineages, suggesting that this mechanism is important in vivo. These studies extend our understanding of PF4's negative paracrine effect in megakaryopoiesis and its potential clinical implications as well as provide insights into the biology of LRP1, which is transiently expressed during megakaryopoiesis.
Introduction
Although the predominant cytokine regulating platelet count is thrombopoietin (TPO), during megakaryopoiesis, many other cytokines have been implicated, including interleukin-6 (IL-6), which increases TPO expression in the liver1 ; stromal-derived factor-1, which enhances megakaryocyte chemotaxis2 ; and IL-11, which directly stimulates megakaryocyte development.3 A pathway by which megakaryopoiesis is auto–down-regulated has been suggested based on in vitro studies of platelet factor 4 (PF4) and later by studies of other chemokines that are also stored in α-granules, including the related CXC chemokines, neutrophil activating peptide-2 and IL-8,4,5 and the more distantly related CC chemokines, regulated upon activation, normal T-cell expressed and secreted6 and macrophage inflammatory peptide-1α.5,6 More recently, in vivo studies have demonstrated the importance of the PF4-negative paracrine loop under steady-state conditions and in chemotherapy-induced thrombocytopenia (CIT).7
PF4 is a 7.8-kDa protein that is produced primarily in megakaryocytes, expressed in platelets as a tetramer, and comprises 2.5% on a molar basis of the α-granular releasate.8 The biologic role(s) of PF4 is not fully understood. In addition to previous in vitro studies demonstrating an effect on megakaryocyte development, we have recently shown that PF4 can play a biologically relevant role in vivo in regulation of steady-state platelet count and in recovery after chemotherapy.7 Unlike other chemokines that have clearly defined chemokine receptors, PF4 appears to function by binding with high affinity to glycosaminoglycans (GAGs) on cell surfaces and to negatively charged domains of several membrane receptors.9-11 Recently, PF4 has been shown to activate endothelial cell expression of E-selectin through the low-density lipoprotein receptor–related protein-1 (LRP1) in an NFκB-dependent fashion.12 These studies provided the impetus for examining LRP1 as a potential candidate receptor of PF4 in megakaryocyte development.
Herein, we present evidence that demonstrates that LRP1 is transiently expressed during megakaryopoiesis with peak levels on large polyploid megakaryocytes and that this subpopulation of cells is susceptible to regulation by PF4. Blocking PF4's interaction with this receptor system increases megakaryopoiesis in vitro and platelet counts in vivo, suggesting the potential of additional clinical strategies for modifying platelet counts.
Methods
Transgenic mice and platelet counting
Animal lines have been described previously, and include mPF4−/− mice generated by replacing the entire coding region for mouse (m) Cxcl4 (also known as Pf4 or Scyb4, LOC56744; 1.2 kb) with a 1.8-kb neomycin resistance gene13 and 2 transgenic mouse lines that overexpress human (h) PF4.14 The hPF4High animals used in most of the described studies are transgenic for a 14-kb fragment of the human PF4 (also known as CXCL4, SCYB4 or MGC138298, LOC5196) locus that contains 10.2-kb upstream and 3-kb downstream sequence from the coding region. Previous analysis of multiple tissues using immunohistochemistry and reverse-transcription–polymerase chain reaction (RT-PCR) showed that hPF4 was expressed exclusively in megakaryocytes in these mice,15 and that platelets from hPF4High mice have 6 times the human PF4 content of 4 human controls concurrently studied.15 A second hPF4-expressing transgenic mouse line (hPF4Mid) with a 10-kb fragment of the human PF4 locus with 5.4-kb upstream and 3.8-kb downstream sequence contains 2 times the amount of PF4 as human controls.15 The genomic type of all animals was determined by PCR as previously described.13,14 All PF4 variant animals were backcrossed onto a C57BL/6J background for more than 10 generations and comparative studies were done using littermate controls.
The mice were housed at the Children's Hospital of Philadelphia animal facility. Animals were anesthetized, and 50 μL EDTA-anticoagulated whole blood was obtained by retro-orbital puncture for complete blood counts measured in an automatic cell counter (HEMAVET; Drew Scientific) set for mouse parameters. All procedures were performed after approval by the Institutional Animal Care and Use Committee (IACUC) of the Children's Hospital of Philadelphia.
Recombinant PF4 preparation
In some in vitro studies, recombinant hPF4 was included.16 Briefly, affinity chromatography using a HiTrap heparin high-performance affinity column (GE Healthcare) was used to purify hPF4 from the supernatant of lysed BL21DE30 pLysS bacterial cells expressing recombinant hPF4 from a pT7-7 plasmid containing the appropriate cDNA. These proteins were further purified by fast protein liquid chromatography using a Resource RPC FPLC column (Amersham Pharmacia Biotech). Purity was assessed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis followed by silver staining. An immunoblot after electrotransfer to polyvinylidenedifluoride was used to confirm the identity of the protein. Primary antibodies were rabbit polyclonal anti–human or anti–murine PF4 antibodies (both prepared by Sigma-Genosys). Detection was performed with horseradish peroxidase (HRP)–conjugated swine anti–rabbit antibody (DAKO) using an electrochemiluminescence kit (Perkin Elmer Life Sciences).
Bone marrow cell cultures, RT-PCR, flow cytometry, and Western blot analysis
Human CD34+ bone marrow–derived cells (University of Pennsylvania Stem Cell Core) were cultured in liquid serum-free media as previously described.18 Briefly, 4 × 105 purified human CD34+ cells were cultured in media consisting of IMDM with 10 mg/mL BSA (Sigma-Aldrich), 740 μg/mL partially saturated transferrin (Sigma-Aldrich), 10 μg/mL human insulin (Sigma-Aldrich), 40 μg/mL low-density lipoprotein (Calbiochem), 2 mM l-glutamine (Fisher), and 100 ng/mL recombinant human TPO (R&D Systems). Cells were incubated at 37°C and medium was replaced on days 4, 7, 11, and 14 of culture. Murine cells were harvested from wild-type (WT) animals and cultured under similar conditions with recombinant murine TPO (R&D Systems).
