Abstract
Hereditary thrombotic thrombocytopenic purpura is caused by mutations in adisintegrin and metalloprotease with thrombospondin motifs (ADAMTS13) resulting in defective processing of von Willebrand factor (VWF) that causes intravascular platelet aggregation culminating in thrombocytopenia with shistocytic anemia. In this study the functional and structural role of a recently identified ADAMTS13 metalloprotease domain mutation S119F was investigated. Secretion from heterologous cells was hampered but not completely eliminated. Secreted S119F was active toward multimeric VWF and FRETS-VWF73 but with abnormal kinetics, having a significantly reduced overall catalytic rate (kcat; 0.88 ± 0.04 s−1 vs 2.78 ± 0.11 s−1) and slightly smaller Michaelis constant (KM; 1.4 ± 0.2μM vs 2.3 ± 0.3μM). A computational model of the metalloprotease domain demonstrates both steric and polar interaction effects caused by S119F. Interestingly, mutant S119A has properties similar to S119F (kcat = 0.82 ± 0.03 s−1 and KM = 1.1 ± 0.1μM), allowing to assign distorted kinetics to the loss of the H-bond with conserved residue W262. We conclude that the S119-W262 H-bond is crucial for maximal turnover.
Introduction
Thrombotic thrombocytopenic purpura (TTP) is an acute thrombotic disorder characterized by consumptive thrombocytopenia and microangiopathic hemolytic anemia.1 In most cases TTP is acquired, but a small number of cases bears mutations in the adisintegrin and metalloprotease with thrombospondin motifs (ADAMTS13) gene, resulting in the same phenotype.2,3
ADAMTS13 is a metalloprotease specifically hydrolyzing the Y1605-M1606 peptide bond in multimeric von Willebrand factor (VWF). VWF multimer size reduction is pivotal in preserving normal hemostasis because uncleaved multimers spontaneously bind platelets causing occlusion of microvasculature, the TTP hallmark.
More than 50 ADAMTS13 gene mutations have been reported since its identification in 2001.4 Most of these were not investigated by in vitro mutagenesis and expression analysis, but those that were cause mainly secretion defects.3 This is in line with the observation that ADAMTS13 antigen is absent in plasma samples from patients with hereditary TTP.5 Nonetheless, some ADAMTS13 mutants are secreted3,6,7 but are less or not functional.
Recently, high-resolution structures of the metalloprotease domains of ADAMTS1, ADAMTS4, and ADAMTS5 were published, providing insight in active-site and small molecule inhibitor orientation.8,9 The structures of the metalloprotease domains of ADAM/ADAMTS proteins generally superimpose with little variation except in the variable loops making up domain periphery.10
In this study, we characterize a recently identified ADAMTS13 metalloprotease domain missense mutation that causes a partial secretion defect and a 3-fold reduction in the turnover number (kcat). We correlate kinetic data with rational assumptions from a homology model of the ADAMTS13 metalloprotease domain.
Methods
Patient characterization
This study was approved by the Katholieke Universiteit Leuven Ethical Committee of the Katholieke Universiteit Leuven and its hospitals. Informed consent was obtained in accordance with the Declaration of Helsinki. The propositus is a 14-year-old white pediatric patient with typical congenital TTP successfully treated with prophylactic plasma infusion therapy. ADAMTS13 exons with exon-intron boundaries were amplified from genomic DNA using polymerase chain reaction with primers described elsewhere.11 Sequencing reactions were outsourced (GATC Biotech AG).
Citrated plasma was used for analysis of ADAMTS13 activity and antigen. Normal human pooled plasma (NHP, n = 20) was used as a control. Antigen was measured by enzyme-linked immunosorbent assay (ELISA) as described5 and by immunoblotting with monoclonal antibody (mAb) 17B10 binding to the third ADAMTS13 thrombospondin-1 repeat. Bound mAb was detected by horseradish peroxidase labeled goat anti–mouse antiserum (GAM-HRP; Jackson Immunoresearch).