Aliquots of cells were taken on day 14 (human) or 7 (murine) of the study and analyzed by flow cytometry after staining with anti-LRP1 light chain antibody labeled with Alexa 647 (Invitrogen) for human studies and biotin-labeled anti-LRP1 light chain antibody with a streptavidin-Alexa 647 secondary antibody (Molecular Innovations) in the case of murine cells and a phycoerythrin-labeled anti-CD41 antibody (BD Bioscience) against either human or murine CD41 as appropriate. Briefly, cells were collected and washed with PBS. Anti-CD16/CD32 (5 μL/106 cells, Fc block; BD Bioscience) was added to cells suspended in 50 μL PBS. After a 10-minute incubation at room temperature, cells were stained with 2 μL of the appropriate antibodies species and incubated for 30 minutes at room temperature. Cells were washed with PBS and resuspended in 300 μL fresh PBS and analyzed immediately on a FACSCalibur machine (BD Biosciences). An isotype antibody (mouse IgG2; BD Bioscience) labeled with Alexa647 (Invitrogen) was used in some experiments as a control. Data were analyzed using FlowJo software (TreeStar).
For ploidy analysis, bone marrow was harvested fresh from murine femurs and prepared as previously described.17 Briefly, bones were flushed with CATCH buffer (0.38% sodium citrate, 2 mM theophylline, 1 mM adenosine, 3.5% BSA in PBS). Cells were spun at 70g for 10 minutes. Cells were resuspended in 500 μL CATCH buffer and incubated with 10 μL Fc-blocking antibody for 10 minutes. Cells were then split into 2 aliquots. One was labeled with anti–CD41-FITC (BD Bioscience) for 30 minutes. Both aliquots were then washed with 10 volumes CATCH buffer and spun at 70g for 10 minutes. Afterward, both aliquots of cells were resuspended in 400 μL hypotonic propidium iodide solution (0.1% sodium citrate with 50 μg/mL propidium iodide) and incubated at 4°C overnight. Thirty minutes before flow cytometry, RNAse A was added (50 μg/mL) at room temperature. Data were collected using a FACSCalibur machine and analyzed using FlowJo software.
In addition, aliquots of cells from day 10 or 14 were pelleted, and RNA was extracted using the RNAeasy kit (QIAGEN) with DNAse treatment on the column. Using the SuperScript II First-Strand Synthesis System (Invitrogen), cDNA was generated from megakaryocyte RNA as well as from RNA similarly extracted from human and murine platelets isolated from peripheral blood samples as previously described.7 Primers for human and murine LRP1 were designed using Primer-BLAST19 (murine LRP1 forward: 5′-gaggtgttggacacagatgg-3′, murine LRP1 reverse: 5′-acaggagaactggttgggtg-3′; human LPR1 forward: 5′-tgaccccgccgttgctcctg-3′, human LRP1 reverse: 5′-tgaaggagccgtctgtgttg-3′). Primer pairs for both human and murine integrin beta-2 (ITGB2) and PF4 were as previously described14 and used to assess white blood cell (WBC) contamination, and RNA quality and specificity, respectively. NIH-3T3 cells and 293T cells were used as positive controls for LRP1 for murine and human RT-PCR, respectively.20,21 PCR from cDNA was performed (after denaturing DNA at 94°C for 5 minutes) for 30 cycles with an annealing temperature of 55°C for 30 seconds per cycle. Extension temperature was 72°C for 1 minute per cycle. Final extension time at 72°C was 5 minutes. Products were run on a 1% Tris, boric acid, EDTA agarose gel and visualized after staining with ethidium bromide. To confirm the identity of the PCR product, DNA from bands of interest was also extracted using GeneClean (Invitrogen), and sequenced by the Nucleic Acid/Protein Research Core Facility at the Children's Hospital of Philadelphia using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) and analyzed on a 3730 DNA Analyzer (Applied Biosystems) following the manufacturer's protocol.
For Western blots, human CD34+ cells were cultured as in the first paragraph of this section. After 14 days, cells were collected by centrifugation at 90g for 10 minutes. They were then frozen at −80°C. After freezing, cells were thawed and resuspended in cell lysis buffer (QIAGEN protein extraction kit). Human platelets were prepared from PRP as previously described7 and similarly frozen and then lysed for protein extraction. Finally, human WBCs were prepared by Ficoll gradient as previously described.22 The cell pellet was then similarly frozen and lysed for protein extraction. Protein concentrations were approximately 2 to 3 mg/mL. Equal amounts were loaded and separated on a 4% to 12% SDS-PAGE gel. The gel was then blotted onto nitrocellulose and 5% milk was used to block. After blocking, primary antibody (mouse anti-LRP1 [Lifespan Biosciences; 1:1000] or mouse antiactin [Sigma-Aldrich; 1:2000]) was added and blots were incubated for 2 hours at room temperature. They were then washed and goat anti–mouse IgG conjugated to horseradish peroxidase (GE Healthcare; 1:5000) was added as a secondary antibody for 1 hour at room temperature. The blot was washed and developed with Western Lighting plus enhanced chemiluminescence (PerkinElmer).
In vitro bone marrow mononuclear cell culture studies
Bone marrow from the tibias and femurs of 6- to 12-week-old male mice was isolated and used for in vitro cell culture as previously described.23 Briefly, Iscove modified Dulbecco medium (IMDM; Invitrogen) without modification was used to flush the marrow cavity of bones harvested from killed mice, and the cells were then passed over a 100-μm nylon filter (BD Biosciences). After centrifugation at 240g, the cell pellet was resuspended in IMDM plus 1% penicillin-streptomycin (Invitrogen) at a concentration of 2.2 × 106 cells/mL. These cells were then cultured in a commercially available murine, serum-free culture system (MegaCult-C; StemCell Technologies). No supportive stromal cells were included in the cultures. In some studies, either hPF4 (20 μg/mL, final concentration) or 1 of the 2 rabbit anti-PF4 antibodies or rabbit pretreatment IgG control (Sigma-Genosys; each at 25 μg/mL, final concentration) was included. In addition, in some studies, receptor-associated protein (RAP) containing low endotoxin (Molecular Innovations; 0.02 μM) or anti-LRP1 light chain antibody (MA5A6; Molecular Innovations) or anti-LRP1 heavy chain antibody (Molecular Innovations) or an anti–cluster II LRP1 antibody (R&D Systems; each at 25 μg/mL, final concentration) was added.
Double-chamber slides were cultured at 37°C under high humidity for 10 days. Afterward, the slides were dried per MegaCult-C kit protocol and stained for acetylcholinesterase to mark megakaryocytes as previously described.24 The slides were analyzed by counting the total number of colonies per chamber in addition to the number of colonies showing positive staining for megakaryocytes. A ratio of megakaryocyte-containing colonies–total colonies was calculated for each well and results from duplicate wells were averaged.
LRP1 shRNA studies
Three shRNAs against murine LRP1 were created in a lentivirus incorporating both puromycin resistance and the anti-LRP1 sequence (EZBiolab). Plasmid DNA for vectors was produced in DH5α bacteria and, using a 3-vector lentivirus system, viral particles were generated in 293T cells as previously described.25 Virus was titered using the QuickTiter Lentivirus Quantitation Kit (Cell Biolabs). shRNA lentiviral vectors were tested in NIH-3T3 cells (a murine fibroblast cell line), which expresses LRP1 constitutively.26 NIH-3T3 cells were cultured in DMEM containing 1% penicillin-streptomycin and 1% l-glutamine as well as 10% fetal bovine serum. Cells were transfected with 40 μL lentivirus using spin transfection.27 Media were replaced with fresh media 6 hours after transfection and then cells were incubated for an additional 48 hours. After this time period, media were again replaced, but now with media containing 1 μg/mL puromycin (Sigma-Aldrich). Cells were cultured for an additional 4 days, harvested, and resuspended in PBS. Cells were incubated for 10 minutes with Fc-blocking antibody and then labeled with an anti-LRP1 light chain antibody (Molecular Innovations) labeled with Alexa 647 (Invitrogen; 2 μL antibody/106 cells for 30 minutes at room temperature) and analyzed using a FACSCalibur machine and FlowJo software.
Similar studies were done using primary murine bone marrow cells obtained from hPF4High and mPF4−/− mice. Cells were pelleted and resuspended at a concentration of 2 × 106 cells/mL in unmodified IMDM. Approximately 500 μL of the cell suspension was placed in a 24-well plate (Corning), and plates were spun at 340g for 5 minutes to form a monolayer of cells. Then, 4 μL polybrene (2 mg/mL; Millipore) was added to cells with 50 μL concentrated virus. Cells were spun again at 1340g for 90 minutes at 20°C and then placed at 37°C for 3 to 4 hours. Afterward, 100 μL of cells was transferred to MegaCult-C media and plated according to the MegaCult-C kit instructions. Colonies were assessed by acetylcholinesterase staining.
Alternatively, cells were spun down and resuspended in serum-free media (IMDM, 1% pen/strep, 1% l-glutamine, 1% Nutridoma-SP [Roche], and 50 μM β-mercaptoethanol) at 106 cells/mL and cultured for 48 hours. Afterward, 1 μg/mL puromycin was added. Cells were further cultured in serum-free media with 50 ng/mL recombinant mouse TPO with puromycin for an additional 5 days and then analyzed by flow cytometry.
G1ME cell studies
G1ME and Plat-E cells (used to generate GATA-1/MIGR1 or empty MIGR1 retroviruses)27,28 were a generous gift from Dr Mitchell Weiss, University of Pennsylvania. Cells were cultured and transfected with GATA-1–containing retrovirus as described.27,28 Briefly, Plat-E cells were seeded in 10-cm dish (Corning), cultured in Plat-E media (DMEM, 1% l-glutamine, 1 mM sodium pyruvate, 10% fetal bovine serum, 1% penicillin/streptomycin, 1 μg/mL puromycin, and 10 μg/mL blastocidin [Invitrogen]), and incubated at 37°C and 5% CO2. Once cells reached 90% to 95% confluence, they were transfected with 24 μg GATA-1/MIGR1 viral DNA or empty MIGR1 vector using Lipofectamine 2000 (Invitrogen). They were incubated at 37°C and 5% CO2. After 24 hours, the media were replaced with G1ME culture media (78% alpha-MEM, 20% fetal bovine serum, 1% phosphate-buffered saline, 1% l-glutamine, and ∼ 50 ng/mL mouse TPO). After another 24 hours, 10 mL media was collected from each 10-cm plate for immediate transfection of G1ME cells.
G1ME cells were then cultured in a serum-free media, StemSpan Serum Free Media (StemCell Technologies) with 1% penicillin/streptomycin, 1% l-glutamine, and 50 ng/mL murine TPO) with or without 25 μg/mL PF4 at 37°C and 5% CO2 for 4 days before transfection with GATA-1/MIGR1 or an empty vector. After 4 days, 2 to 4 million G1ME cells were resuspended in 5 mL of the retroviral supernatant with 8 μg/mL polybrene (Millipore), 50 μL HEPES (Invitrogen), and 50 ng/mL mouse TPO. Cells were plated in 6-well tissue culture–treated plates (Corning) and spun at 2400g for 90 minutes at room temperature. Plates were then placed at 37°C and 5% CO2 for 3 to 4 hours. After this time, cells were spun to remove the supernatant and resuspended in 6 to 10 mL StemSpan with or without 25 μg/mL PF4. Cells were removed on days 0, 1, 2, 3, 4, 5, 6, and 8 for flow cytometric analysis. Rat anti–mouse CD42 antibody labeled with phycoerythrin (Emfret) and rat anti–mouse CD41 (BD Bioscience) were used to identify the proportion of eGFP+ cells that were differentiating along the megakaryocyte lineage. Cells were separated into relatively large or small CD42+ cells based on forward scatter.
In vivo studies of platelet counts and response to RAP infusion
Eight- to 12-week-old hPF4High or mPF4−/− mice were anesthetized with isoflurane (Baxter), and a subcutaneous infusion pump (DURECT) was placed in the posterior nuchal subcutaneous space.29 Infusion pumps were filled with 4 to 7 ng/μL endotoxin-free RAP containing low endotoxin (Molecular Innovations). Platelet counts were then measured on blood obtained by retro-orbital bleed.