Mutagenesis and expression of recombinant proteins
Mutagenesis was performed with the QuikChange XL Site-Directed Mutagenesis kit (Stratagene) using primers bearing the desired mutations. All mutants were constructed using the inducible expression plasmid pGene/V5-His (Invitrogen) containing the ADAMTS13 cDNA.5 Vectors were lipofected in Chinese hamster ovary GeneSwitch cells and human embryonic kidney 293 (HEK293) GeneSwitch cells (Invitrogen) for stable and transient expression, respectively. Serum-free conditioned medium was purified by ion exchange (Q-sepharose; GE Healthcare).12
ADAMTS13 activity and kinetic measurements
Activity toward multimeric VWF was measured using mild denaturants as described previously.13 Reactions contained 10 μg/mL purified VWF (CSL Behring) and 0.6nM recombinant ADAMTS13 (rADAMTS13) and were incubated for 16 hours. Proteolysis was analyzed by residual collagen binding.14 Proteolytic fragments were visualized by Western blotting with anti-VWF monoclonal antibodies RG46, 52K-2, and 52K-8 (Dr Zaverio M. Ruggeri, Scripps Research Institute, La Jolla, CA)15 and GAM-HRP.
ADAMTS13 metalloprotease domain homology model
MODELLER 9v418 was used to generate a homology model based on the structures of ADAMTS1, ADAMTS4, and ADAMTS5 (respective Protein Data Bank codes: 2V4B, 3B2Z, and 3B8Z).19 Sequence identity between ADAMTS13 and templates varied between 25% and 30%. Modeling preserved both the active-site zinc ion and the zinc-coordinating water molecule. The dead-end elimination method20 was used to optimize orientation of all side chains and polar hydrogens. Idealization of bond geometry and removal of unfavorable nonbonded contacts was performed by a conjugate gradient energy minimization with the GROMACS Version 3.3.1 software package21 in the presence of explicit water and counter ions. We applied the GROMOS96 43a1 force field. Mutant structure S119F was generated using the BRUGEL package22 and least squares fitting was done using PyMOL (DeLano Scientific).
Results
Homozygous mutation 356C>T in the ADAMTS13 metalloprotease coding sequence
ADAMTS13 antigen was severely reduced to 5% in remission phase and undetectable (< 5%) in acute phase plasma as determined by Western blot (Figure 1A left) and ELISA. Both samples contained no measurable VWF73 cleaving activity (not shown). A homozygous missense mutation 356C>T and a known single nucleotide polymorphism 1342C>G were identified, encoding a serine to phenylalanine substitution (S119F) and a glutamine to glutamic acid exchange (Q448E), respectively. Meyer et al recently reported S119F as the first congenital TTP case on the African continent.23 Importantly, the serine residue is conserved among ADAMTSs except in ADAMTS16 and ADAMTS1824 where a threonine resides at the corresponding position, alluding to a role of the side-chain hydroxyl group.
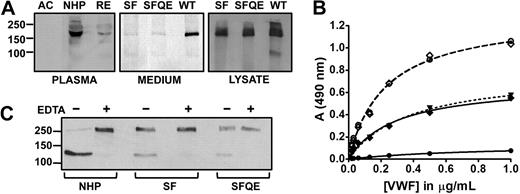
Expression levels, activity, and site specificity of S119F. (A left) Representative Western blot of endogenous ADAMTS13 in acute (AC) and remission (RE) phase of the propositus and in NHP. Blotted protein was detected with anti-ADAMTS13 mAb17B10 and GAM-HRP. Secondary antibody al1 was negative (not shown). (A middle and right) Transient expression of wild-type rADAMTS13 (WT), and mutants S119F (SF) and S119F/Q448E (SFQE). Protein in medium (middle) and cell lysate (right) was detected with peroxidase-labeled anti-V5 antibodies. (B) VWF collagen binding ELISA to assess residual VWF activity. Purified multimeric VWF (10 μg/mL) was incubated with 0.6nM ADAMTS13 in NHP (●), or 0.6nM rADAMTS13 S119F (♦) or S119F/Q448E (▾, dotted line) for 16 hours in 1M urea. All reactions were inhibited by 10mM ethylenediaminetetraacetic acid (EDTA; dashed lines) as shown for NHP (○) and S119F (◇). (C) Analogous samples as in panel B, in the presence (+) or absence (−) of EDTA, were analyzed in Western blot under reducing conditions where VWF product bands were detected by anti-A1 domain mAbs and GAM-HRP.