Statistics
Differences between groups were compared using the Student t test. Statistical analyses were performed using STATA (Stata) or Microsoft Excel (Microsoft). Differences were considered significant at P values less than .05.
Results
LRP1 expression on megakaryocytes and platelets
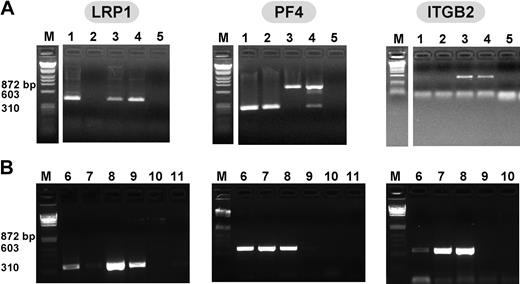
RT-PCR using species-specific LRP1 primers beginning with total RNA extracted from human and murine megakaryocytes revealed a band at the expected size for LRP1 in both (Figure 1A-B). These bands were extracted from the gel and the identity was confirmed by sequencing (data not shown); the data are consistent with a recent report describing LRP1 on the surface of human megakaryocytes published while our studies were under way.18 RT-PCR beginning with megakaryocytes did not show a band consistent with the myeloid marker ITGB2,30 but showed a strong PF4 band supporting that the observed LRP1 band was from megakaryocytes and not contaminating LRP1-expressing WBCs (Figure 1A). On the other hand, platelet RNA extracts showed either no band or a faint band of consistent size to LRP1 cDNA from both human and mouse sources. These findings suggest that there is significant LRP1 RNA in megakaryocytes, and either no to little LRP1 RNA in platelets. Platelets intermittently demonstrated a weak band for ITGB2, but consistently showed a strong PF4 band, supporting that the platelet RNA was intact and that the low to no detected LRP1 was indeed due to either little LRP1 in platelets or to minor WBC contamination.
RT-PCR of human and murine megakaryocytes and platelets. (A) Murine RT-PCR studies of total RNA from (1) megakaryocyte, (2) platelets, (3) WBCs, (4) NIH-3T3 cells, known to express LRP1,20 and (5) water for LRP1 (first gel), PF4 (second gel), and ITGB2 (third gel). Expected band size is approximately 400 base pair (bp) for LRP1 and ITGB2, and approximately 300 bp for PF4. White space indicates where lanes were removed for ease of presentation. (B) Human RT-PCR studies of total RNA from (6) megakaryocytes, (7) platelets, (8) WBCs, (9) 293T cells, embryonic kidney line known to express LRP1,21 (10) water, and (11) no reverse transcriptase controls for LRP1 (first gel), PF4 (second gel), and ITGB2 (third gel).
RT-PCR of human and murine megakaryocytes and platelets. (A) Murine RT-PCR studies of total RNA from (1) megakaryocyte, (2) platelets, (3) WBCs, (4) NIH-3T3 cells, known to express LRP1,20 and (5) water for LRP1 (first gel), PF4 (second gel), and ITGB2 (third gel). Expected band size is approximately 400 base pair (bp) for LRP1 and ITGB2, and approximately 300 bp for PF4. White space indicates where lanes were removed for ease of presentation. (B) Human RT-PCR studies of total RNA from (6) megakaryocytes, (7) platelets, (8) WBCs, (9) 293T cells, embryonic kidney line known to express LRP1,21 (10) water, and (11) no reverse transcriptase controls for LRP1 (first gel), PF4 (second gel), and ITGB2 (third gel).
Next, we performed flow cytometry to assess surface expression of LRP1 on both megakaryocytes and platelets. Megakaryocytes were isolated from cultured human or murine bone marrow. Flow cytometry of murine cells showed a shift in fluorescence in cells stained with anti-LRP1 antibody compared with cells stained using only secondary antibody, whereas platelets showed little to no shift (Figure 2A). Similarly in human cells, a population of LRP1-positive cells was seen in cultured megakaryocytes (Figure 2B top), but not in human platelets (Figure 2B bottom). Thus surface LRP1 level is parallel to the observed LRP1 message level in these cells. Western blot analysis of primary human megakaryocytes and platelets was consistent with the flow cytometric studies, in that isolated megakaryocytes expressed LRP1 (Figure 2C) as do primary WBCs, but platelets had little to no LRP1 present.
Flow cytometry and Western blot of human and murine megakaryocytes and platelets. (A) Representative examples of flow cytometry of murine bone marrow–derived megakaryocytes (top) stained with a biotin labeled anti-hLRP1 antibody known to cross-react with mouse LRP131 and then stained with streptavidin, PE–Alexa 647 secondary antibody. The gray line represents unstained cells. The broken black line represents secondary antibody alone. The solid black line is megakaryocytes with both antibodies. The bottom graph shows flow cytometry of platelets similarly performed. (B) As in panel A but for human cultured megakaryocytes and human peripheral blood platelets. LRP1 antibody was directly labeled with Alexa 647 for these experiments. As in panel A, the solid gray line represents unstained cells. The broken black line represents cells with isotype control antibody. The solid black line represents cells stained with the LRP1 antibody. (C) Western blot for LRP1 and actin as a control for protein loading. (1) Megakaryocytes, (2) platelets, and (3) WBCs. LRP1 band is expected at approximately 85 kDa and actin at approximately 25 kDa.
Flow cytometry and Western blot of human and murine megakaryocytes and platelets. (A) Representative examples of flow cytometry of murine bone marrow–derived megakaryocytes (top) stained with a biotin labeled anti-hLRP1 antibody known to cross-react with mouse LRP131 and then stained with streptavidin, PE–Alexa 647 secondary antibody. The gray line represents unstained cells. The broken black line represents secondary antibody alone. The solid black line is megakaryocytes with both antibodies. The bottom graph shows flow cytometry of platelets similarly performed. (B) As in panel A but for human cultured megakaryocytes and human peripheral blood platelets. LRP1 antibody was directly labeled with Alexa 647 for these experiments. As in panel A, the solid gray line represents unstained cells. The broken black line represents cells with isotype control antibody. The solid black line represents cells stained with the LRP1 antibody. (C) Western blot for LRP1 and actin as a control for protein loading. (1) Megakaryocytes, (2) platelets, and (3) WBCs. LRP1 band is expected at approximately 85 kDa and actin at approximately 25 kDa.
In vitro studies of the effect of RAP and anti-LRP1 antibodies
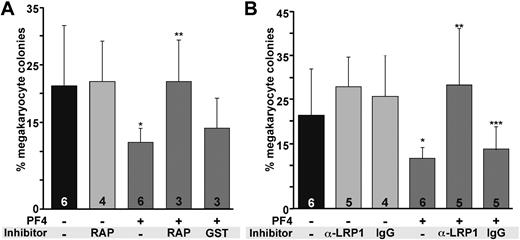
Previous studies have shown that supplementing in vitro cultures of marrow progenitor cells with 25 μg/mL PF4 decreased the number of megakaryocyte-containing colonies by approximately 50%.7,32 Similarly, blocking anti-PF4 antibodies were able to restore colony formation.7 If PF4 inhibits megakaryopoiesis through LRP1, we would expect that the addition of the anti-LDL receptor, RAP,33 or anti-LRP1 would block the PF4 effect as well. Addition of 25 μg/mL recombinant mPF4 to WT murine hematopoietic cells inhibited megakaryocyte-containing colonies by approximately 40% (P = .004, Figure 3A). Addition of RAP (0.7 μg/mL) reversed the inhibitory effect of the supplemental PF4 (P < .003, Figure 3A). An anti-LRP1 light chain also prevented the decrease in megakaryocyte-containing colonies (P < .003) in the presence of recombinant PF4 (Figure 3B). Addition of 2 other specific anti-LRP1 antibodies, an anti-LRP1 heavy chain antibody and an anti-LRP1 cluster II antibody showed similar restoration of colony formation (data not shown), whereas addition of isotype control antibody did not block the PF4 effect (Figure 3B).
In vitro studies of the effect of RAP and anti-LRP1 antibodies on megakaryopoiesis. (A) The effect of RAP on megakaryocyte colony formation. GST indicates empty GST without conjugated RAP. Graphed is the mean percentage of megakaryocytes per well plus 1 SD. Number of experiments, each performed in duplicate, is indicated in each bar. *P = .004 versus WT cultures without PF4; **P < .003 compared with WT culture with PF4. (B) The effect of anti-LRP1 antibody (MA5A6). Ig is isoimmune control for the anti-LRP1 antibody. Mean percentage of megakaryocytes per well plus 1 SD is graphed. Number of experiments done in duplicate is indicated in each bar. *P = .004 compared with WT without PF4; **P < .003 compared with WT with PF4; ***P = .04 compared with WT without PF4.
In vitro studies of the effect of RAP and anti-LRP1 antibodies on megakaryopoiesis. (A) The effect of RAP on megakaryocyte colony formation. GST indicates empty GST without conjugated RAP. Graphed is the mean percentage of megakaryocytes per well plus 1 SD. Number of experiments, each performed in duplicate, is indicated in each bar. *P = .004 versus WT cultures without PF4; **P < .003 compared with WT culture with PF4. (B) The effect of anti-LRP1 antibody (MA5A6). Ig is isoimmune control for the anti-LRP1 antibody. Mean percentage of megakaryocytes per well plus 1 SD is graphed. Number of experiments done in duplicate is indicated in each bar. *P = .004 compared with WT without PF4; **P < .003 compared with WT with PF4; ***P = .04 compared with WT without PF4.
shRNA suppression of LRP1 in megakaryocytes
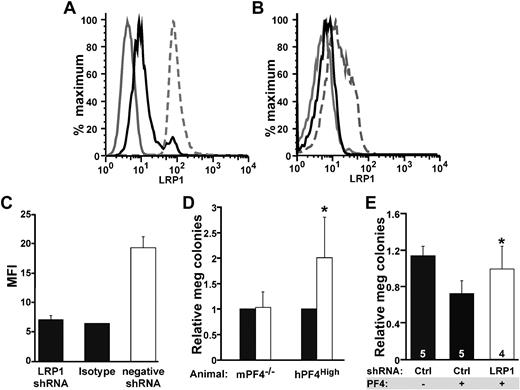
To confirm these in vitro culture findings, shRNA studies were used to inhibit LRP1 expression by megakaryocytes and to observe the effect of PF4 on megakaryopoiesis in this setting. Lentiviral vectors designed to express shRNAs for LRP1 were first tested on NIH-3T3 cells to see how effective they were at decreasing surface LRP1 expression. Cells transfected with the LRP1.1 vector showed an approximately 43% decrease in LRP1-positive cells after 72 hours of selection with puromycin (Figure 4A), whereas LRP1.2 and LRP1.3 had little to no effect on LRP1 expression (data not shown). Based on these findings, we hypothesized that the LRP1.1 shRNA (henceforth called LRP1 shRNA) would be most effective in mitigating the effect of PF4.
shRNA suppression of LRP1 and megakaryocyte colony formation. (A) Representative flow cytometry of 3T3 cells (positive control for LRP1) stably transfected with different shRNA viral vectors. The solid gray line represents cells that were unstained. The broken gray line is LRP1 expression on cells expressing the empty lentiviral vector. Solid black line shows the decrease in surface LRP1 expression after stable transfection with the LRP1 shRNA virus. (B) Same as panel A except for murine bone marrow cells after culture in media containing TPO and puromycin for 5 days. The solid gray line represents isotype control. The broken gray line is cells transfected with the negative viral vector. The solid dark line represents cells transfected with LRP1 shRNA. (C) Quantitation of change in mean fluorescence index (MFI) in murine bone marrow cells transfected with virus. Data represent mean +1 SD for 3 independent experiments. Viral titers were between 1 to 2 × 1011 viral particles/mL. (D) Effect of LRP1 shRNA on megakaryopoiesis (meg) using mPF4−/− bone marrow and hPF4High bone marrow expressed relative to megakaryopoiesis with the control empty vector. ■ is relative level of megakaryocyte seen after transfection with the empty lentiviral vector, and □ is relative level after transfection with the LRP1 shRNA vector. Data represent mean +1 SD for 4 experiments, each performed in duplicate. *P < .006 for LRP1 versus negative control for hPF4. shRNA had no effect on colony formation in mPF4−/− bone marrow. (E) Effect of LRP1 shRNA megakaryocyte colony formation in mPF4−/− bone marrow treated with exogenous PF4 (25 μg/mL). Percentage of meg colonies were normalized as in panel D. Ctl indicates control studies with empty vector. Data represent mean +1 SD. Numbers in bars represent times experiments were performed (each in duplicate). *P < .008 for LRP1 versus empty virus in the presence of PF4.