Expression levels, activity, and site specificity of S119F. (A left) Representative Western blot of endogenous ADAMTS13 in acute (AC) and remission (RE) phase of the propositus and in NHP. Blotted protein was detected with anti-ADAMTS13 mAb17B10 and GAM-HRP. Secondary antibody al1 was negative (not shown). (A middle and right) Transient expression of wild-type rADAMTS13 (WT), and mutants S119F (SF) and S119F/Q448E (SFQE). Protein in medium (middle) and cell lysate (right) was detected with peroxidase-labeled anti-V5 antibodies. (B) VWF collagen binding ELISA to assess residual VWF activity. Purified multimeric VWF (10 μg/mL) was incubated with 0.6nM ADAMTS13 in NHP (●), or 0.6nM rADAMTS13 S119F (♦) or S119F/Q448E (▾, dotted line) for 16 hours in 1M urea. All reactions were inhibited by 10mM ethylenediaminetetraacetic acid (EDTA; dashed lines) as shown for NHP (○) and S119F (◇). (C) Analogous samples as in panel B, in the presence (+) or absence (−) of EDTA, were analyzed in Western blot under reducing conditions where VWF product bands were detected by anti-A1 domain mAbs and GAM-HRP.
S119F kinetic parameters kcat and KM are reduced
Transient expression of both the single (S119F) and the double (S119F/Q448E) mutant revealed normal expression but markedly impaired secretion (Figure 1A right). VWF multimers were completely cleaved by 0.6 nM of ADAMTS13 in NHP, whereas an identical concentration of either single or double mutant only partially cleaved the substrate (Figure 1B). Western blotting of the reduced VWF N-terminal reaction products revealed no obvious size difference between VWF fragments generated by S119F and wild type (WT), indicating site specificity was unaltered (Figure 1C).
Importantly, kcat of S119F (0.88 ± 0.04 s−1, mean ± SD) was significantly smaller than WT (2.78 ± 0.11 s−1), whereas KM was only slightly smaller (1.4 ± 0.2μM vs 2.3 ± 0.3μM) as determined by FRETS-VWF73 kinetic measurements (Figure 2A). A reduced kcat indicates that substrate turnover and/or product release is affected by the mutation. Furthermore, KM is only slightly different, indicating ground-state binding is comparable with WT or somewhat better. As a consequence, the specificity constant (kcat/KM) reduces by half from 1.2μM−1s−1 (WT) to 0.6μM−1s−1 (S119F). This parameter is inversely related to the free energy of transition state complex formation.25 Therefore, more energy is required to attain transition state in S119F following the enzyme substrate binding step. Hence, higher concentrations of S119F are needed for optimal VWF processing and TTP may precipitate at enzyme levels that would be safe if no mutation were present.
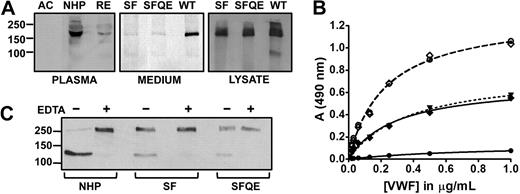
Model of the ADAMTS13 metalloprotease domain and kinetics of S119 mutants. (A) Michaelis-Menten kinetics of WT rADAMTS13 (○), mutants S119F (▿) and S119A (♦). Initial velocity in function of FRETS-VWF73 concentration ([S]) at fixed enzyme concentration (0.1nM) is shown. (B) Surface representation of the ADAMTS13 metalloprotease model. Depicted in red and green are the S1′ and S2′ loop, respectively. The loops constitute part of the active-site cleft. Mutated residue serine 119 (magenta) is located to the bottom left, facing the C-terminus of the S1′ loop. (C) Cartoon representation zoomed into the S1′ loop (red) and inner molecule. Depicted in sticks are S119, M249 (Met-turn), and residues making up the hydrophobic patch to which the W262 indole side chain is oriented (V248, L120, and L114). Hydrogen bonds are shown in dashes. Mutation of residue S119 to phenylalanine (overlaid in orange) disrupts the hydrogen bond and causes a steric clash between the S1′ loop for all possible rotamers at fixed backbone positions (1 rotamer shown).