shRNA suppression of LRP1 and megakaryocyte colony formation. (A) Representative flow cytometry of 3T3 cells (positive control for LRP1) stably transfected with different shRNA viral vectors. The solid gray line represents cells that were unstained. The broken gray line is LRP1 expression on cells expressing the empty lentiviral vector. Solid black line shows the decrease in surface LRP1 expression after stable transfection with the LRP1 shRNA virus. (B) Same as panel A except for murine bone marrow cells after culture in media containing TPO and puromycin for 5 days. The solid gray line represents isotype control. The broken gray line is cells transfected with the negative viral vector. The solid dark line represents cells transfected with LRP1 shRNA. (C) Quantitation of change in mean fluorescence index (MFI) in murine bone marrow cells transfected with virus. Data represent mean +1 SD for 3 independent experiments. Viral titers were between 1 to 2 × 1011 viral particles/mL. (D) Effect of LRP1 shRNA on megakaryopoiesis (meg) using mPF4−/− bone marrow and hPF4High bone marrow expressed relative to megakaryopoiesis with the control empty vector. ■ is relative level of megakaryocyte seen after transfection with the empty lentiviral vector, and □ is relative level after transfection with the LRP1 shRNA vector. Data represent mean +1 SD for 4 experiments, each performed in duplicate. *P < .006 for LRP1 versus negative control for hPF4. shRNA had no effect on colony formation in mPF4−/− bone marrow. (E) Effect of LRP1 shRNA megakaryocyte colony formation in mPF4−/− bone marrow treated with exogenous PF4 (25 μg/mL). Percentage of meg colonies were normalized as in panel D. Ctl indicates control studies with empty vector. Data represent mean +1 SD. Numbers in bars represent times experiments were performed (each in duplicate). *P < .008 for LRP1 versus empty virus in the presence of PF4.
Confirmatory analysis of knockdown of LRP1 expression in primary murine cultured bone marrow cells was shown for the LRP1 shRNA lentivirus. Cells transfected with the negative lentivirus (containing a puromycin selection cassette but not the LRP1 shRNA) showed the expected level of LRP1 expression after 7 days in culture with TPO (5 days of puromycin; Figure 4B). Data shown are a representative flow cytometry histogram, but quantitative analysis of mean fluorescence intensity (MFI) consistently shows LRP1 surface expression knockdown (Figure 4C).
We have previously shown an inverse relationship between the level of PF4 content in developing megakaryocytes and megakaryocyte colony numbers for WT, mPF4−/−, and hPF4High mice in vitro when grown in the absence of serum or exogenous PF4.7 Bone marrow from hPF4High animals develop 40% to 50% fewer megakaryocyte colonies when cultured than littermate WT bone marrow, whereas mPF4−/− marrow generates approximately 10% more megakaryocyte colonies than WT.7 Bone marrow from mPF4−/− and from hPF4High animals was transfected with LRP1 shRNA lentivirus, and megakaryocyte colony formation was followed. Because there was some decrease in megakaryocyte colony formation with the empty viral vector, results were normalized to empty virus in each experiment. Transfection of bone marrow with the LRP1 shRNA showed a dramatic increase in megakaryocyte colony formation in hPF4High bone marrow compared with controls (P < .006, Figure 4D). On the other hand, expression of LRP1 shRNA had no effect on colony formation in the mPF4−/− mice, consistent with the absence of a PF4 inhibitory loop in these cultures. However, when mPF4−/− bone marrow was cultured in the presence of PF4, LRP1 shRNA was able to restore megakaryocyte colony formation (Figure 4E).
G1ME cell expression of LRP1 and sensitivity to PF4 inhibition
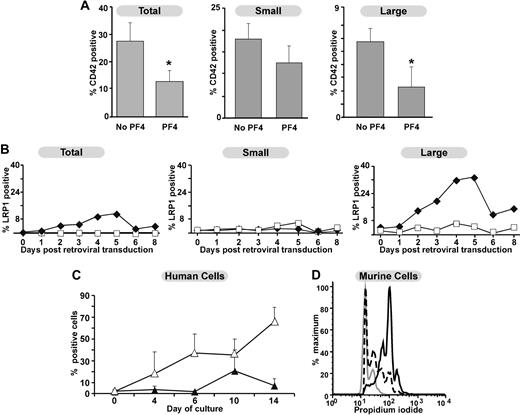
Since megakaryocytes express LRP1, but platelets do not, we wished to further explore the specific point during megakaryopoiesis in which PF4 inhibited this process. G1ME cells represent a MEP-like cell line derived from GATA-1null embryonic stem cells.27 Upon re-expression of GATA-1, they undergo development into both megakaryocytes and erythroid cells. We anticipated that these cells would allow us to examine the common megakaryocyte precursor and a fairly synchronous population of maturing megakaryocytes for PF4 sensitivity and LRP1 expression. Addition of 25 μg/mL PF4 to G1ME coincident with GATA-1 expression reduced the percentage of megakaryocytes by approximately 50% (Figure 5A right panel). This effect was predominantly on the large CD42+ megakaryocytes, with much less of an effect on the smaller CD42+ megakaryocytes (Figure 5A middle and left panels). When LRP1 expression was examined in these cells, G1ME cells prior to GATA-1 expression did not have detectable surface LRP1 (Figure 5B right panel). Surface LRP1 increased after GATA-1 expression and peaked on day 5 (Figure 5B). This LRP1 was primarily found on the large CD42+ cells (Figure 5B middle and left panels) based on forward scatter. Confirmatory time course experiments were performed on human bone marrow–derived CD34+ cells cultured in TPO. Expression of CD41 increased, as expected, with days in culture. A population of cells that was positive for both LRP1 and CD41 appeared after 7 days and peaked around 10 days in culture consistent with our findings that LRP1 expression begins on terminally differentiated megakaryocytes (Figure 5C). In addition, murine bone marrow studies examining the relationship of ploidy to LRP1 expression suggest that those CD41+ cells expressing LRP1 are higher ploidy than CD41+ cells that do not express LRP1 (Figure 5D).
Effect of PF4 on G1ME cells and expression of LRP1. (A) The effect of PF4 on percentage of CD42+ cells after re-expression of GATA-1 by the introduction of a GATA-1-IRES-eGFP MIGR1 retrovirus with and without 25 μg/mL PF4 in serum-free media. Results are for eGFP+-transfected cells. On the left are total CD42+ cells; middle are small, CD42+ cells; and right are large, CD42+ cells (based on forward scatter on flow cytometry). Data are shown as mean +1 SD of 4 experiments. *P < .008 comparing with and without PF4 added. (B) LRP1 expression in G1ME cells both before and after transfection with either a MIGR1 empty retrovirus (□) or MIGR1 retrovirus containing GATA-1 (♦). Figure organized as in panel A. Shown is a representative experiment of 3. (C) LRP1 expression on human cultured megakaryocytes derived from adult CD34+ bone marrow cells. Open triangles show total CD41+ cells, whereas closed triangles show LRP1+/CD41+ cells. Mean +1 SD is shown for 4 independent experiments. (D) Ploidy analysis in relation to LRP1 expression. The gray line represents cells that are CD41−. The broken line is cells that are CD41+ but LRP1−. The solid, dark line represents the cells that are positive for both LRP1 and CD41. Data are from a single experiment, but are representative of results from 5 independent experiments.
Effect of PF4 on G1ME cells and expression of LRP1. (A) The effect of PF4 on percentage of CD42+ cells after re-expression of GATA-1 by the introduction of a GATA-1-IRES-eGFP MIGR1 retrovirus with and without 25 μg/mL PF4 in serum-free media. Results are for eGFP+-transfected cells. On the left are total CD42+ cells; middle are small, CD42+ cells; and right are large, CD42+ cells (based on forward scatter on flow cytometry). Data are shown as mean +1 SD of 4 experiments. *P < .008 comparing with and without PF4 added. (B) LRP1 expression in G1ME cells both before and after transfection with either a MIGR1 empty retrovirus (□) or MIGR1 retrovirus containing GATA-1 (♦). Figure organized as in panel A. Shown is a representative experiment of 3. (C) LRP1 expression on human cultured megakaryocytes derived from adult CD34+ bone marrow cells. Open triangles show total CD41+ cells, whereas closed triangles show LRP1+/CD41+ cells. Mean +1 SD is shown for 4 independent experiments. (D) Ploidy analysis in relation to LRP1 expression. The gray line represents cells that are CD41−. The broken line is cells that are CD41+ but LRP1−. The solid, dark line represents the cells that are positive for both LRP1 and CD41. Data are from a single experiment, but are representative of results from 5 independent experiments.
In vivo studies with RAP
To assess the in vivo role of LRP1 in megakaryopoiesis, we examined the effect of continuous infusion of RAP in hPF4High mice, which already have a lower baseline platelet count than their WT littermates.7,14 The dose of RAP given (785 μg/kg per day) was calculated to achieve an approximate concentration of 0.02 μM (0.7 μg/mL) in the plasma. Mice receiving an infusion of RAP showed an increase in platelet count over baseline, whereas mice receiving PBS diluent alone did not. An increase in platelet count was evident by day 5 (P = .01 vs PBS infusion) and this increase remained through the end of the infusion on day 7 (P = .02 versus PBS infusion, 121% ± 27% of baseline platelet count; Figure 6A). In contrast, when the same experiment was performed in mPF4−/− mice, no significant differences in platelet counts were seen (Figure 6B). No other hematologic parameter was either increased or decreased to a significant degree (data not shown).
The in vivo effect of RAP infusion in hPF4High and mPF4−/− animals. Both hPF4High mice (left) and mPF4−/− mice (right) were treated with approximately 800 μg/kg per day by continuous 7-day infusion of RAP or PBS diluent using an Alzet osmotic pump. Mice receiving only PBS are shown as □ and those receiving a RAP infusion are shown as  . For hPF4High mice, 10 animals were studied per arm and for the mPF4−/− mice, 5 per arm. *P < .02 versus PBS-treated animals on days 5 and 7 after pump placement.
. For hPF4High mice, 10 animals were studied per arm and for the mPF4−/− mice, 5 per arm. *P < .02 versus PBS-treated animals on days 5 and 7 after pump placement.  represents time of RAP infusion.
represents time of RAP infusion.
The in vivo effect of RAP infusion in hPF4High and mPF4−/− animals. Both hPF4High mice (left) and mPF4−/− mice (right) were treated with approximately 800 μg/kg per day by continuous 7-day infusion of RAP or PBS diluent using an Alzet osmotic pump. Mice receiving only PBS are shown as □ and those receiving a RAP infusion are shown as  . For hPF4High mice, 10 animals were studied per arm and for the mPF4−/− mice, 5 per arm. *P < .02 versus PBS-treated animals on days 5 and 7 after pump placement.
. For hPF4High mice, 10 animals were studied per arm and for the mPF4−/− mice, 5 per arm. *P < .02 versus PBS-treated animals on days 5 and 7 after pump placement.  represents time of RAP infusion.
represents time of RAP infusion.