Model of the ADAMTS13 metalloprotease domain and kinetics of S119 mutants. (A) Michaelis-Menten kinetics of WT rADAMTS13 (○), mutants S119F (▿) and S119A (♦). Initial velocity in function of FRETS-VWF73 concentration ([S]) at fixed enzyme concentration (0.1nM) is shown. (B) Surface representation of the ADAMTS13 metalloprotease model. Depicted in red and green are the S1′ and S2′ loop, respectively. The loops constitute part of the active-site cleft. Mutated residue serine 119 (magenta) is located to the bottom left, facing the C-terminus of the S1′ loop. (C) Cartoon representation zoomed into the S1′ loop (red) and inner molecule. Depicted in sticks are S119, M249 (Met-turn), and residues making up the hydrophobic patch to which the W262 indole side chain is oriented (V248, L120, and L114). Hydrogen bonds are shown in dashes. Mutation of residue S119 to phenylalanine (overlaid in orange) disrupts the hydrogen bond and causes a steric clash between the S1′ loop for all possible rotamers at fixed backbone positions (1 rotamer shown).
Importance of the S119-W262 H-bond
In our metalloprotease domain model (Figure 2B), S119 resides on a short α-helix juxtaposed to the C-terminus of the S1′ loop (residues 247-261; Figure 2C).9,26 This loop lacks secondary structure and is important for substrate recognition.10 The side-chain hydroxyl group from S119 accepts an H-bond from the backbone amine of conserved residue W262. Furthermore, the W262 indole side chain is oriented in a hydrophobic pocket that is conserved among ADAMTSs and compiled of aliphatic residues from nearby strands including the short α-helix harboring S119 (Figure 2C).8,9 Both the H-bond and the optimal orientation of the short α-helix are hence affected in S119F. To distinguish between these mechanisms underlying the abnormal kinetics, a mutant S119A was constructed. This mutant prevents H-bond formation but exerts no steric strain on the helix bearing S119. Interestingly, kinetic parameters were not different between S119A and S119F (kcat = 0.82 ± 0.03 s−1 and KM = 1.1 ± 0.1μM). This indicates that steric interference in S119F is not the leading cause of decreased catalytic efficiency. The data show that the S119-W262 H-bond is crucial for optimal turnover (reduced kcat), although not required for substrate recognition or binding (equal KM). Alternatively, disruption of the H-bond induces structural changes in the active-site shape that would concomitantly explain the poor secretion. In conclusion, as the C- and N-terminus of the S1′ loop meet, it adopts an Ω shape, which is in part sustained by the hydrophobic interactions including C-terminal W262 keeping the ends of the flexible strand together in the pocket. Loss of the W262 H-bond with S119 might render the backbone of the S1′ loop more flexible, influencing overall structure and/or optimal alignment for transition state complex formation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Zaverio M. Ruggeri (Scripps Research Institute) for monoclonal antibodies against VWF.
H.B.F. and K.V. are fellows and C.V.G. is clinical investigator of the Research-Foundation Flanders (Fonds voor Wetenschappelijk Onderzoek-Vlaanderen [FWO]). R.V. is a graduate student of the FWO. This work is supported by grants from the FWO (G.0299.06) and the Katholieke Universiteit Leuven (GOA/2004/09).
Authorship
Contribution: H.B.F. designed and performed research, analyzed and interpreted data, and wrote the paper; I.P. performed research and interpreted data; R.V. performed research and critically reviewed the manuscript; M.D.M., H.D., and C.V.G. provided essential reagents and/or patient material and critically reviewed the manuscript; and K.V. designed research, analyzed and interpreted data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Karen Vanhoorelbeke, Laboratory for Thrombosis Research, Katholieke Universiteit Leuven Campus Kortrijk, E Sabbelaan 53, 8500 Kortrijk, Belgium; e-mail: karen.vanhoorelbeke@kuleuven-kortrijk.be.