Discussion
PF4 is an unusual CXC chemokine in that it does not appear to have its biologic effects through a G protein–coupled CXCR, but rather binds as a tetramer with an equatorial band of positively charged residues to large negatively charged molecules such as membrane surface GAGs.34,35 A previous report has described the binding of PF4 to a variant splice form of CXCR3 in humans with high affinity, but such a variant does not exist in mice and the concentration range over which PF4 inhibits megakaryopoiesis is 103-fold higher than saturating concentrations for this receptor.36 PF4 does bind to several specific proteins.37 In these cases, it is unclear whether the PF4 is binding directly to the proteins or to side chain modifications such as the chondroitin sulfate side chain of the thrombomodulin GAG domain. As mentioned in “Introduction,” PF4 appears to activate endothelial cells through LRP1, so we explored the possible role of this receptor in PF4 regulation of megakaryopoiesis. This receptor is a member of the superfamily of low-density lipoprotein receptors and has been described to have diverse roles in lipid metabolism, uptake of coagulation and other proteases, and cellular entry of bacterial toxins and viruses (see Lillis et al33 for review). LRP1 has been shown to undergo endocytosis after ligand binding, but can also directly lead to signal transduction in cells without endocytosis.38-40 In blood vessels, LRP1 has been shown to play an important (but not fully defined) role in TGFβ-dependent signaling mediating cell proliferation.39,41 Thus, the interaction of LRP1 and PF4 may result in direct inhibition of cellular proliferation. Alternatively, binding of PF4 to LRP1 may prevent interaction with another ligand that would then promote proliferation. Our shRNA and anti-LRP1 blocking antibody in vitro megakaryocyte colony-formation studies data suggest that the former mechanism is more likely given that directly blocking LRP1 results in an increase in megakaryocyte colony formation.
While our studies were under way, Bouchard et al reported the presence of LRP1 on human megakaryocytes and its role in the endocytosis of factor V.18 Our studies confirm their observation that LRP1 is present on the surface of megakaryocytes. In addition, we extended these findings to show that this receptor is only transiently expressed during megakaryopoiesis. LRP1 is not present on progenitor MEP cells, is present at low levels on CD41+/CD42+ small cells, becomes readily detectable on large, polyploidy megakaryocytes, and then is absent to near absent on circulating platelets. The near absence of LRP1 on platelets is also associated with a near absence of detectable LRP1 message RNA in platelets. This disappearance of the LRP1 on the platelet surface may be due to LRP1 message instability and/or LRP1 endocytosis.18 Clearly, further studies on the biology of LRP1 and megakaryopoiesis are needed, including those that address the following questions: Is LRP1 uniquely restricted to factor V uptake or is it also involved in the endocytosis of other proteins from the milieu that then are stored in alpha-granules? Is PF4 also endocytosed and is this how PF4 inhibits megakaryopoiesis or does PF4 binding result in intracellular signaling without concurrent endocytosis? Could PF4 and factor V (and other proteins) compete for uptake and does an excess of one in the microenvironment limit the uptake and/or effects of the other?
Given the narrow window during which LRP1 is expressed on developing megakaryocytes, PF4 released into the microenvironment would only be able to inhibit, in a paracrine fashion, other nearby megakaryocytes undergoing terminal differentiation. In addition, LRP1 surface expression varied greatly even on large megakaryocytes derived from G1ME cells. On G1ME cells 20% to 35% of these cells have high levels of LRP1 (Figure 5B), in contrast to only 14% of primary human megakaryocytes at day 10 of culture (Figure 5C). The absence of detectable LRP1 on a significant portion of megakaryocytes may explain why even under optimal conditions, all groups that have studied the inhibitory effect of PF4 either in vitro or in vivo have observed the same approximately 50% maximal inhibition of megakaryocyte-containing colony formation by PF4.5,7,32
What are the clinical implications of these findings? We believe that although TPO is the predominant regulator of platelet production, PF4 inhibition may have an important role in settings associated with increased intramarrow megakaryocyte turnover such as in CIT, radiation-induced thrombocytopenia, and in some patients with immune thrombocytopenia purpura with intramedullary apoptosis and lysis of maturing megakaryocytes.42 We have already demonstrated this for CIT,7 and in unpublished data (M.P.L., March 2009) found the same for radiation-induced thrombocytopenia. We propose that the 25% of patients showing less than acceptable responses to TPO mimetic therapy43-47 may have release an excess of intramedullary PF4.
In summary, the studies reported here extend our understanding of the underlying receptor and target cell in PF4's negative paracrine effect on megakaryopoiesis and extend the potential therapeutic targets to limit thrombocytopenia secondary to intramedullary PF4 release. These studies also highlight the transient presence of LRP1 on developing megakaryocytes and extend the range of activities in which this receptor participates during megakaryopoiesis.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Li Zhai for production of recombinant PF4. We also thank Mitchell J. Weiss at the University of Pennsylvania and the Children's Hospital of Philadelphia for providing the G1ME cells, the Plat-E cells, and GATA-1 and control MIGR1 retroviruses.
This work was supported in part by funding from the American Heart Association (M.P.L.) and the National Institutes of Health (PO1 HL40387, M.P.; K23 HL092164, M.P.L.).
National Institutes of Health
Authorship
Contribution: M.P.L. was the primary investigator and carried out most of the experiments, developed experimental direction, and wrote the paper; Y.W. performed the G1ME studies and assisted in the LRP1 shRNA experiments; K.H.B. helped design the RAP studies and provided RAP for initial experiments; Y.N. performed the LRP1 shRNA and part of the in vivo RAP studies; M.A.K. helped plan the bone marrow studies and taught M.P.L. the techniques involved and assisted in data interpretation and paper revision; and M.P. provided overall scientific direction, and assisted in data interpretation and paper preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michele P. Lambert, The Children's Hospital of Philadelphia, 3615 Civic Center Blvd, ARC, Rm 316C, Philadelphia, PA 19104; e-mail: lambertm@email.chop.edu.









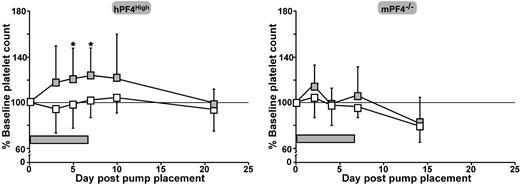
 . For hPF4High mice, 10 animals were studied per arm and for the mPF4−/− mice, 5 per arm. *P < .02 versus PBS-treated animals on days 5 and 7 after pump placement.
. For hPF4High mice, 10 animals were studied per arm and for the mPF4−/− mice, 5 per arm. *P < .02 versus PBS-treated animals on days 5 and 7 after pump placement.  represents time of RAP infusion.
represents time of RAP infusion.