![Figure 2. Model of the ADAMTS13 metalloprotease domain and kinetics of S119 mutants. (A) Michaelis-Menten kinetics of WT rADAMTS13 (○), mutants S119F (▿) and S119A (♦). Initial velocity in function of FRETS-VWF73 concentration ([S]) at fixed enzyme concentration (0.1nM) is shown. (B) Surface representation of the ADAMTS13 metalloprotease model. Depicted in red and green are the S1′ and S2′ loop, respectively. The loops constitute part of the active-site cleft. Mutated residue serine 119 (magenta) is located to the bottom left, facing the C-terminus of the S1′ loop. (C) Cartoon representation zoomed into the S1′ loop (red) and inner molecule. Depicted in sticks are S119, M249 (Met-turn), and residues making up the hydrophobic patch to which the W262 indole side chain is oriented (V248, L120, and L114). Hydrogen bonds are shown in dashes. Mutation of residue S119 to phenylalanine (overlaid in orange) disrupts the hydrogen bond and causes a steric clash between the S1′ loop for all possible rotamers at fixed backbone positions (1 rotamer shown).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/21/10.1182_blood-2009-07-230615/4/m_zh89990945040002.jpeg?Expires=1769160492&Signature=JHFkfohb7jUNx3EGEDUCgXLaZZwPSL1nokC9JoIp-3kYDxSB8upvJVzinhRwcj16Ddl0munhZTSFvisz0Hu5UtVAgTBtdWI~nAXL71EawdV5THXjJFdJKcz-UaFdQqj8Q1DiNOyDZi9tsF80SoU3zVj2tWYPTTsCjn-YZXzYHljgNshrlSC0QnIj~puw8OjsAAG39CRx4ZV-h-d~jf09oKPGEF0yaKOp54JswQ5Z79t~ik4dW951lZEMwF0oaqgaeUPqmn~lsC2T1311WU6e5vcZg4RRe2XTOkg7RuSE2tCQpZHotlpD2PjluXQpUx71kXMKf3vKvXY0~gXTzUIdJA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 2. Model of the ADAMTS13 metalloprotease domain and kinetics of S119 mutants. (A) Michaelis-Menten kinetics of WT rADAMTS13 (○), mutants S119F (▿) and S119A (♦). Initial velocity in function of FRETS-VWF73 concentration ([S]) at fixed enzyme concentration (0.1nM) is shown. (B) Surface representation of the ADAMTS13 metalloprotease model. Depicted in red and green are the S1′ and S2′ loop, respectively. The loops constitute part of the active-site cleft. Mutated residue serine 119 (magenta) is located to the bottom left, facing the C-terminus of the S1′ loop. (C) Cartoon representation zoomed into the S1′ loop (red) and inner molecule. Depicted in sticks are S119, M249 (Met-turn), and residues making up the hydrophobic patch to which the W262 indole side chain is oriented (V248, L120, and L114). Hydrogen bonds are shown in dashes. Mutation of residue S119 to phenylalanine (overlaid in orange) disrupts the hydrogen bond and causes a steric clash between the S1′ loop for all possible rotamers at fixed backbone positions (1 rotamer shown).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/21/10.1182_blood-2009-07-230615/4/m_zh89990945040002.jpeg?Expires=1769160493&Signature=1aaM1vUjHSrHSANFb~Tm6xgLGlDww9n-206MVXJxzr9tUJq2HPlIu3DdQ4kBdlWAG6JgPWTYkhu~kBt2qR2ugzhWCmtEHmi0lTlsqscyDL7wUN4w8VHx6u63Zfhsn5fOdHpPmeLcy3UzE6xyaD91GX60d81RW2NyQm~wzsJSw0Y~D58uB2Yw2OaKz7sNr1YoBi1RDM1865ZsIwUDpwQGUgtDvqa7KHUonwudJhGDdb8DBa0qxk52GTG3aJXLRoIhPYF0qAP18K13xK6SNEiHdi1co8G-a-AZVa3IJKXrQllQgvRJn66eZj8vYfYTO5yld0sUHNkWyc7qpUQZf6SJQA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)